Abstract
Signaling through the Notch pathway controls cell growth and differentiation in metazoans. Following binding of its ligands, the intracellular part of the cell surface Notch1 receptor (Notch1-IC) is released and translocates to the nucleus, where it alters the function of the DNA-binding transcription factor CBF1/RBP-Jκ. As a result, CBF1/RBP-Jκ is converted from a repressor to an activator of gene transcription. Similarly, the Epstein Barr viral oncoprotein EBNA2, which is required for B-cell immortalization, activates genes through CBF1. Moreover, the TAN-1 and int-3 oncogenes represent activated versions of Notch1 and Notch4, respectively. Here, we show that the adenoviral oncoprotein 13S E1A also binds to CBF1/RBP-Jκ, displaces associated corepressor complexes, and activates CBF1/RBP-Jκ–dependent gene expression. Our results suggest that the central role of the Notch–CBF1/RBP-Jκ signaling pathway in cell fate decisions renders it susceptible to pathways of viral replication and oncogenic conversion.
Keywords: Oncogene, transformation, transcription, tumor virus, differentiation, development
The cell surface Notch receptors participate in intercellular signaling events that mediate cell fate specification and influence a broad spectrum of development processes. Notch receptors are expressed in uncommitted cells during development, and Notch signaling is believed to maintain the proliferative capacity of immature cells in the adult organism (Egan et al. 1998; Artavanis-Tsakonas et al. 1999). Activation of Notch by ligand-expressing cells results in the proteolytic cleavage, release, and translocation of the intracellular domain (Notch-IC) in the nucleus followed by the induction of its target genes (Honjo 1996; Kopan et al. 1996; Kidd et al. 1998; Struhl and Adachi 1998). Constitutive activation of Notch inhibits differentiation of various cell lineages and has been associated with malignancies. In human T-cell lymphoblastic leukemia, the translocation t(7:9) (q34q34.3) generates truncated Notch1 proteins (Notch1-IC/TAN-1; Ellisen et al. 1991) and mice expressing ectopically Notch1-IC develop T-cell neoplasms (Pear et al. 1996). The Notch4 gene is also frequently rearranged by mammary tumor virus (MMTV) proviral DNA insertion, and expression of the truncated-activated protein (Notch4-IC/int-3) in mice leads to the development of mammary adenocarcinoma (Robbins et al. 1992; Gallahan et al. 1996).
Genetic evidence identifies CBF1, also named Su(H) in Drosophila melanogaster, Lag-1 in Caenorhabditis elegans, and RBP-Jκ or KBF2 in vertebrates, as the essential transcription factor downstream from Notch (Matsunami et al. 1989; Furukawa et al. 1992; Schweisguth and Posakony 1992; Fortini and Artavanis-Tsakonas 1994; Honjo 1996). In D. melanogaster, deletion of CBF1 or Notch leads to a lethal expansion of neural tissues at the expense of epidermal tissue in the developing embryo (Furukawa et al. 1992; Schweisguth and Posakony 1992). In mice, CBF1 gene knock-out reveals that the protein is essential for postimplantation development, in particular for the somite formation and the differentiation of the nervous system (Oka et al. 1995). CBF1 binds to the DNA sequence GTGGGAA and functions in vertebrate cells as an inducible transcriptional factor (Hsieh and Hayward 1995; Waltzer et al. 1995; Hsieh et al. 1996; Kao et al. 1998). CBF1 regulates expression of a group of basic helix-loop-helix proteins encoded by the enhancer of split [E(Spl)] locus in D. melanogaster or the homologous HES genes in vertebrates. The E(Spl)/HES proteins serve as transcriptional repressors involved in cell fate decisions (Egan et al. 1998; Artavanis-Tsakonas et al. 1999).
