Abstract
Poxviruses are large DNA viruses that encode a multisubunit RNA polymerase, stage-specific transcription factors, and enzymes that cap and polyadenylate mRNAs within the cytoplasm of infected animal cells. Genome-wide microarray and RNA-seq technologies have been used to profile the transcriptome of vaccinia virus (VACV), the prototype member of the family. Here, we adapted tag-based methods in conjunction with SOLiD and Illumina deep sequencing platforms to determine the precise 5′ and 3′ ends of VACV early mRNAs and map the putative transcription start sites (TSSs) and polyadenylation sites (PASs). Individual and clustered TSSs were found preceding 104 annotated open reading frames (ORFs), excluding pseudogenes. In the majority of cases, a 15-nucleotide consensus core motif was present upstream of the ORF. This motif, however, was also present at numerous other locations, indicating that it was insufficient for transcription initiation. Further analysis revealed a 10-nucleotide AT-rich spacer following functional core motifs that may facilitate DNA unwinding. Additional putative TSSs occurred in anomalous locations that may expand the functional repertoire of the VACV genome. However, many of the anomalous TSSs lacked an upstream core motif, raising the possibility that they arose by a processing mechanism as has been proposed for eukaryotic systems. Discrete and clustered PASs occurred about 40 nucleotides after an UUUUUNU termination signal. However, a large number of PASs were not preceded by this motif, suggesting alternative polyadenylation mechanisms. Pyrimidine-rich coding strand sequences were found immediately upstream of both types of PASs, signifying an additional feature of VACV 3′-end formation and polyadenylation.
INTRODUCTION
High-throughput cDNA sequencing has enabled the genome-wide profiling of the transcriptomes of eukaryotic (58) and microbial organisms (57) and of complex DNA viruses (37, 61). We recently applied RNA-seq technology for whole-transcriptome analysis of vaccinia virus (VACV) (61), a poxvirus that replicates and transcribes its 195-kbp DNA genome within the cytoplasm of infected cells (42). Early transcripts, synthesized before viral DNA replication, were mapped to 118 closely spaced open reading frames (ORFs), and additional transcripts, synthesized only after DNA replication, were mapped to 93 ORFs. Whole-transcriptome analysis, however, may not delineate the ends of RNAs to high precision or delineate overlapping transcripts. Here, we adapted tag-based RNA-seq methods (27, 39, 56) to map the 5′ and 3′ ends of early VACV transcripts and determine putative regulatory sequences for transcription start sites (TSSs) and polyadenylation sites (PASs).
VACV and other poxviruses package a complete virus-encoded transcription system, which allows the early class of mRNAs to be synthesized immediately after entry into the cytoplasm (42). De novo synthesis of proteins and DNA are required to transcribe additional genes, which are subdivided into intermediate and late postreplication (PR) classes. The three categories of genes have distinctive promoters (7, 16, 17) and are transcribed by the viral multisubunit DNA-dependent RNA polymerase in concert with early-, intermediate-, and late-stage-specific transcription factors (1, 9, 13, 34, 49). Early mRNAs contain a transcription termination signal comprised of five consecutive U's followed by any nucleotide and then another U (U5NU) (53, 64) that is not functional in PR mRNAs, which mostly have heterogenous 3′ ends (15, 38). Methylated caps (60) and poly(A) tails (33) are added to the 5′ and 3′ ends of VACV mRNAs by virus gene-encoded enzymes (8, 40, 43). In addition, PR mRNAs and a few early mRNAs have a 5′ poly(A) tract immediately following the cap structure, which apparently arises from RNA polymerase slippage on consecutive T's (2, 6, 31, 47, 50). Because of the structural differences between the 5′ and 3′ ends of early and PR mRNAs and distinct methods needed for their analysis, the present study focused on the early transcriptome. We mapped hundreds of TSSs and PASs, which greatly extend previous knowledge and reveal new features and complexity of VACV transcription.
MATERIALS AND METHODS
Cells and virus.
HeLa S3 cells were cultured in minimum essential medium with spinner modification (Quality Biological, Gaithersburg, MD) and with 5% equine serum (HyClone, Logan, UT) at 37°C in a 5% CO2 atmosphere. Preparation, sucrose gradient purification, and titration of the Western Reserve (WR) strain of vaccinia virus (VACV) (American Type Culture Collection VR-1354) have been described previously (21, 23). HeLa S3 cells suspended at 107 cells/ml were infected with 20 PFU of VACV per cell. The cells were diluted to 5 × 105 cells/ml with spinner medium containing 5% serum after 30-min adsorption at 37°C. The cells were harvested for RNA preparation at the desired time postinfection. Cycloheximide (CHX) was added to the medium at 100 μg/ml where indicated.
RNA isolation.
Total RNA was isolated from VACV-infected cells using Trizol (Invitrogen, Carlsbad, CA). Polyadenylated mRNA was isolated with Dynabeads mRNA direct kit (Invitrogen) and treated with DNase I to remove DNA.
Generation of the SOLiD oCAGE library.
The SOLiD (sequencing by oligonucleotide ligation and detection) oCAGE (oligonucleotide cap analysis of gene expression) library was generated as described previously (27) with modifications (see Fig. S1A in the supplemental material). The RNA (100 μg) was treated with bacterial alkaline phosphatase (TaKaRa, Shiga, Japan) to remove the 5′ phosphate on fragmented RNA and then treated with tobacco acid pyrophosphatase (Epicenter, Madison, WI) to remove the cap structure and expose the phosphate at the 5′ end of the mRNA. The RNA was ligated to an RNA linker containing the EcoP15I recognition site (5′-CUGCCCCGGGUUCCUCAUUCUCUCAGCAG-3′) with T4 RNA ligase (TaKaRa) at 16°C for 4 h or overnight. The polyadenylated RNA was isolated with Dynabeads mRNA kit (Invitrogen) and reverse transcribed (SuperScript II; Invitrogen) with dT adapter-primer [5′-GCGGCTGAAGACGGCCTATGTGCAGCAG(T)17-3′] at 12°C for 1 h and then at 42°C for another hour. RNA was degraded after first-strand synthesis, and the cDNA was amplified by 10 cycles of PCR using 5′ and 3′ primers. The 5′ primer was 5′-biotin-TEG-CTGCCCCGGGTTCCTCATTCT −3′ where TEG is a tetraethylene glycol spacer, and the 3′ primer was 5′-GCGGCTGAAGACGGCCTATGT-3′. The product was digested with EcoP15I endonuclease (New England BioLabs, Ipswich, MA), and the 5′-terminal cDNA fragments were isolated with streptavidin-coated magnetic beads (Dynal, Oslo, Norway) and ligated to a SOLiD sequencing linker (5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′ annealed to 5′-ATCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGGTT-3′) with T4 DNA ligase (Invitrogen) at 16°C for at least 2 h. The DNA was amplified by PCR using primers 5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ and 5′-CTGCCCCGGGTTCCTCATTCT-3′, and the products were purified for sequencing with the SOLiD 3+ system (Applied Biosystems, Carlsbad, CA).
Generation of the SOLiD FL-CAGE library.
