Abstract
Cdc13 is a single-strand telomeric DNA-binding protein that positively regulates yeast telomere replication by recruiting telomerase to chromosome termini through a site on Cdc13 that is eliminated by the cdc13-2 mutation. Here we show that Cdc13 has a separate role in negative regulation of telomere replication, based on analysis of a new mutation, cdc13-5. Loss of this second regulatory activity results in extensive elongation of the G strand of the telomere by telomerase, accompanied by a reduced ability to coordinate synthesis of the C strand. Both the cdc13-5 mutation and DNA polymerase α mutations (which also exhibit elongated telomeres) are suppressed by increased expression of the Cdc13-interacting protein Stn1, indicating that Stn1 coordinates action of the lagging strand replication complex with the regulatory activity of CDC13. However, the association between Cdc13 and Stn1 is abolished by cdc13-2, the same mutation that eliminates the interaction between Cdc13 and telomerase. We propose that Cdc13 participates in two regulatory steps—first positive, then negative—as a result of successive binding of telomerase and the negative regulator Stn1 to overlapping sites on Cdc13. Thus, Cdc13 coordinates synthesis of both strands of the telomere by first recruiting telomerase and subsequently limiting G-strand synthesis by telomerase in response to C-strand replication.
Keywords: Telomere, telomerase, Cdc13, Stn1, DNA replication
Telomeres are guanine-rich, simple repeat sequences that constitute the physical ends of eukaryotic chromosomes. The sequence and structure of these specialized DNA termini permits extension of the G strand by the enzyme telomerase, thereby counteracting sequence loss due to incomplete replication or degradative activities (Lingner et al. 1995; Greider 1996). In yeast and human cells, a telomerase deficiency leads to a progressive decline in telomere length that inhibits the proliferative capacity of the cell and heralds replicative senescence (Lundblad and Szostak 1989; Bodnar et al. 1998). Analysis of telomerase-deficient mice has also revealed an essential role for telomere maintenance in the long-term viability of high-renewal organ systems (Lee et al. 1998). Furthermore, loss of telomerase in the absence of checkpoint function can also promote genetic instabilities and carcinogenesis, especially in highly regenerative tissues (Chin et al. 1999; Artandi et al. 2000), illustrating the importance of telomeres in maintaining genomic stability.
Increasing attention has been directed at the importance of coordinating G-strand synthesis by telomerase with replication of the C strand of the telomere. Following extension of the 3′ terminus of the G-rich strand by telomerase, fill-in synthesis of the other strand, presumably by the machinery that normally performs lagging strand DNA replication, is thought to be necessary to prevent elongated single-stranded regions at chromosome termini. Several recent studies indicate that disruption of C-strand DNA synthesis can impair telomere length control (for review, see Price 1997; Evans and Lundblad 2000). In the ciliate Euplotes, inhibition of DNA polymerases α and δ during de novo telomere synthesis alters the length of the telomeric C strand and also causes an increase in the length and heterogeneity of the G strand (Fan and Price 1997). Similarly, propagation of S. cerevisiae strains defective for components of the lagging strand DNA replication machinery cause a telomerase-dependent increase in telomere length (Carson and Hartwell 1985; Adams and Holm 1996; Adams Martin et al. 2000). In addition, de novo telomere formation at a newly created double-strand break requires not only telomerase but also DNA polymerase α and δ (Diede and Gottschling 1999). Collectively, these results indicate that in both yeast and ciliates, G-strand and C-strand synthesis are tightly coregulated, although the molecular mechanism for this coordination has not been elucidated.
In yeast, a critical contributor to several aspects of telomere function is the protein Cdc13, which exhibits high affinity sequence-specific binding for single-stranded telomeric DNA (Lin and Zakian 1996; Nugent et al. 1996; Hughes et al. 2000a). Consistent with this substrate specificity, Cdc13 is associated with the telomere during S phase (C.I. Nugent and V. Lundblad, in prep.), which correlates with the transient increased single-strand G-rich extension observed at yeast telomeres (Wellinger et al. 1993, 1996). CDC13 performs an essential activity necessary to protect chromosome termini, as loss of CDC13 function is accompanied by immediate and extensive loss of one strand of the telomere (Garvik et al. 1995; Diede and Gottschling 1999). In addition to this end protection function, CDC13 positively regulates telomere replication by recruiting telomerase to the telomere, an activity that is abolished by the cdc13-2 missense mutation. The cdc13-2 strain, which is normal for telomeric end protection, exhibits a telomere replication defect similar to that of a strain that lacks telomerase (Nugent et al. 1996), although telomerase activity in cell-free cdc13-2 extracts is unaffected (Lingner et al. 1997). This defect is partially suppressed by increased expression of the telomerase-associated Est1 protein (Nugent et al. 1996), and a complex containing both proteins can be coimmunoprecipitated in yeast, using overexpressed recombinant versions of Est1 and Cdc13 (Qi and Zakian 2000). Furthermore, the cdc13-2 mutation can be reciprocally suppressed by a specific missense mutation in EST1 (Pennock et al. 2001), thereby demonstrating that positive regulation of telomere replication by Cdc13 is due to a direct interaction between Cdc13 and a subunit of telomerase, resulting in recruitment of the enzyme to the telomere.