Analysis of CBF1 structure and function showed that its central part is required for DNA binding and gene regulation (Chung et al. 1994; Hsieh and Hayward 1995; Waltzer et al. 1995; Kao et al. 1998). Several proteins, such as the deacetylase complex SMRT/sin3/HDAC (Hsieh and Hayward 1995; Hsieh et al. 1996; Kao et al. 1998), CIR (Hsieh et al. 1999), SKIP (Zhou et al. 2000), or TFIIA and TFIID (Olave et al. 1998), were found to mediate gene repression in conjunction with CBF1. Binding of Notch-IC was proposed to displace corepressor complexes from CBF1 and to turn CBF1 into a transcriptional activator (Hsieh et al. 1996; Kao et al. 1998; Kidd et al. 1998; Struhl and Adachi 1998). The Notch–CBF1 growth control pathway is exploited by the EBNA2 oncoprotein of the Epstein-Barr virus (EBV) to activate cellular and viral genes (Grossman et al. 1994; Henkel et al. 1994; Zimber-Strobl et al. 1994; Hsieh and Hayward 1995; Johannsen et al. 1995; Waltzer et al. 1995). Moreover, additional cellular and viral proteins such as Hairless (Brou et al. 1994; Schweisguth and Posakony 1994), KyoT2 (Taniguchi et al. 1998), and the Epstein-Barr viral proteins EBNA3A,C (Robertson et al. 1995; Zhao et al. 1996) also modulated CBF1 activity.
Functional CBF1 binding sites have been identified in various adenoviral promoters. This let us explore whether the adenoviral proteins E1A, similarly to EBNA2, may target CBF1. The early adenoviral proteins E1A have been studied intensively to dissect proliferation, differentiation, and transformation mechanisms. Comparison of adenoviral proteins from various serotypes highlights three conserved regions (CR): CR1, CR2, and CR3. CR1 and CR2 are present in 12S and 13SE1A splice product variants and are essential for fibroblast transformation (Flint and Shenk 1997). They bind to and modulate the activity of various cellular proteins, such the pocket-binding proteins or the pCAF and CBP/p300 acetyl-transferases (Mymryk 1996; Flint and Shenk 1997). CR3, specific for 13SE1A, displays transactivation potential. Its C-terminal sequence binds to transcription factors (Liu and Green 1990, 1994; Scholer et al. 1991; Webster and Ricciardi 1991; Mymryk 1996; Flint and Shenk 1997) and anchors the protein to promoters, whereas the N-terminal zinc finger contacts the transcription machinery (Geisberg et al. 1994; Boyer et al. 1999). 13SE1A is important for viral gene activation and viral life cycle progression. In addition, CR3 may enhance transformation (Mymryk 1996) and apoptosis (Teodoro et al. 1995). Here, we show that the 13SE1A protein binds to CBF1 and activates gene expression through CBF1 sites. Our results suggest that the function of the transcription factor CBF1, similarly to the Rb and p53 tumor suppressors, is modulated by various transforming proteins.
Results and Discussion
To test whether CBF1 is a target of E1A, we first examined the ability of E1A to modulate the expression of various CBF1-responsive reporter genes. As shown in Figure 1, E1A genomic DNA, encoding the 13S and 12S isoforms, 13SE1A, or Notch1-IC, but not 12SE1A, activate CBF1-responsive promoters derived from the cellular HES1 or the viral LMP2A genes, as well as HES5 and LMP1 promoters (data not shown). Mutation or deletion of the binding sites for CBF1 abrogated both Notch1-IC and E1A-mediated reporter activation (Fig. 1A,B). Furthermore, transfer of the CBF1 response element from the LMP2A promoter to a minimal β-globin promoter conferred repression of the basal promoter activity and both Notch-IC and E1A-inducible reporter activation (Fig. 1C, left panel). The CBF1 response element also conferred reporter activation in fibroblasts in presence of E1A (data not shown) and in the 293 cell line that harbors a single chromosomal copy of the E1A region expressing both E1A isoforms (Fig. 1C, right panel). Moreover, inducible reporter activity is enhanced by CBF1 binding site multimerization and abrogated by mutation or deletion of CBF1 response elements (Figs. 1C and 3). These data show that 13SE1A activates gene expression through CBF1 response elements.
Figure 1.