Components from the SuperScript full-length cDNA library construction kit (Invitrogen) were used in generating the SOLiD FL-CAGE (full-length CAGE) library unless otherwise specified (see Fig. S1B in the supplemental material). Purified polyadenylated RNA was reverse transcribed using a primer [5′-GCGGCTGAAGACGGCCTATGTGCAGCAG(T)17-3′], and the cDNA-RNA hybrids were treated with RNase I to degrade truncated cDNA-RNA hybrids. The full-length cDNA was selected using RNA 5′ cap antibody-conjugated magnetic beads and ligated with a SOLiD sequencing linker containing an EcoP15I recognition site (5′-TCCGCCCTGCCCCGGGTTCCTCATTCTCTCAGCAG-3′ annealed to 5′-p-CTGCTGAGAGAATGAGGAACCCGGGGCAGG-amine-3′). The cDNA was amplified by 10 cycles of PCR using a 5′ PCR primer (5′ [biotin-TEG]-CTGCCCCGGGTTCCTCATTCT-3′) and 3′ PCR primer (5′-GCGGCTGAAGACGGCCTATGT-3′). The product was digested with the EcoP15I endonuclease, and the 5′-terminal cDNA fragments were bound to streptavidin magnetic beads and ligated to a SOLiD sequencing linker (5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′ annealed to 5′-p-NNATCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGG-amine-3′). The product was amplified using primers (5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ and 5′-CTGCCCCGGGTTCCTCATTCT-3′) and purified for sequencing by using SOLiD sequencing technology.
Generation of the Illumina CAGE library.
The Illumina CAGE library was generated as described previously (56) with modifications (see Fig. S2 in the supplemental material). Total RNA (100 μg) was treated with bacterial alkaline phosphatase and tobacco acid pyrophosphatase and ligated with RNA oligonucleotide (5′-ACACUCUUUCCCUACACGACGCUCUUCCGAUCUGG-3′) using T4 RNA ligase (TaKaRa). Polyadenylated RNA was isolated with Dynabeads mRNA kit. First-strand cDNA was synthesized using reverse transcriptase (SuperScript II; Invitrogen) with random hexamer primer (5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTNNNNNNC-3′) at 12°C for 1 h and then at 42°C for 3 h. RNA was degraded after first-strand synthesis, and the cDNA was amplified by PCR using primers 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and 5′-CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT-3′. PCR products of 200 to 500 bp were purified for Illumina GA IIx sequencing.
Generation of SOLiD 3′-end library.
The procedure for generating the SOLiD 3′-end library was modified from the method described by Wei et al. (59) for SOLiD sequencing (see Fig. S3 in the supplemental material). Polyadenylated mRNA was isolated by using the Dynabeads mRNA kit. Double-stranded cDNA was prepared using the SuperScript double-stranded cDNA synthesis kit as suggested by the manufacturer (Invitrogen). An anchored oligo(dT)16 primer with a GsuI restriction site [5′-biotin-TEG-GAGAGAGAGACTGGAG(T)16VN-3′], where V is A, C, or G and N is any nucleotide, was used for first-strand synthesis with 0.5 mM methyl-dCTP instead of dCTP to prevent the digestion of DNA by GsuI at sites other than the one in the primer. The second strand was synthesized according to the manufacturer's recommendation except that excess dCTP (0.6 mM) was used. The cDNAs were purified with Invitrogen PCR column and bound to magnetic streptavidin beads followed by GsuI digestion. The released fraction was recovered, purified, and ligated to a linker with EcoP15I recognition site and biotin-TEG (5′-p-CTGCTGAGAGAATGAGGAACCCGGGGCAG-3′ annealed to 5′-biotin-TEG-CCACTGCCCCGGGTTCCTCATTCTCTCAGCAGTT-3′) using T4 DNA ligase (Invitrogen). The ligated DNA was digested with EcoP15I endonuclease, and the 50- to 60-bp product was recovered from a 10% Tris-borate-EDTA (TBE) gel (Invitrogen), bound to streptavidin beads, and ligated with a SOLiD sequencing linker (5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT-3′ annealed to 5′-p-NNATCACCGACTGCCCATAGAGAGGAAAGCGGAGGCGTAGTGG-amine-3′). The resulting DNA was then amplified using primers (5′-CCACTACGCCTCCGCTTTCCTCTCTATG-3′ and 5′-CTGCCCCGGGTTCCTCATTCT-3′) and purified for sequencing.
Sequencing and data processing.
Sequencing was performed with the SOLiD 3+ system (Applied Biosystems, Carlsbad, CA) or the GA IIx system (Illumina, San Diego, CA) according to the manufacturer's instructions. The SOLiD reads were mapped to the VACV genome (NCBI accession no. NC_006998) using a modified version of Applied Biosystems small RNA analysis tool. The Illumina reads were trimmed of the leading poly(A) sequence and then mapped to the VACV genome using ZOOM (Bioinformatics Solutions Inc., Waterloo, Canada). Uniquely mapped reads and reads that mapped to two loci were analyzed. The reads that mapped to two loci were used to compensate for duplication of VACV genes in the inverted terminal repetition. The reads mapped to VACV genome were used for transcription start site (TSS) and polyadenylation site (PAS) determination. The TSSs and PASs were visualized with Mochiview (28). The sequencing data have been deposited in the Sequence Read Archive (SRA) of NCBI under accession number SRA029884.
Annotation of TSSs of VACV early ORFs.
The following rules for annotation of SOLiD oCAGE data were applied. (i) TSSs with at least 10 counts (the number of sequence reads that map to a specific TSS are referred to as counts) for SOLiD oCAGE and Illumina CAGE or 5 counts for SOLiD FL-CAGE separated by less than 10 nucleotides (nt) were considered a cluster. (ii) The priority order of usage of SOLiD oCAGE data for annotation and analysis was as follows: 1 h in the presence of cycloheximide (CHX), 2 h in the presence of CHX, and 4 h in the presence of CHX. (iii) For TSS patterns, the peaks were defined as the TSSs with highest counts and with more than 25% of the latter. (iv) The closest TSS cluster before the start codon was annotated in Table S2 in the supplemental material or multiple TSS clusters within 100 nt upstream of the start codon were annotated in a few cases. (v) Peaks separated by more than 25 nt were treated as separate TSS clusters. (vi) Previously reported start codons for VAC WR 046, VAC WR 061, and VAC WR 174 and an internal start site for VAC WR 101 (61) were used in place of annotated ones. (vii) VAC WR 008 and VAC WR 033 were classified as postreplicative genes but are expressed at low levels before DNA replication (61). The TSS annotation of genes with Illumina CAGE and SOLiD FL-CAGE were the same as SOLiD oCAGE except that the annotation was limited to the locations between the start codon of a gene and 100 nt further upstream of the TSS annotated in SOLiD oCAGE, and 5 counts were used as the minimum read counts for SOLiD FL-CAGE data.
Annotation of PAS sites.
The following rules were applied. (i) PASs from cells infected with virus for 2 h in the absence of CHX and cells infected with virus for 2 h in the presence of CHX were used. (ii) The highest PAS count in a cluster had to be at least 4 to be considered. (iii) PASs with at least 4 counts separated by less than 10 nt were considered in one cluster. (iv) PASs with the highest counts in clusters were used for analysis. (v) Reads with at least six consecutive A's before PASs were removed, since they might be derived from internal priming during reverse transcription.
Consensus motif generation and searches.
The consensus motif of VACV early promoters was generated using the online motif discovery tool Multiple Expectation maximization for Motif Elicitation (MEME) program with the assumption that there is zero or one motif per sequence (4). The searching of motif occurrences was performed with the Find Individual Motif Occurrences (FIMO) program using the motif generated from MEME as the input motif (4).
RESULTS
Comparison of capped 5′ ends determined by alternative methods and platforms.