In addition to this positive regulatory role, Cdc13 has been implicated in negative control of telomere length. The thermolabile cdc13-1 mutant exhibits greatly elongated telomeres when grown at intermediate temperatures (Grandin et al. 1997). However, this mutant strain is also severely impaired for the essential end protection function of CDC13 (Garvik et al. 1995; Diede and Gottschling 1999); therefore, analysis of the cdc13-1 mutation has not clarified whether the proposed role in negative length control is distinct from the essential function of CDC13. An additional factor that contributes to negative length regulation is the Cdc13-interacting protein Stn1. These two proteins associate in a two-hybrid assay and display a number of genetic interactions, suggesting that Stn1 and Cdc13 function together at the telomere as a complex (Grandin et al. 1997). Furthermore, a mutation in STN1 increases telomere length by ∼1 kb, leading Charbonneau and colleagues to propose that Stn1 is a negative regulator of telomere length (Grandin et al. 1997, 2000). Cdc13 is also associated with DNA polymerase α, although mutations that disrupt this association have only a modest (50–150 bp) increase in telomere length (Qi and Zakian 2000); therefore, the contribution of the Cdc13-Pol α association to telomere length control is unclear.
We describe here the identification of a mutation of CDC13, cdc13-5, that exhibits a striking defect in negative regulation of telomere length, with no impact on the essential function of CDC13. In this strain, telomere length is increased by ≥1000 bp as a result of unregulated elongation of the G-rich strand by telomerase. This is accompanied by a reduced ability to coordinate synthesis of the C-rich strand, resulting in chromosome termini with greatly extended single-strand extensions of the G strand. The cdc13-5 defect is dependent on the ability to recruit telomerase to the telomere, as cdc13-5-mediated telomere elongation is blocked by the cdc13-2 mutation. Genetic interactions between the cdc13-5 mutation and DNA polymerase α also implicate misregulation of C-strand synthesis in this mutant phenotype. Therefore, cdc13-5 defines a role for Cdc13 in coordination of the synthesis of the two strands of the telomere, which is distinct from the telomerase recruitment activity defined by the cdc13-2 mutation. We propose that these two regulatory activities of Cdc13 correspond to two distinct steps in telomere replication that coordinate and regulate synthesis of the two strands of the telomere. In the first step, Cdc13 initiates telomere replication by recruiting telomerase, which extends the 3′ terminus of the G strand of the telomere. In the second step, fill-in synthesis of the C strand by lagging strand synthesis machinery acts to limit extension of the G strand by telomerase, an inhibitory activity that is defective in the cdc13-5 mutant. We further suggest that these two steps are the consequence of successive binding of the telomerase-associated Est1 subunit and the negative regulator Stn1 to overlapping binding sites on Cdc13. By relying on a single protein as the regulatory center, this proposed mechanism provides a simple means for coordinating synthesis of the two strands of the telomere.
Results
Cdc13 is a negative regulator of telomerase
As part of our investigation of the Cdc13 protein, we uncovered two alleles of CDC13, cdc13-3 and cdc13-5, that result in a pronounced defect in telomere length maintenance. The telomeric tract, which is normally ∼350 bp in a wild-type strain, was elongated by ∼500–1000 bp in cdc13-5 and cdc13-3 strains, along with an accompanying increase in telomere length heterogeneity (Fig. 1A). The cdc13-3 mutation was the result of an alanine scan mutagenesis of CDC13 (Hughes 1998) and mapped to residues 695–699 at the C-terminal boundary of the Cdc13 DNA-binding domain. Western analysis, using an N-terminally epitope-tagged version (HA3–Cdc13-3), indicated that the mutant Cdc13-3 protein apparently existed in two forms in yeast: the full-length 105-kD protein and a truncated version that migrated at ∼80 kD, with both variants present in substantially reduced levels relative to the wild-type full-length Cdc13 protein (data not shown). We surmised that the cdc13-3 mutation resulted in an unstable protein that was readily proteolyzed to remove ∼20 kD from the C-terminal region of the protein. To test directly whether removal of the C terminus of Cdc13 was responsible for telomere elongation, an additional allele, cdc13-5, was constructed by the introduction of a stop codon following amino acid 694, which truncated the Cdc13 protein by 230 amino acids. This truncation occurs at the boundary of the DNA-binding domain, which can be expressed separately as a stable polypeptide (Hughes et al. 2000a). The cdc13-5 mutant yeast strain exhibited extensive telomere elongation to a degree roughly comparable to that observed with the cdc13-3 strain (Fig. 1A). As described in more detail later in this article, the cdc13-5 strain also grew as well as a wild-type strain. The Cdc13-5 protein was also produced at levels comparable to wild-type protein (Fig. 1B) in contrast to the situation with the cdc13-3 mutant strain. Therefore, unless otherwise noted, the experiments described in this article were conducted with the cdc13-5 allele.