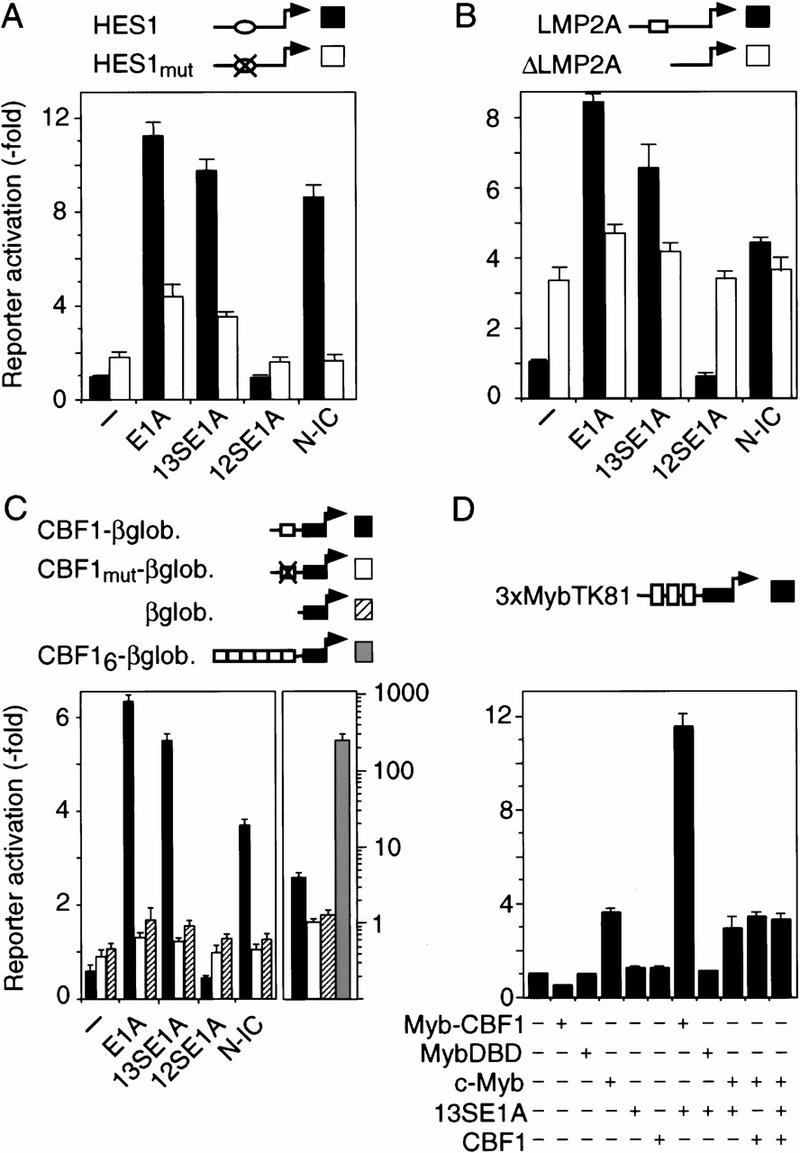
13SE1A activates gene transcription via CBF1. (A) Murine HES1 promoter reporter construct (black bars) or a promoter construct that carries a mutation in the HES1–CBF1 binding site (white bars) was transfected in HD3 cells together with genomic E1A, 13SE1A, 12SE1A, or Notch1-IC (N-IC), as indicated. (B) Epstein-Barr viral LMP2A promoter (black bars) or a construct with a deletion encompassing the 54 bp LMP2A–CBF1 response element (white bars) was transfected together with E1A or Notch1-IC as in A. (C, left) Artificial CBF1 responsive promoters that carry one (black bars), six wild-type (grey bars), or one mutated LMP2A–CBF1 response element (white bars), respectively, fused to the minimal β-globin core promoter (hatched bars), were transfected together with E1A or Notch1-IC as in A. (Right) Reporter activation of the same constructs in the 293 cells that carries a chromosomally integrated single copy of genomic E1A. (D) CBF1 was fused to the c-Myb DNA-binding domain (DBD) and cotransfected with 13S E1A in HD3 cells. Activation of reporter expression from a Myb-responsive promoter was determined following transfection of effector plasmid combinations as indicated.
Figure 3.
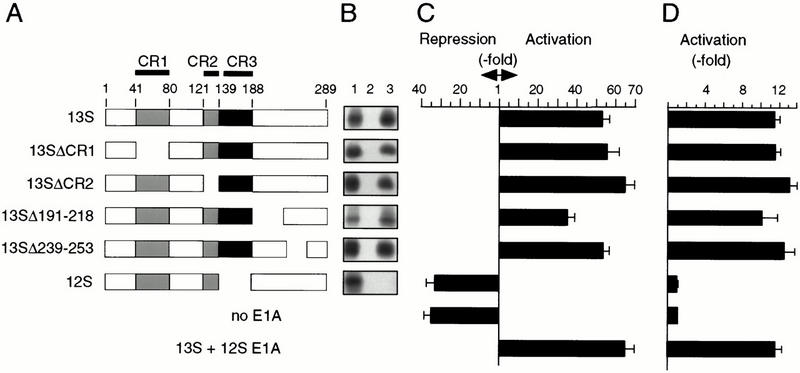
E1A binding and CBF1 activity. (A) Schematic representation of E1A proteins indicating conserved regions (CR) and amino acid numbers on the top. (B) Binding of radiolabeled E1A proteins as depicted on the left (lane 1; input, 10% of reaction) to GST (lane 2; specificity control) or GST–CBF1 fusion protein (lane 3) comprising the N-terminal 205 amino acids of CBF1. (C) Reporter expression from the β-globin-promoter with 6 LMP2A–CBF1 response elements (as in Fig. 1C and 2D). (D) Activation of a Myb–CBF1 fusion protein by E1A was examined on a Myb responsive promoter, similarly to Fig. 1D. Reporter activation through Myb–CBF1 (or MybDBD as a control) in presence of E1A proteins was examined. Ratio of reporter activities in presence of Myb–CBF1 versus MybDBD is represented.