We used SOLiD (sequencing by oligonucleotide ligation and detection) oCAGE (oligonucleotide cap analysis of gene expression), SOLiD full-length CAGE (FL-CAGE), and Illumina CAGE technologies to sequence the 5′ capped ends of polyadenylated mRNAs from vaccinia virus (VACV)-infected cells. The procedures involved tagging the capped ends of mRNA and accommodated the relatively short sequence reads obtained with the SOLiD and Illumina sequencing platforms (see Fig. S1 and S2 in the supplemental material). In the SOLiD oCAGE method, enzymatic cap removal was followed by attachment of a linker oligonucleotide encoding a site recognized by the EcoP15I type III endonuclease, which cleaves DNA 25 to 27 bp distal. FL-CAGE included steps in which the full-length cDNA was isolated along with mRNA by cap antibody-conjugated beads. The Illumina CAGE method used enzymatic cap removal and attachment of a linker oligonucleotide, like SOLiD oCAGE, but this was followed by random priming instead of EcoP15I digestion and provided 76-nt length reads, allowing detection of nontemplated as well as templated sequences downstream of the cap. Results obtained by the three different methods and two sequencing platforms were compared below.
Previously, we determined by RNA-seq that the viral mRNAs detected from 0.5 to 2 h after infection of HeLa cells with VACV in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX) or the DNA replication inhibitor cytosine arabinoside belong exclusively to the early regulatory class and that additional postreplication (PR) viral mRNAs are present at 4 h in the absence of inhibitors (61). For the present study, we used SOLiD oCAGE technology to sequence short cDNAs derived from the 5′-capped ends of polyadenylated mRNAs of HeLa cells infected with VACV for 1, 2, and 4 h in the presence of CHX and 2 h in the absence of CHX. For comparison, we also used the SOLiD FL-CAGE and Illumina CAGE methods to sequence samples from cells infected with virus with CHX for 2 h and cells infected with virus without CHX for 2 h. In the presence of CHX, the number of SOLiD oCAGE VACV-specific sequence reads increased from 357,000 at 1 h to 1.6 million at 2 h and 4.3 million at 4 h. Approximately 102,000 sequence reads were obtained by SOLiD FL-CAGE from a sample of cells infected with virus for 2 h in the presence of CHX. For cells infected with VACV for 2 h in the absence of CHX, we obtained 1.2 million VACV reads by SOLiD oCAGE and 708,000 reads by Illumina CAGE. These short sequence reads were aligned with the VACV genome, and the 5′ nucleotides were considered putative transcription start sites (TSSs). However, as will be discussed later, not all 5′ ends determined by CAGE analysis may be true TSSs.
The number of sequence reads that map to a specific TSS are referred to as counts. In order to compare the data from different protocols and platforms, the number of counts for individual TSSs was expressed as a percentage of the total number of counts for all TSSs from the same sample. The TSSs that contained at least 0.01% of the total viral TSS counts were aligned with respect to the top and bottom strands of the VACV genome. Very similar results were obtained regardless of the infection time, presence or absence of CHX, CAGE method, and sequencing platform (Fig. 1). The densities of TSSs were highest at the right side of the top strand and the left side of the bottom strand, which is consistent with whole-transcriptome mapping of early mRNAs (61).
Fig. 1.
Comparison of vaccinia virus (VACV) genome-wide transcription start sites (TSSs) determined at different times and by various CAGE (cap analysis of gene expression) methods. The sequences were from cells that were infected with virus for 1 h, 2, or 4 h in the presence of cycloheximide (1h/CHX, 2h/CHX, or 4h/CHX, respectively) or cells infected with virus for 2 h in the absence of cycloheximide (2h). The number of viral reads that mapped to each nucleotide were divided by the total number of reads for that sample and the ones with at least 0.01% of the total were aligned with the top and bottom DNA strands. The genome nucleotide numbers are below each strand in kilobases (kb).
TSS patterns.
We considered TSSs with at least 10 counts each (or 5 counts for FL-CAGE) that occurred within 10 nt of another as belonging to a cluster. The TSSs with the largest number of reads in a cluster and those with at least 25% of that were defined as peaks. Three common cluster patterns were defined: the single-peak (SP) pattern has only one peak in a cluster; the multiple-peak (MP) pattern has more than one peak located within a 4-nt window and no other peaks; the broad-region (BR) pattern has multiple peaks with some more than 4 nt from each other. Examples of the three patterns are shown in Fig. 2A. In agreement with previous studies, which showed that most VACV early mRNAs initiated with a G or A (12, 16), we found G or A at the majority of TSSs for the annotated ORFs (Fig. 2B; see Table S1 in the supplemental material). However, T was predominant in TSS clusters with a broad pattern (Fig. 2B), suggesting a difference in TSS selection or 5′-end formation.
Fig. 2.
TSS patterns and initiator nucleotide usage. (A) TSS patterns. The vertical bars indicate the percentages of counts relative to the highest in the cluster at each position in a 64-nt window. The number in the top left corner of each panel indicates the genome nucleotide location of the TSS with the highest counts in the cluster. Examples of single-peak (SP), multiple-peak (MP) and broad-region (BR) patterns are shown. (B) Initiator nucleotide usage. The frequencies of A, C, G, and T at TSSs were determined from the highest peak in each cluster. The SOLiD oCAGE data set was used.
Genome-wide mapping of TSSs.
VACV ORFs are continuous, closely spaced, mostly nonoverlapping, and located on both DNA strands. The putative TSSs from the 2-h CHX SOLiD oCAGE were plotted along the genome (Fig. 3). Counts above and below the line represent putative TSSs with rightward and leftward transcription, respectively, corresponding to the top and bottom strands of Fig. 1. Most TSSs mapped to regions preceding or within ORFs, with only a minority antisense to ORFs. The ORFs expressed early (depicted by red arrows) are clustered in 50- to 60-kbp regions near each end of the genome and are mostly transcribed toward the nearer terminus. The black arrows represent the ORFs that are transcribed after viral DNA replication. Although the majority of TSSs are associated with ORFs previously annotated as early (61), some align within PR ORFs. There was a wide range in the number of counts obtained at individual sites, so that those with less than 50 at a single nucleotide may be difficult to see in Fig. 3. In view of the multistep procedure, including PCR amplification, and the wide range in the length of transcripts, the number of counts cannot be confidently used for quantification.
Fig. 3.
VACV genome-wide TSS map. RNA was isolated at 2 h postinfection in the presence of CHX and processed by the SOLiD oCAGE method. The TSS counts mapping to the top and bottom DNA strands are displayed above and below the black horizontal line, respectively. The highest counts are off the scale for display purposes. The red and black arrowheads and arrows indicate the direction of transcription of early and postreplication (PR) ORFs, respectively, as described previously (61). The genome nucleotide numbers (in kilobases) are shown below each panel.
Annotation of TSSs preceding VACV early ORFs.
We first analyzed the reads preceding ORFs, as they are likely to depict true TSSs rather than locations of RNA processing. Using 10 counts as the lower limit for significance with SOLiD oCAGE, 74 TSSs were identified at 1 h after infection in the presence of CHX; 96 TSSs were identified at 2 h and 110 were identified at 4 h (more than one TSS was present within 100 nt of some ORFs). In the absence of CHX, 101 TSSs were detected at 2 h. Data from the SOLiD FL-CAGE (lower limit 5 counts) and Illumina CAGE (lower limit 10 counts) yielded TSSs for most of the early genes. In this manner, we annotated the TSSs for 102 early genes and 2 postreplicative genes with low-level expression at early times by SOLiD oCAGE, 87 by Illumina CAGE, and 77 by SOLiD FL-CAGE.