Figure 1.
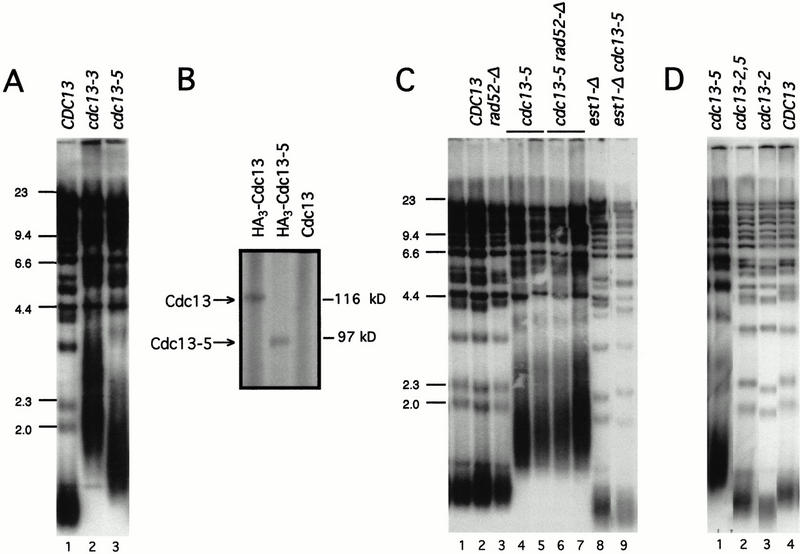
Cdc13 is a negative regulator of telomere elongation. (A) Telomere length of CDC13+ (lane 1), cdc13-3 (lane 2), and cdc13-5 (lane 3). The cdc13-3 and cdc13-5 haploid strains were derived from DVL233 and DVL326, respectively, by tetrad dissection; DNA was prepared after ∼35 generations of growth. (B) Anti-HA immunoblot of anti-HA immunoprecipitates from equal amounts of whole cell extract prepared from a cdc13-Δ strain expressing HA3–Cdc13 (lane 1; pVL841), HA3–Cdc13-5 (lane 2; pVL903) and untagged Cdc13 (lane 3; pVL440); all plasmids were single-copy CEN vectors, with CDC13 expressed from its native promoter. Arrowheads indicate the full-length Cdc13 protein (∼116 kD) and the truncated Cdc13-5 protein (∼80 kD). (C) Telomere length of CDC13/cdc13-5 EST1/est1-Δ RAD52/rad52-Δ (lane 1), CDC13+ (lane 2), rad52-Δ (lane 3), cdc13-5 (lanes 4,5), cdc13-5 rad52-Δ (lanes 6,7), est1-Δ (lane 8), and cdc13-5 est1-Δ (lane 9). The haploid strains shown in lanes 2–9 were obtained from DVL328 (lane 1) by tetrad dissection; DNA was prepared after ∼35 generations of growth, except for lanes 5 and 7, which were grown for an additional 10 generations. (D) Telomere length of cdc13-5 (lane 1), cdc13-2,5 (lane 2), cdc13-2 (lane 3), and CDC13+ (lane 5); strains were generated by transforming cdc13-Δ/pVL438 with pVL1033 (cdc13-5), pVL1360 (cdc13-2,5), pVL690 (cdc13-2), and pVL440 (CDC13), followed by eviction of pVL438 on 5-FOA. Although a cdc13-2,5 strain exhibits a senescence phenotype characteristic of a telomere replication defect, telomere length of the cdc13-2,5 strain is reproducibly not quite as short as the cdc13-2 strain at any given time point (cf. lanes 2 and 3). This may be caused by residual recruitment activity of the Cdc13-2 mutant protein (which is consistent with the prior demonstration that the senescence phenotype of a cdc13-2 strain is somewhat delayed relative to telomerase null mutations; Lendvay et al. 1996), thereby allowing partial manifestation of the cdc13-5 defect.
To determine whether the increase in telomere length was mediated by telomerase or recombination, telomerase-defective and recombination-defective cdc13-5 double mutant strains were examined. In a cdc13-5 strain that lacked telomerase (cdc13-5 tlc1-Δ or cdc13-5 est1-Δ), telomere elongation was abolished, and the double mutant strain showed the senescence phenotype characteristic of telomerase null strains (Fig. 1C; data not shown). In contrast, telomere length in the cdc13-5 strain was unaffected by loss of the major pathway for homologous recombination, as elongation occurred to the same degree in cdc13-5 rad52-Δ and cdc13-5 RAD52 strains (Fig. 1C). Therefore, the greatly elongated telomeres observed in the cdc13-5 strain are the result of action by telomerase. The cdc13-5 mutation was recessive to CDC13+, indicating that the ability to negatively regulate telomere length is a property of the wild-type protein (Fig. 1C).