A chimeric CBF1 protein with altered DNA-binding specificity was constructed to confirm that reporter activation by E1A depended on CBF1. To do so, the CBF1 coding region was fused to the c-Myb DNA-binding domain (DBD) and tested on a Myb responsive promoter (Ness et al. 1989). Figure 1D shows that E1A enhanced reporter activity in the presence of the Myb–CBF1 chimera, yet not in its absence. This shows that E1A-mediated transactivation occurs through CBF1.
Coimmunoprecipitation analysis was performed to determine whether E1A proteins associate with CBF1 in cells. As shown in Figure 2A, CBF1 coprecipitated with 13SE1A but not with 12SE1A. These data suggest that the 13S-specific CR3 is required for direct CBF1 binding. Immunoprecipitation of E1A from 293 cells revealed that resident CBF1 also forms a complex with the endogenous oncoprotein (Fig. 2B). GST-pull-down assays, as shown in Figure 2C, further revealed that 13SE1A binds to the N-terminal one third of the CBF1 protein.
Figure 2.
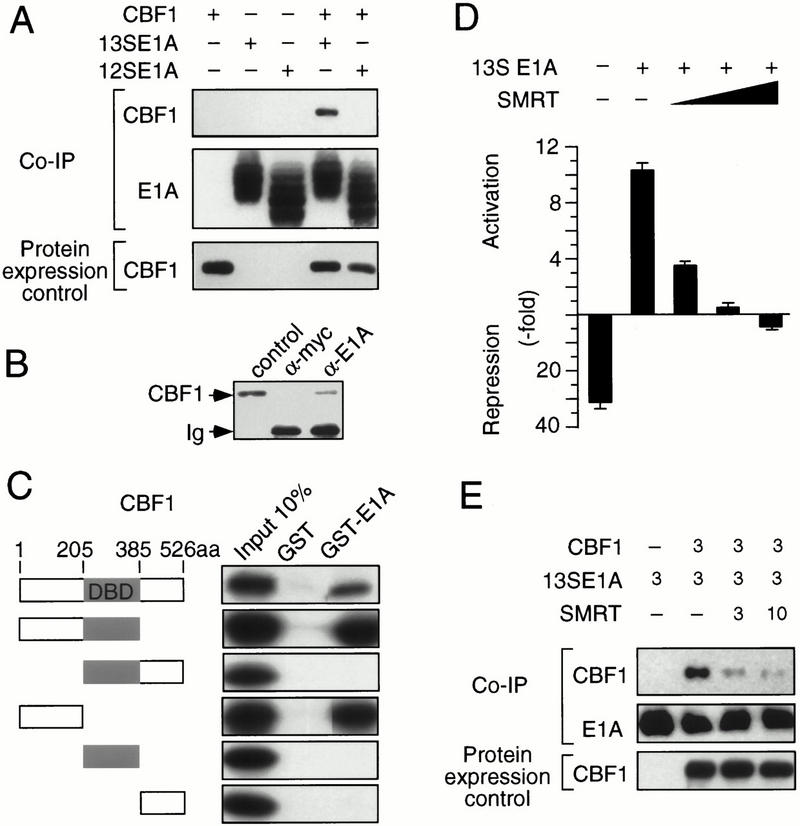
13SE1A binds to CBF1 and releases an associated corepressor complex. (A) QT6 fibroblasts were transfected with E1A and/or Flag-tagged CBF1 cDNA expression vectors as indicated. Cellular protein complexes were immunoprecipitated with monoclonal E1A antibody. Coimmunoprecipitated (CoIP) and ectopic expression of CBF1 were revealed by immunoblotting with Flag M2 antibody. (B) E1A interacts with endogenous CBF1. Protein extracts from 293 cells were immunoprecipitated with anti-c-myc or anti-E1A antibodies as indicated. CoIP CBF1 was revealed by immunoblotting using anti-CBF1. Fibroblasts transfected with the murine CBF1 protein were used as control (lane 1). (C) Various in vitro translated CBF1 fragments (left) were subjected to binding to GST or GST–13SE1A fusion proteins as indicated on the top. (D) SMRT abolishes 13SE1A mediated transactivation via CBF1. Reporter expression from the hexamerized LMP2A–CBF1 reporter construct was determined as described in Fig. 1C. A limiting amount of 13SE1A effector plasmid (50 ng) was transfected. SMRT effector plasmid concentrations were 0.1, 0.4, and 0.8 μg, respectively. The ratio between reporter activity obtained with the hexamerized CBF1 response element in relation to the core promoter is shown. (E) Interaction between E1A and CBF1 is inhibited by SMRT. QT6 fibroblasts were transfected with expression vectors encoding Flag-tagged CBF1 (3 μg), 13S E1A (3 μg), and SMRT (3 and 10 μg), as indicated. Immunoprecipitation and CBF1 detection were performed as described in A.