Data obtained by each CAGE method for the TSSs previously identified by conventional high-resolution methods (primer extension and S1 nuclease mapping) and for the entire early transcriptome (omitting 17 pseudogenes near the left end of the genome) are provided in Table 1 and Table S1 in the supplemental material, respectively. Both tables also indicate the initiator nucleotide, the lengths of the 5′ untranslated regions (5′ UTRs), the patterns of the TSSs, and the presence of a conserved core promoter motif, which will be discussed later. Of 21 previously determined TSSs, 20 were in agreement with the present data or differed by only a few nucleotides (Table 1), providing confidence for the much larger number of TSSs that were not previously analyzed (see Table S1 in the supplemental material). We also observed good agreement between the three methods used in the present study: 77 out of the 87 annotated TSS clusters by Illumina CAGE agreed closely with the TSSs by SOLiD oCAGE, and 64 out of the 77 annotated TSS clusters by SOLiD FL-CAGE agreed closely with the TSSs by SOLiD oCAGE (Table S1). Most of the differences occurred with TSSs with pyrimidine initiators and/or broad cluster patterns. The initiator nucleotide and length of the 5′ UTR were determined from the TSS with the highest counts. The lengths of the 5′ untranslated regions varied from 3 to 601 nt with a median of 21 nt. The TSS for the F2L ORF was −2 nt relative to the annotated first codon, suggesting that the second ATG located 8 nt downstream may be the actual translation initiation site. Protein analyses will be needed to verify that ORFs associated with apparently short untranslated leaders use a downstream initiation codon.
Table 1.
Comparison of TSSs determined by genome-wide CAGE and conventional methods
| ORF WRa | ORF COPa | TSS in literaturea | SOLiD oCAGEb | Illumina CAGEb | SOLiD FL-CAGEb | TSS patternc | Motifd | 5′ UTR lengthe | 5′ nte |
|---|---|---|---|---|---|---|---|---|---|
| 001 | C23L | 4447; 4449 | 4447; 4449 | 4447; 4449 | 4450; 4455 | BR | HM | 74 | G |
| 009 | C11R | 7278 | 7276; 7277; 7278; 7279 | 7277; 7278; 7279 | 7271; 7272; 7278 | MP | HM | 56 | C |
| 010 | C10L | 8908–8911 | 8909 | 8908 | 8908; 8909 | SP | 4 | G | |
| 029 | N2L | 22848; 22849 | 22829; 22845 | 22845 | 22851 | BR | HM | 9 | G |
| 060 | E4L | 49209–49211 | 49208 | 49205; 49208 | 49202; 49206; 49212 | SP | HM | 21 | G |
| 065 | E9R | 56658 | 56658; 56660; 56661 | 56659; 56675; 56677 | MP | HM | 4 | A | |
| 072 | I3L | 61869 | 61867; 61868; 61869 | 61867–61869 | 61869 | MP | HM | 27 | A |
| 073 | I4L | 64255; 64256 | 64255 | 64255 | 64255 | SP | HM | 15 | A |
| 080 | G2R | ∼600 bp upstream | 70477 | 70477/70642 | 70471 | SP | HM | 601 | G |
| 094 | J2R | 80718 | 80718; 80721 | 80718 | 80712 | MP | HM | 6 | G |
| 096 | J4R | 82040; 82043 | 82043 | 82039–82043 | 82042 | SP | HM | 196 | A |
| 098 | J6R | 83298 | 83298 | 83296; 83298 | 83292 | SP | HM | 67 | A |
| 101 | H3L | 83620 | 88617; 88619 | MP | HM | NA | G | ||
| 103 | H5R | 91794 | 91793 | 91793 | 91786; 91793 | SP | HM | 78 | G |
| 106 | D1R | 93935–93937 | 93935; 93936 | 93935; 93936 | 93929 | MP | HM | 13 | A |
| 109 | D4R | 97559–97562 | 97094 | SP | T | ||||
| 110 | D5R | 98269–98274; 98299 | 98271; 98269–98276 | 98272–98274 | 98273 | BR | HM | 4 | C |
| 112 | D7R | 102604–102610 | 102610; 102606–102611 | 102610 | BR | HM | 3 | T | |
| 114 | D9R | 103972; 104007; 104008; 104010 | 104008; 104014; 104017 | 104008; 104014 | 104001; 104007 | BR | HM | 3 | A |
| 117 | D12L | 108197–108201; 108204 | 108191; 108199; 108198–108201 | 108187; 108198 | 108188; 108197; 108199 | BR | HM | 4 | G |
| 138 | A18R | 125782–125784 | 125946; 125949/125785f | BR/SPf | 260/424f | C/Gf |
Vaccinia virus (VACV) Western Reserve (WR) and corresponding Copenhagen (COP) ORF nomenclature. Table S1 in the supplemental material contains the complete list of TSSs for all ORFs and literature references.
Annotation follows the rules in Materials and Methods. The TSS with the highest count in a cluster is underlined. TSS annotation with Illumina CAGE and SOLiD FLCAGE was limited to the locations between the start codon of the gene and 100 nt upstream of TSS annotated in SOLiD oCAGE.
TSS pattern abbreviations: BR, broad range; MP, multiple peaks; SP, single peak. The TSS patterns were determined from data of the earliest time point available in SOLiD oCAGE for cells infected with virus in the presence of CHX.
Highly conserved motifs (HM) have a P value of <0.0001 by the FIMO or MEME program.
Based on TSSs with the highest counts annotated in SOLiD oCAGE.
The 5′ nucleotide identified from a sample of cells infected with virus for 4 h in the presence of CHX analyzed by SOLiD oCAGE.
5′ poly(A) sequences.
The TSSs of characterized PR genes have a TTT in the template strand sequence, which apparently results in RNA polymerase slippage and a 5′ poly(A) leader. There are, however, two reported examples of VACV early mRNAs with poly(A) sequences at their 5′ ends (2, 31). The 76-nt sequence reads obtained by Illumina CAGE were long enough to detect 5′ poly(A) sequences and still align the remainder of the sequence with the genome, whereas this could not be done with the shorter SOLiD reads. We identified 18 early mRNAs with 5′ poly(A) sequences, including the two previously found (see Table S2 in the supplemental material). Up to 13 consecutive A's were detected; however, most had fewer than 8 A's. Examination of the published gene sequences of VACV WR indicated that 16 had at least three T's in the template strand at the TSS, but in two cases, there were only two T's. Presumably, the 5′ poly(A) sequences of early RNAs result from transcriptional slippage, which evidently is not restricted to intermediate and late genes but is probably a characteristic of the VACV RNA polymerase per se.
VACV early promoter motif.
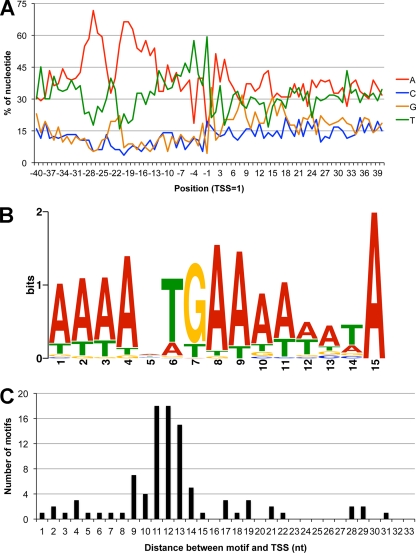
An optimal core promoter sequence, deduced by comprehensive mutagenesis, corresponded with a multiple alignment of sequences upstream of representative early genes (16) and a motif derived from our previous whole-transcriptome analysis (61). The accurate mapping of more than 100 TSSs now allowed us to carry out a more comprehensive analysis. The occurrences of the four bases at each position surrounding the peak TSS upstream of ORFs, determined by SOLiD oCAGE, were plotted (Fig. 4A). This representation revealed a high frequency of A's interrupted by TG on the coding strand from positions −13 to −29 relative to the peak TSSs. In addition, there was a high incidence of T residues just upstream of the TSS. We then extracted the 50-nt sequence upstream of the TSS of each ORF and generated a consensus motif using the MEME program (4) (Fig. 4B). Interesting features of the 15-nt motif include the TG at positions 6 and 7 and the high frequency of A's at positions 4, 8, 9, and particularly 15. In addition, the second most frequent residue in place of A is T throughout the motif. We determined that the sequences upstream of 84 out of the 111 ORF TSSs matched the motif with a P value of <0.0001 based on the FIMO (5) or MEME program. The remaining ORF TSSs had short runs of A's in the location of the core motif. The distances from the conserved core motifs to the annotated 84 TSSs are depicted in Fig. 4C. The median distance after position 15 of the motif was 12 nt. TSSs preceded by highly conserved promoter motifs were less likely to have a BR pattern and more likely to have a purine (76.5%) than those with less conserved motifs (23.1%) (see Table S1 in the supplemental material).