Both Notch1-IC or EBNA2 are thought to displace CBF1-bound repressor complex by competitive binding to the central part of CBF1 (Hsieh and Hayward 1995; Waltzer et al. 1995; Hsieh et al. 1996; Kao et al. 1998). As 13SE1A binds to the CBF1 N terminus (Fig. 2C), we wondered whether CBF1 activation through 13SE1A also comprises a derepression step. As shown in Figure 2D, SMRT inhibited E1A-mediated CBF1 reporter activation in a dose-dependent fashion. Moreover, SMRT also inhibited coimmunoprecipitation of CBF1 by E1A (Fig. 2E). This shows that SMRT and 13SE1A interact with CBF1 in a competitive fashion, although binding sites differ. These data can be explained by intramolecular interactions between N-terminal and central protein parts that were previously revealed by mutational analysis of CBF1 (Waltzer et al. 1995). In any case, our data show that corepressor displacement is a common step in CBF1-mediated gene activation by Notch1-IC, EBNA2, or 13S E1A.
Mutant E1A proteins were examined for CBF1 binding and for reporter activation through CBF1 sites or through the Myb–CBF1 fusion protein, as shown in Figure 3. Western blot analysis confirmed that all E1A proteins were expressed at comparable levels (data not shown). Removal of the major protein interaction sites in E1A, including CR1, CR2, or parts of the C terminus (Δ191–218; Δ239–253), did not affect its binding to CBF1 (Fig. 3A,B). Likewise, reporter activation through CBF1 sites (Fig. 3C) or through the Myb–CBF1 chimera (Fig. 3D) was not affected. Similarly, point mutations that selectively abolish interaction of E1A with Rb, CBP/p300, or pCAF (Babiss et al. 1986; Lundblad et al. 1995; Condorelli and Giordano 1997; Reid et al. 1998) also did not affect reporter activation (data not shown). In contrast, removal of CR3 (12SE1A) abrogated binding to CBF1 and CBF1-mediated reporter activation (Fig. 3). These results show that CR3 is responsible for both CBF1 binding and CBF1-mediated gene activation.
The function of CR3 is bipartide: Its N-terminal zinc finger interacts with the preinitiation complex (Geisberg et al. 1994, 1995; Mazzarelli et al. 1997) or with the polymerase holoenzyme via mediator binding (Boyer et al. 1999), whereas its C-terminal part interacts with DNA-bound transcription factors (Liu and Green 1990, 1994; Scholer et al. 1991; Webster and Ricciardi 1991). We therefore examined zinc finger and C-terminal E1A mutants for CBF1 effects. As shown in Figure 4A, disruption of the CR3 zinc finger structure (13S E1AC157S) or deletion of a C-terminal part of CR3 (13S E1AΔ184/187) abrogate both, activation through CBF1 sites (left panel) or activation through the Myb–CBF1 fusion protein (right panel). Coimmunoprecipitation revealed that wild type or the zinc finger E1A mutant precipitated CBF1, whereas the C-terminal CR3 mutant failed to do so (Fig. 4B). This result suggested that the C-terminal part of CR3 binds to CBF1, whereas the N-terminal zinc finger mediates CBF1-dependent gene activation. This conclusion was further supported by results obtained with E1A V147L and R177K mutations, shown previously to annihilate zinc finger functions (Geisberg et al. 1994), which also strongly reduced CBF1-mediated reporter activation (data not shown). Next, we replaced the CBF1 interacting domain of Notch-IC, the so-called RAM domain, by E1A CR3 (Fig. 4C) and examined CBF1-dependent reporter activation as shown in Figure 4. It is evident that reporter activation by Notch-IC was entirely dependent on the RAM–CBF1 interaction. However, CR3 or the C-terminal part of CR3, lacking the zinc finger, functionally replaced the Notch RAM domain (Fig. 4D,E). These results corroborate the notion that the C-terminal part of E1A mediates CBF1 interaction and the N-terminal E1A zinc finger mediates gene activation once 13SE1A is bound to CBF1. These experiments do not exclude the remote possibility that CR3 sequences positively modulate Notch-IC ankyrin repeats to interact with CBF1 (Kato et al. 1997).
Figure 4.
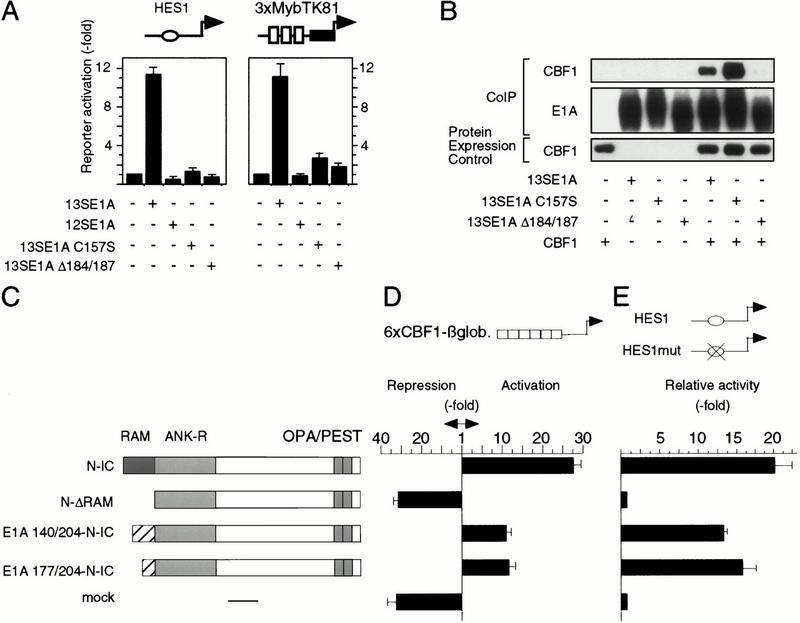
CR3 C-terminal and N-terminal sequences are required for CBF1 activation. (A, left) Activation of the murine HES1 promoter reporter construct (as in Fig. 1A). (Right) Activation of the Myb–CBF1 fusion protein (as in Fig. 1D) through E1A mutants. Ratio of reporter activities in presence of Myb–CBF1 versus MybDBD is represented. (B) CR3 C-terminal sequence binds to CBF1. QT6 fibroblasts were transfected with Flag-tagged CBF1 and E1A cDNA expression vectors as indicated. Immunoprecipitation and CBF1 detection were performed as described in Fig. 2A. (C) Functional replacement of the CBF1 interacting RAM domain of Notch-IC by E1A CR3. Schematic representation of the constructs used in D and E. (N-IC) Notch-IC; (N-ΔRAM) Notch-IC deleted of its RAM domain; (E1A-N-IC) the RAM domain of Notch has been replaced by CR3 (residues 140–204 or 177–204). (D) Reporter expression from 6 LMP2A–CBF1 response elements, as in Fig. 2C. (E) Reporter expression from the murine HES1 promoter reporter construct or from a promoter construct that carries a mutation in the CBF1 binding site, as in Fig. 1A. The ratio between wild type versus mutant promoter is shown.