Fig. 4.
Early promoter motif. (A) Plot of A, C, G, and T frequencies at each position from 40 nt upstream and 40 nt downstream of all annotated VACV TSSs from SOLiD oCAGE samples. (B) The core promoter motif generated from 50-nt sequences upstream of the annotated VACV TSSs by the MEME program with the assumption that there is zero or one motif in each sequence. (C) The distances after position 15 of the motif to the transcription site were plotted. In each panel, the nucleotide (nt) at the highest peak in a cluster was used as the TSS for the calculations.
AT-rich spacer between the conserved promoter motif and TSS.
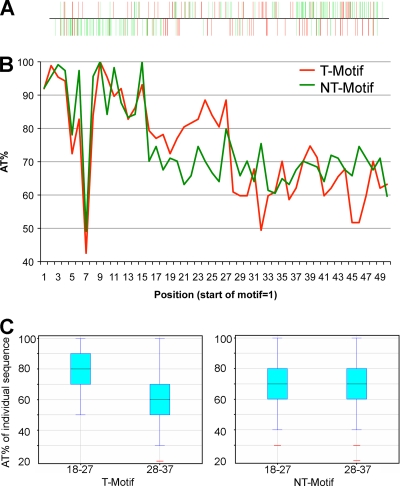
We were curious as to whether a highly conserved core promoter motif was sufficient for transcription and therefore searched for copies throughout the VACV genome. Using the FIMO program with a P value output threshold of <0.0001, we found 318 occurrences of the highly conserved A-rich motif (Fig. 5A). Of these, 114 motifs were not associated with an identified TSS, suggesting that this sequence is insufficient to initiate early transcription. We refer to the highly conserved core motifs associated with TSSs of annotated ORFs as T-motifs and those not associated with TSSs as NT-motifs, respectively. When the sequences downstream of T- and NT-motifs were analyzed, we found a 10-nt sequence with a higher AT content in the T-motifs than in the NT-motifs (Fig. 5B). The AT contents of the sequences preceding the TSS (nt 18 to 27) and following the TSS (nt 28 to 37) of individual T- and NT-motifs are shown in box-and-whisker plots (Fig. 5C). The median AT content was 80% and 60% for nt 18 to 27 and nt 28 to 37, respectively, associated with T-motifs, whereas both were 70% for the NT-motifs. Thus, the presence of an AT-rich spacer, in addition to a core motif, is characteristic of a functional TSS.
Fig. 5.
AT frequency following the core motif correlates with transcription. (A) Distribution of the 318 core motifs on the VACV genome (P < 0.0001 by the FIMO program). The motifs with no or only one TSS count were colored red, and the ones with more than one count were colored green. (B) Incidence of A and T at each position 50 nt downstream of the T-motif and NT-motif. (C) Box-and-whisker plots of AT frequency of the 10 nt from positions 18 to 27 and positions 28 to 37 of individual T-motifs and NT-motifs, respectively. In the box-and-whisker plots, the first and third quartiles are indicated by the bottom and top of the box, respectively. The median is indicated by the line in the middle of the box. The “whiskers” extend to the farthest points that are within 1.5 times the interquartile range.
Anomalous TSSs.
Until now, we have mainly considered TSSs preceding annotated ORFs. However, with each method, many anomalous RNA 5′ ends were mapped within ORFs and a smaller number antisense to ORFs. Since the majority of TSSs upstream of ORFs are preceded by a highly conserved promoter motif, we analyzed the sequences preceding the anomalous TSSs. Thirty-three putative TSSs with highly conserved core promoter motifs (P < 0.0001), from the sample of cells infected with virus for 2 h in the presence of CHX that was analyzed by the SOLiD oCAGE, are listed in Table S3 in the supplemental material. The majority of such TSSs was located in or between ORFs and initiated with a purine. SP, MP, and BR TSS patterns were found. Some TSSs were positioned to provide additional upstream TSSs for annotated ORFs and would not result in a new or altered protein. TSSs within ORFs were predicted to encode a shorter in-frame protein. The median length of such shortened ORFs was 641 nt, and seven were predicted to encode proteins greater than 24 kDa.
The two antisense TSSs listed in Table S3 in the supplemental material were closely spaced and predicted to encode a novel 52-amino-acid protein. Additional putative antisense TSSs without a conserved core promoter motif were found. Thirty-five TSSs identified in the sample of cells infected with virus for 2 h in the presence of CHX and analyzed by oCAGE and these TSSs are located antisense to ORFs are listed in Table S4 in the supplemental material. Some of the antisense TSSs occurred in clusters with five antisense to the I6L ORF, five to the A4L ORF, and three to the A55R ORF.
Determination of genome-wide PASs.
We also analyzed the sequences adjacent to the 3′ poly(A) tails of VACV early RNAs, as no genome-wide analysis of the 3′ ends of VACV mRNAs had been reported. The scheme used for isolation of ∼25-nt PAS sequence tags from the 3′ ends of RNAs is shown in Fig. S3 in the supplemental material. An important feature was the use of a biotinylated poly(dT) dinucleotide-anchored primer with a GsuI type IIs restriction endonuclease site for reverse transcription. The dinucleotide anchor ensured that reverse transcription did not initiate within the poly(A) tail, which would have resulted in sequences too long for analysis with the SOLiD platform. After second-strand synthesis, the cDNA was cleaved with GsuI to leave two A's marking the original poly(A) site. After additional steps, including attachment and subsequent digestion of a linker with an EcoP15I site, the short DNA fragments were sequenced.
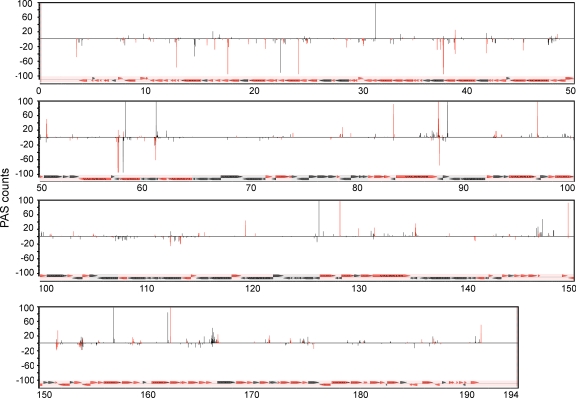
HeLa cells were infected with VACV in the presence and absence of CHX for 2 h, and polyadenylated RNA was isolated. The cDNAs derived from the 3′ ends were sequenced as described above, and 15,516 and 16,574 VACV reads were generated from the samples infected in the presence and absence of CHX, respectively. The data from the no-CHX sample are shown aligned with the VACV genome in Fig. 6. Because of the close spacing of VACV genes, PASs frequently occurred within downstream ORFs. Previous in vitro studies had shown that a U5NU sequence (transcription termination signal comprised of five consecutive U's followed by any nucleotide and then another U) signals transcription termination of VACV early mRNAs (53, 64), and the mapping of representative mRNAs from infected cells supports this mechanism (22, 36). PASs within 100 nt after a T5NT (corresponding to U5NU in the RNA) sequence are shown in red, and those without such a sequence are shown in black (Fig. 6). The median distance of T5NTs to the closest PAS after the stop codon of an ORF was determined to be 40 nt (Fig. 7A; see Table S5 in the supplemental material).
Fig. 6.
VACV genome-wide polyadenylation site (PAS) map. RNA was isolated at 2 h after VACV infection in the absence of CHX and processed to determine PASs as outlined in Fig. S3 in the supplemental material. The counts mapping to individual nucleotides in the top and bottom DNA strands are displayed above and below the black horizontal line, respectively. PASs that are preceded within 100 nt by T5NT are colored red, and those without T5NT are black. The red and black arrows indicate the directions of transcription of the early and postreplicative ORFs, respectively. The genome nucleotide numbers (in kilobases) are shown below each panel.
Fig. 7.
Relation of PASs to upstream sequences. (A) Distance between T5NT and the nearest downstream PAS. (B) Examples of discrete and cluster PASs. In the top two panels, the T5NT sequences are marked by black horizontal lines. The black vertical bars indicate the PASs. No T5NT sequences were present within 100 nt upstream in the bottom two panels. The numbers indicate the start and end nucleotide of each displayed sequence.
Approximately 70% of PASs were discrete, i.e., a single nucleotide, while others consisted of small clusters (Fig. 7B). Some of the clusters followed consecutive or overlapping T5NT sequences as shown by an example (Fig. 7B). Both discrete and clustered PASs also occurred in the absence of a T5NT sequence (Fig. 7B).
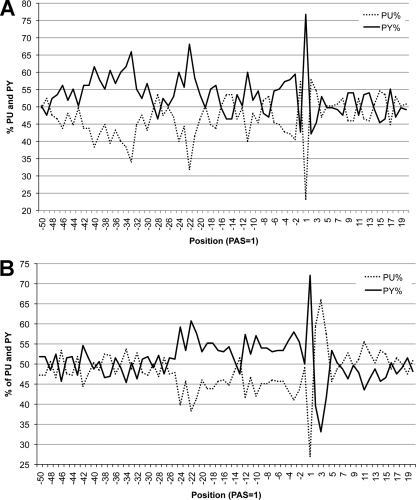
Taking into account all PASs with 4 or more reads per nucleotide, only 185 of the 508 PASs were preceded by T5NT. However, if we consider only the closest PAS after the stop codon of an ORF, the majority (67 of 86) had a T5NT within 100 nt upstream (see Table S5 in the supplemental material). The large number of PASs that did not follow a T5NT motif suggested the existence of alternative mechanisms of polyadenylation site selection. In searching for other correlates of termination, we first considered the possibility of other T-rich sequences and therefore plotted the frequency of the four bases in the regions flanking PASs that are preceded by T5NT or are not preceded by T5NT. No enrichment of T's or any other base was apparent upstream of the anomalous PASs (data not shown). However, an intriguing feature was noted when the percentages of purines and pyrimidines were plotted (Fig. 8). In the case of the U5NU-associated PASs, the region up to position −50 has a pyrimidine-rich coding strand with peaks at position −34, which includes the U5NU motif, and at position −22 (Fig. 8A). The coding strand upstream of those PASs that are not associated with U5NU is also pyrimidine rich up to position −25 (Fig. 8B). These data suggested a possible role for the proximal pyrimidine-rich region in polyadenylation site selection both with and without the canonical U5NU motif.
Fig. 8.
Pyrimidine (PY) and purine (PU) frequencies surrounding PASs. (A) Frequencies for PASs preceded within 100 nt by T5NT; (B) frequencies for PASs not preceded by T5NT.
DISCUSSION
Poxviruses are unusual in that they carry out the entire replication cycle within the cytoplasm and encode a multisubunit RNA polymerase and specific transcription factors as well as enzymes that form the 5′ cap and 3′ poly(A) tail. Gene expression profiles of vaccinia virus (VACV) have been determined with tiling microarrays (3, 48) and by RNA-seq technology (61). The main objectives of the present study were to define the transcriptional start and stop sites and analyze cis-acting signals regulating VACV early gene expression. Relative to studies with eukaryotic genomes, mapping was simplified by the absence of splicing or cleavage steps in early mRNA formation and the ∼200,000-bp size of the genome, which made alignments accurate even for relatively short sequence reads. To reduce bias in the 5′ analysis, we used three different CAGE (cap analysis of gene expression) methods, two sequencing platforms, and isolated RNAs at different times and in the absence and presence of cycloheximide (CHX), which prevented the formation of viral proteins, including enzymes for decapping mRNAs, viral DNA synthesis, and postreplication (PR) transcription. Overall, similar data were obtained by each method and condition of infection. We concentrated our analysis on the 5′ ends that closely precede the translation initiation codon, as these should be directly involved in VACV gene expression.
TSSs preceding 104 ORFs were identified, including 101 early ORFs, one internal ORF, and two PR ORFs with low-level expression at the early stage. In some cases, there was a single predominant 5′-end nucleotide, whereas one or more clusters of 5′ ends were found in others. The lengths of the untranslated leaders varied from 3 to 601 nt with a median of 21 nt. It is possible that an unannotated downstream ATG is used as the initiation codon for transcripts with apparent leader sequences that are very short. Having defined the TSSs of essentially all early mRNAs, we extracted a 15-nt consensus promoter motif that is enriched in A residues at specific locations and has a characteristic TG near the center, consistent with the requirements for gene expression determined by promoter mutations and binding of the VACV gene-encoded early transcription factor (16, 63). Sequences upstream of 84 out of the 111 VACV early TSSs (for 104 ORFs) matched the motif with a P value of <0.0001, indicating high significance. Interestingly, this motif is similar to one found upstream of mimivirus early genes (37). Mimiviruses and poxviruses belong to the group of nucleocytoplasmic large DNA viruses, which are believed to be distantly related (35). Unlike the situation with eukaryotic transcription, there is no evidence for the use of enhancer elements in VACV.
The TSSs occurred at a single predominant nucleotide in some cases and in clusters in others, similar to findings with eukaryotic TSSs (14). Earlier studies had indicated that the VACV caps are predominantly m7GpppGm and m7GpppAm, implying that transcription starts with a G or A residue (12). We found G or A at the majority of TSSs, but T was frequent in TSS clusters with a broad pattern and a less-conserved core promoter motif. On the other hand, we found a large number of highly conserved motifs that were not associated with a TSS, suggesting that functional promoters may have another previously unrecognized feature. We found that the motifs associated with TSSs had a 20% higher AT content immediately before the TSS than immediately after it, whereas this characteristic was not present at the same location after motifs not associated with a TSS. The conserved core motif (P < 0.0001) followed by a high AT-rich sequence was also found preceding 46 of 62 ORFs of molluscum contagiosum virus, a distantly related poxvirus with a high overall GC content (Z. Yang, unpublished data). We speculate that the AT-rich “spacer region” between the core motif and the TSS is a conserved feature of poxvirus promoters and may facilitate unwinding of the DNA strands to form the open complex for transcription initiation.