Oncoviral proteins disrupt ordered cell growth and differentiation through interaction with critical cellular growth regulatory proteins (Flint and Shenk 1997). We found that the transforming adenoviral protein 13SE1A, but not 12SE1A, binds to the transcription factor CBF1 and converts CBF1 from a repressor into an activator of genes. Similarly, genomic E1A, encoding both isoforms activate CBF1-driven reporter expression in transfected cells or in 293 cells (Fig. 1A–C). This indicates that activation of CBF1 by 13SE1A is dominant over 12SE1A inhibitory functions (Reid et al. 1998; Chakravarti et al. 1999; Hamamori et al. 1999), potentially through recruitment of mediator/RNA Pol II to CBF1 target genes (Boyer et al. 1999). Significantly, CBF1 is the major downstream transcriptional regulator of the developmentally important Notch pathway of gene regulation that controls many differentiation and proliferation processes (Artavanis-Tsakonas et al. 1995). CBF1 functions are also modified by the EBNA2 oncoprotein of EBV that is essential for the activation of EBV promoters and for B-cell immortalization (Zimber-Strobl et al. 1996). Because oncogenic versions of Notch receptors such as TAN-1 and int-3 have been identified as cellular oncogenes of the lymphoid lineage, it will be interesting to determine whether hematopoietic cells of, for example, the B- and/or T-cell lineage are targets of 13SE1A specific transformation. Although E1A transformation potential resides in the 12SE1A product (Flint and Shenk 1997), our results hint at a novel function of the 13SE1A-specific CR3 in distinct cell types. Moreover, the result that CBF1 is activated by various viral and cellular oncogenes places CBF1 into a category established by Rb-type pocket binding proteins or p53, fundamental cellular growth regulatory proteins whose functions are modified by multiple viral early gene products.
Materials and methods
Plasmid constructs and transfection
Murine Flag-tagged CBF1, E1A, Notch, and E1A-Notch cDNAs were constructed by PCR and cloned in pcDNA3 or in pGEX 4T1. Amino acids (aa) 38–80 and 123–142 were deleted in ΔCR1 and ΔCR2 13SE1A mutants, respectively. Notch constructs consist of aa 1751–2531 (N-IC) or aa 1851–2531 (N-ΔRAM). In the E1A–Notch chimeras (E1A–N-IC), the RAM domain was replaced by CR3 (aa 140–204 or 177–204). Chimeras were Flag-tagged at their N terminus. Plasmid constructs were sequenced, and protein expression was confirmed by immunoblotting using monoclonal M73 anti-E1A (Calbiochem) or anti-Flag M2 antibody (Integra Bioscience). In Myb-chimeras, avian c-Myb (aa 18–192) was fused to murine CBF1 protein. SMRT expression vector is described in Dhordain et al. (1997). CBF1 and Myb responsive reporters, HES1 and HES5 luciferase constructs were described previously (Ness et al. 1989; Laux et al. 1994; Zimber-Strobl et al. 1994; Nishimura et al. 1998). Avian HD3 erythroblasts and quail QT6 fibroblasts were transfected as described in Kowenz-Leutz et al. (1994). CBF1 reporter assays were performed 24 h posttransfection, using 2 μg of reporter plasmid and 0.2 μg of E1A or Notch plasmid. In assays using c-Myb–CBF1 chimeras, 3 μg of reporter, 0.2 μg of c-Myb–CBF1 and 0.05 μg of E1A expression vectors were used, and cells were harvested 18 h posttransfection. E1A-transformed 293 cells (106 cells) were transfected (1.5 μg of reporter plasmid) using the transfection reagent exgen 500 (Fermentas).
Coimmunoprecipitation experiments
Immunoprecipitation was performed in 100 mM NaCl, 50 mM Hepes (pH 7), 0.5% NP-40 supplemented with protease inhibitors. Briefly, the lysate was incubated with 1 μg of M73 anti-E1A antibody (Calbiochem) at 4°C for 3 h. Protein-A sepharose (Pharmacia) was added and incubated at 4°C for 1 h. Beads were washed six times and proteins separated by SDS-PAGE. Immunoblots were visualized by ECL (Amersham). Immunoprecipitation in 293 cells (5 × 106 cells) was performed as described previously, in 200 mM NaCl, 50 mM Hepes (pH 7), 0.2% NP-40, either with the monoclonal anti-c-myc Ab-2 antibody (Calbiochem) or with the monoclonal M73 anti-E1A antibody. Endogenous CBF1 protein was revealed with a polyclonal antibody (Brou et al. 2000).
GST pull-down assay
GST-fusion proteins expressed in BL21 bacteria (Wulczyn et al. 1992) were extracted in NETN buffer (50 mM Tris at pH 8, 100 mM NaCl, 0.1% NP40, 1 mM EDTA, 1 mM DTT, 10% glycerol). In vitro translation was performed in presence of [35S]methionine (TNT, Promega). Bacterial extracts were incubated with glutathione agarose beads and in vitro translated product at 4°C for 4 h. Beads were washed six times with NETN and separated by SDS-PAGE.