Consistent with recent studies of the eukaryotic transcriptome (11, 29, 32, 44), we detected a large number of VACV 5′ ends by both oCAGE and FL-CAGE technologies that could not be assigned to sites immediately upstream of annotated ORFs. The majority of these anomalous RNAs were mapped within positive-sense ORFs. A small number of these (less than 5%) were associated with highly conserved core promoter motifs and could allow translation of shorter protein isoforms. In two cases, Western blotting was performed and smaller proteins were identified (62) (Yang, unpublished). The majority of the anomalous 5′ ends, however, were not associated with conserved promoter motifs and had a higher frequency of broad and wide TSSs with a pyrimidine in the +1 position. Similarly, in Drosophila, the 5′ capped ends that mapped within ORFs are less likely to have the canonical TATA box promoter element and more likely to have a broad pattern with C in the +1 position (44). These characteristics raise the question of whether such capped 5′ ends were generated by nonstringent transcription initiation, posttranscriptional mechanisms, or imperfect CAGE methodology. With regard to posttranscriptional mechanisms, there is evidence for secondary recapping of processed RNAs in the cytoplasm (24, 41, 45) and a 5′-phosphate polynucleotide kinase was isolated from VACV cores (54). Few TSSs with relatively low read counts were mapped antisense to VACV ORFs. However, since only polyadenylated RNAs were analyzed in the present studies, additional nonpolyadenylated antisense RNAs may be unrecognized.
Polyadenylation of newly synthesized RNA polymerase II transcripts is carried out by a multiprotein complex that cleaves the nascent RNA usually 10 to 30 nt downstream of AAUAAA or variants of that sequence and then adds multiple adenylates to the 3′ end (10). A cleavage mechanism involving the H5 protein of VACV has been proposed for processing of VACV late mRNAs (18, 19, 30). However, biochemical studies have shown that polyadenylation of VACV early mRNAs involves other identified virus gene-encoded proteins and occurs after termination at 25 to 50 nt after a U5NU sequence (20, 51, 52, 64). S1 nuclease analysis of several early mRNAs made in VACV-infected cells indicated clusters of 3′ ends (36). For the majority of ORFs that are transcribed early in infection, we found a T5NT (corresponding to U5NU in the RNA) motif about 40 nt before the first PAS. PASs further downstream may provide backup termination or termination of anomalous transcripts or represent repolyadenylation of processed RNAs. The majority of PASs mapped to a single nucleotide, but clusters were also found. In some cases, the cluster followed a tandem or overlapping T5NT sequence. In addition, a large number of PASs were not closely associated with a T5NT sequence, suggesting alternative mechanisms of mRNA 3′-end formation. We noted a pyrimidine-rich sequence in the coding strand immediately upstream of PASs with and without T5NT motifs. The significance of this observation may be related to a recent finding that the VACV gene-encoded nucleoside triphosphate phosphohydrolase II (NPH-II) efficiently unwinds a DNA-RNA hybrid with a purine-rich DNA tracking strand (55). NPH-II was originally characterized as a DNA- and RNA-dependent triphosphate phosphohydrolase (46) and has subsequently been shown to be an RNA helicase (26). Interestingly, NPH-II-defective virions produce RNA that is abnormally long and inefficiently released from the viral core, suggesting a role in termination (25). The enrichment of purines in the tracking strand (corresponding to pyrimidines in the coding strand) upstream of the PASs may facilitate the termination activity of NPH-II.
In summary, this comprehensive, genome-wide, high-resolution analysis enormously extended knowledge of the 5′ capped and 3′ polyadenylated ends of VACV early mRNAs, uncovered an unanticipated degree of transcriptional complexity, and provided a resource for future experiments. The study confirmed and refined previously recognized promoter and termination motifs and identified new sequence elements as shown in Fig. 9. Moreover, the similar arrangement of genes in other members of the chordopoxvirus subfamily and evidence for the interchangeability of promoters (42) suggest that the results will be broadly applicable.
Fig. 9.
Features associated with TSSs and PASs. The TSS is indicated by +1. The most common nucleotide at each position of the 15-nt core promoter motif is indicated. N means that there was no predominant nucleotide. The core motif associated with a TSS is followed by an AT-rich spacer, which distinguishes it from silent core motifs in the genome. The untranslated RNA leader sequence (UTR) preceding the ATG translation initiation codon varies greatly in length with a median size of 21 nt. The RNA U5NU motif (T5NT in DNA) can occur before or after the stop codon and signals transcription termination approximately 40 nt downstream. RNAs with and without the U5NU sequence frequently have a pyrimidine-rich sequence near the PAS.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mariam Quinones and Jeff Skinner (Bioinformatics and Computational Biosciences Branch, National Institute of Allergy and Infectious Diseases) for suggestions on data analysis, Alison McBride (Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases) for technical help with the 2100 bioanalyzer, and Catherine Cotter in our laboratory for cell culture.
This study was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 13 April 2011.
REFERENCES
- 1. Ahn B.-Y., Gershon P. D., Moss B. 1994. RNA polymerase-associated protein RAP94 confers promoter specificity for initiating transcription of vaccinia virus early stage genes. J. Biol. Chem. 269:7552–7557 [PubMed] [Google Scholar]
- 2. Ahn B.-Y., Jones E. V., Moss B. 1990. Identification of the vaccinia virus gene encoding an 18-kilodalton subunit of RNA polymerase and demonstration of a 5′ poly(A) leader on its early transcript. J. Virol. 64:3019–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Assarsson E., et al. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. U. S. A. 105:2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 5. Bailey T. L., Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54 [DOI] [PubMed] [Google Scholar]
- 6. Baldick C. J., Jr., Moss B. 1993. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate stage genes. J. Virol. 67:3515–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldick C. J., Keck J. G., Moss B. 1992. Mutational analysis of the core, spacer, and initiator regions of vaccinia virus intermediate class promoters. J. Virol. 66:4710–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barbosa E., Moss B. 1978. mRNA (nucleoside-2′-)-methyltransferase from vaccinia virus. Purification and physical properties. J. Biol. Chem. 253:7692–7697 [PubMed] [Google Scholar]
- 9. Baroudy B. M., Moss B. 1980. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J. Biol. Chem. 255:4372–4380 [PubMed] [Google Scholar]
- 10. Beaudoing E., Freier S., Wyatt J. R., Claverie J. M., Gautheret D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Birney E., et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boone R. F., Moss B. 1977. Methylated 5′ terminal sequences of vaccinia virus mRNA species made in vivo at early and late times after infection. Virology 79:67–80 [DOI] [PubMed] [Google Scholar]
- 13. Broyles S. S., Yuen L., Shuman S., Moss B. 1988. Purification of a factor required for transcription of vaccinia virus early genes. J. Biol. Chem. 263:10754–10760 [PubMed] [Google Scholar]
- 14. Carninci P., et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38:626–635 [DOI] [PubMed] [Google Scholar]
- 15. Cooper J. A., Wittek R., Moss B. 1981. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J. Virol. 39:733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davison A. J., Moss B. 1989. The structure of vaccinia virus early promoters. J. Mol. Biol. 210:749–769 [DOI] [PubMed] [Google Scholar]
- 17. Davison A. J., Moss B. 1989. The structure of vaccinia virus late promoters. J. Mol. Biol. 210:771–784 [DOI] [PubMed] [Google Scholar]
- 18. D'Costa S. M., Antczak J. B., Pickup D. J., Condit R. C. 2004. Post-transcription cleavage generates the 3′ end of F17R transcripts in vaccinia virus. Virology 319:1–11 [DOI] [PubMed] [Google Scholar]