Acknowledgments
We thank Sabine Krause for excellent technical assistance, Dr. Ryoichiro Kageyama for providing Hes promoter–luciferase constructs, Dr Alain Israel for providing CBF1 antiserum, and Dr. Georg Bornkamm, Dr. Bettina Kempkes, and members of the laboratory of A. Leutz for discussion.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL aleutz@mdc-berlin.de; FAX 49-30-9406-3298.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.189301.
References
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Babiss LE, Liaw WS, Zimmer SG, Godman GC, Ginsberg HS, Fisher PB. Mutations in the E1a gene of adenovirus type 5 alter the tumorigenic properties of transformed cloned rat embryo fibroblast cells. Proc Natl Acad Sci. 1986;83:2167–2171. doi: 10.1073/pnas.83.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Inhibition of the DNA-binding activity of Drosophila suppressor of hairless and of its human homolog, KBF2/RBP-Jκ, by direct protein– protein interaction with Drosophila hairless. Genes & Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- Chung CN, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of suppressor of hairless, RBP-Jκ. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G, Giordano A. Synergistic role of E1A-binding proteins and tissue-specific transcription factors in differentiation. J Cell Biochem. 1997;67:423–431. doi: 10.1002/(sici)1097-4644(19971215)67:4<423::aid-jcb1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan SE, St-Pierre B, Leow CC. Notch receptors, partners and regulators: From conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West C, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Maruyama S, Kawaichi M, Honjo T. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell. 1992;69:1191–1197. doi: 10.1016/0092-8674(92)90640-x. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith GH. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56:1775–1785. [PubMed] [Google Scholar]
- Geisberg JV, Lee WS, Berk AJ, Ricciardi RP. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc Natl Acad Sci. 1994;91:2488–2492. doi: 10.1073/pnas.91.7.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Chen JL, Ricciardi RP. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol Cell Biol. 1995;15:6283–6290. doi: 10.1128/mcb.15.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- Honjo T. The shortest path from the surface to the nucleus: RBP-Jκ/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman SR. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kidd S, Lieber T, Young MW. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes & Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBP beta (NF-M) transcriptional control: Activation through derepression. Genes & Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- Laux G, Dugrillon F, Eckert C, Adam B, Zimber-Strobl U, Bornkamm GW. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Green MR. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- ————— Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- Matsunami N, Hamaguchi Y, Yamamoto Y, Kuze K, Kangawa K, Matsuo H, Kawaichi M, Honjo T. A protein binding to the Jκ recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature. 1989;342:934–937. doi: 10.1038/342934a0. [DOI] [PubMed] [Google Scholar]
- Mazzarelli JM, Mengus G, Davidson I, Ricciardi RP. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAF(II)135. J Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymryk JS. Tumour suppressive properties of the adenovirus 5 E1A oncogene. Oncogene. 1996;13:1581–1589. [PubMed] [Google Scholar]
- Ness SA, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte- specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Isaka F, Ishibashi M, Tomita K, Tsuda H, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and enhancer of split. Genomics. 1998;49:69–75. doi: 10.1006/geno.1998.5213. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, et al. Disruption of the mouse RBP-J κ gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Olave I, Reinberg D, Vales LD. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes & Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JL, Bannister AJ, Zegerman P, Martinez-Balbas MA, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: A member of the notch gene family transforms mammary epithelial cells. J Virol. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson ES, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer HR, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: Implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- Schweisguth F, Posakony JW. Suppressor of hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- ————— Antagonistic activities of suppressor of hairless and hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro JG, Shore GC, Branton PE. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- Waltzer L, Bourillot PY, Sergeant A, Manet E. RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster LC, Ricciardi RP. Trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol. 1991;11:4287–4296. doi: 10.1128/mcb.11.9.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Naumann M, Scheidereit C. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κ B. Nature. 1992;358:597–599. doi: 10.1038/358597a0. [DOI] [PubMed] [Google Scholar]
- Zhao B, Marshall DR, Sample CE. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Hayward SD. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74:1939–4197. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber-Strobl U, Strobl LJ, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm GW. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-Jκ, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber-Strobl U, Kempkes B, Marschall G, Zeidler R, Van Kooten C, Banchereau J, Bornkamm GW, Hammerschmidt W. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 1996;15:7070–7078. [PMC free article] [PubMed] [Google Scholar]