- 19. D'Costa S. M., Bainbridge T. W., Condit R. C. 2008. Purification and properties of the vaccinia virus mRNA processing factor. J. Biol. Chem. 283:5267–5275 [DOI] [PubMed] [Google Scholar]
- 20. Deng L., Shuman S. 1996. An ATPase component of the transcription elongation complex is required for factor-dependent transcription termination by vaccinia RNA polymerase. J. Biol. Chem. 271:29386–29392 [DOI] [PubMed] [Google Scholar]
- 21. Earl P. L., Cooper N., Wyatt L. S., Moss B., Carroll M. W. 1998. Preparation of cell cultures and vaccinia virus stocks, p. 16.16.1–16.16.3 In Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (ed.), Current protocols in molecular biology, vol. 2 John Wiley and Sons, New York, NY [Google Scholar]
- 22. Earl P. L., Hügin A. W., Moss B. 1990. Removal of cryptic poxvirus transcription termination signals from the human immunodeficiency virus type 1 envelope gene enhances expression and immunogenicity of a recombinant vaccinia virus. J. Virol. 64:2448–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Earl P. L., Moss B. 1998. Characterization of recombinant vaccinia viruses and their products, p. 16.18.1–16.18.11 In Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (ed.), Current protocols in molecular biology, vol. 2 Greene Publishing Associates & Wiley Interscience, New York, NY [Google Scholar]
- 24. Fejes-Toth K., et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457:1028–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gross C. H., Shuman S. 1996. Vaccinia virions lacking the RNA helicase nucleoside triphosphate hydrolase II are defective in early transcription. J. Virol. 70:8549–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gross C. H., Shuman S. 1996. Vaccinia virus RNA helicase: nucleic acid specificity in duplex unwinding. J. Virol. 70:2615–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hashimoto S., et al. 2009. High-resolution analysis of the 5′-end transcriptome using a next generation DNA sequencer. PLoS One 4:e4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Homann O. R., Johnson A. D. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoskins R. A., et al. 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 21:182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howard S. T., Ray C. A., Patel D. D., Antczak J. B., Pickup D. J. 1999. A 43-nucleotide RNA cis-acting element governs the site-specific formation of the 3′ end of a poxvirus late mRNA. Virology 255:190–204 [DOI] [PubMed] [Google Scholar]
- 31. Ink B. S., Pickup D. J. 1990. Vaccinia virus directs the synthesis of early mRNAs containing 5′ poly(A) sequences. Proc. Natl. Acad. Sci. U. S. A. 87:1536–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kapranov P., Willingham A. T., Gingeras T. R. 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8:413–423 [DOI] [PubMed] [Google Scholar]
- 33. Kates J., Beeson J. 1970. Ribonucleic acid synthesis in vaccinia virus. I. The mechanism of synthesis and release of RNA in vaccinia cores. J. Mol. Biol. 50:1–18 [DOI] [PubMed] [Google Scholar]
- 34. Keck J. G., Baldick C. J., Moss B. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late transactivator genes. Cell 61:801–809 [DOI] [PubMed] [Google Scholar]
- 35. Koonin E. V., Yutin N. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee-Chen G. J., Bourgeois N., Davidson K., Condit R. C., Niles E. G. 1988. Structure of the transcription initiation and termination sequences of seven early genes in the vaccinia virus HindIII D fragment. Virology 163:64–79 [DOI] [PubMed] [Google Scholar]
- 37. Legendre M., et al. 2010. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 20:664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahr A., Roberts B. E. 1984. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J. Virol. 49:510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mangone M., et al. 2010. The landscape of C. elegans 3′UTRs. Science 329:432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin S. A., Paoletti E., Moss B. 1975. Purification of mRNA guanylyltransferase and mRNA (guanine 7-)methyltransferase from vaccinia virus. J. Biol. Chem. 250:9322–9329 [PubMed] [Google Scholar]
- 41. Mercer T. R., et al. 2010. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 20:1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moss B. 2007. Poxviridae: the viruses and their replication, p. 2905–2946 In Knipe D. M., Howley P. M. (ed.), Fields virology, vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 43. Moss B., Rosenblum E. N., Paoletti E. 1973. Polyadenylate polymerase from vaccinia virions. Nature (New Biol.) 245:59–63 [DOI] [PubMed] [Google Scholar]
- 44. Ni T., et al. 2010. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods 7:521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Otsuka Y., Kedersha N. L., Schoenberg D. R. 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell. Biol. 29:2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paoletti E., Moss B. 1974. Two nucleic acid-dependent nucleoside triphosphate phosphohydrolases from vaccinia virus. Nucleotide substrate and polynucleotide cofactor specificities. J. Biol. Chem. 249:3281–3286 [PubMed] [Google Scholar]
- 47. Patel D. D., Pickup D. J. 1987. Messenger RNAs of a strongly-expressed late gene of cowpox virus contains a 5′-terminal poly(A) leader. EMBO J. 6:3787–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rubins K. H., et al. 2008. Comparative analysis of viral gene expression programs during poxvirus infection: a transcriptional map of the vaccinia and monkeypox genomes. PLoS One 3:e2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanz P., Moss B. 1999. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc. Natl. Acad. Sci. U. S. A. 96:2692–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwer B., Visca P., Vos J. C., Stunnenberg H. G. 1987. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell 50:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shuman S., Broyles S. S., Moss B. 1987. Purification and characterization of a transcription termination factor from vaccinia virions. J. Biol. Chem. 262:12372–12380 [PubMed] [Google Scholar]
- 52. Shuman S., Moss B. 1989. Bromouridine triphosphate inhibits transcription termination and mRNA release by vaccinia virions. J. Biol. Chem. 264:21356–21360 [PubMed] [Google Scholar]
- 53. Shuman S., Moss B. 1988. Factor-dependent transcription termination by vaccinia virus RNA polymerase: evidence that the cis-acting termination signal is in nascent RNA. J. Biol. Chem. 263:6220–6225 [PubMed] [Google Scholar]
- 54. Spencer E., Loring D., Hurwitz J., Monroy G. 1978. Enzymatic conversion of 5′-phosphate terminated RNA to 5′-di and triphosphate-terminated RNA. Proc. Natl. Acad. Sci. U. S. A. 75:4793–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taylor S. D., Solem A., Kawaoka J., Pyle A. M. 2010. The NPH-II helicase displays efficient DNA center dot RNA helicase activity and a pronounced purine sequence bias. J. Biol. Chem. 285:11692–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tsuchihara K., et al. 2009. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 37:2249–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Vliet A. H. 2010. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 302:1–7 [DOI] [PubMed] [Google Scholar]
- 58. Wang Z., Gerstein M., Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wei C. L., et al. 2004. 5′ Long serial analysis of gene expression (LongSAGE) and 3′ LongSAGE for transcriptome characterization and genome annotation. Proc. Natl. Acad. Sci. U SA. 101:11701–11706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wei C. M., Moss B. 1975. Methylated nucleotides block 5′-terminus of vaccinia virus mRNA. Proc. Natl. Acad. Sci. U. S. A. 72:318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang Z., Bruno D. P., Martens C. A., Porcella S. F., Moss B. 2010. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang Z., Moss B. 2009. Interaction of the vaccinia virus RNA polymerase-associated 94-kilodalton protein with the early transcription factor. J. Virol. 83:12018–12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yuen L., Davison A. J., Moss B. 1987. Early promoter-binding factor from vaccinia virions. Proc. Natl. Acad. Sci. U. S. A. 84:6069–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yuen L., Moss B. 1987. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc. Natl. Acad. Sci. U. S. A. 84:6417–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.