CDC13 also has a positive role in maintaining telomere length, which is eliminated by the cdc13-2 mutation (Nugent et al. 1996). One model consistent with both positive and negative regulatory roles for CDC13 proposes that the wild-type Cdc13 protein first recruits telomerase (an activity that is lost in the cdc13-2 mutant) and then subsequently acts to limit the degree of extension of the G-rich strand by telomerase (an activity that is lost in the cdc13-5 mutant). A prediction of this model is that an allele of CDC13 that contains both mutations should fail to recruit telomerase and, therefore, should not result in elongated telomeres. Consistent with this prediction, a cdc13-2,5 strain, with both regulatory mutations introduced into the CDC13 gene, no longer exhibited long telomeres. Instead, telomeres were short (Fig. 1D), and the cdc13-2,5 strain showed a senescence phenotype (data not shown). Therefore, the loss of regulation of telomere length exhibited by the cdc13-5 mutation depends on the telomerase recruitment function of CDC13.
The cdc13-5 mutation results in greatly increased single-strand extension of the G-rich strand of the telomere
One means by which telomerase synthesis could become unregulated could be because of a loss of coordination between G-strand and C-strand synthesis. Impairment of DNA polymerase α results in elongated telomeres, as well as an increase in the extent of the terminal single-strand extension of the G-rich strand (Carson and Hartwell 1985; Adams and Holm 1996; Fan and Price 1997; Adams Martin et al. 2000). To examine whether the cdc13-5 mutant similarly affected the extent of single-stranded G-rich DNA, nondenaturing in-gel hybridization was used to examine chromosome termini. In wild-type yeast, the G-rich strand can only be readily detected as single stranded in late S phase, whereas at other points in the cell cycle, the chromosome terminus is either blunt or the overhang is below the detection limit (<30 bases; Wellinger et al. 1993). Therefore, due to both detection limits and the transient nature of the single strandedness of the G strand during S phase, single-stranded yeast termini cannot be reliably detected in asynchronous cultures of wild-type cells. However, when unsynchronized cultures of both cdc13-3 and cdc13-5 mutant strains were examined, G-rich single-strand telomeric signals were readily detected using a strand-specific probe (Fig. 2A). Treatment with Escherichia coli ExoI, a 3′ → 5′ single-strand exonuclease, abolished the signal, arguing for the presence of a terminal single-stranded G-rich overhang, as opposed to internal gaps or nicks. Moreover, no single strandedness corresponding to the C-rich strand was detected (data not shown). Strand separation gels that measured the length of the C strand showed that this strand was also elongated in a cdc13-5 strain, relative to the length of the C strand in a wild-type strain (data not shown). Therefore, the increased single strandedness of the G strand does not appear to be caused by extensive resection of the C strand. These results indicate that increased elongation of the G strand by telomerase in this mutant strain is accompanied by a loss in coordination of the synthesis of the C strand.
Figure 2.
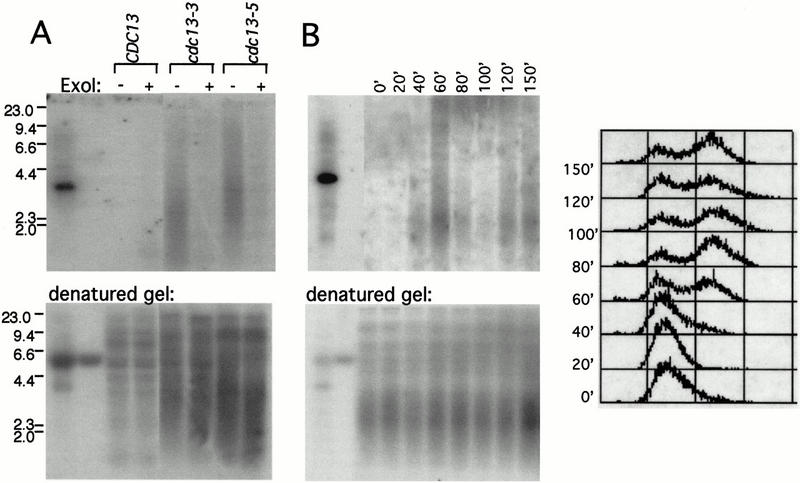
The cdc13-5 strain exhibits an altered end structure during S phase. (A) DNA from CDC13+, cdc13-3, and cdc13-5 strains were mock treated (lanes 3,5,7) or treated with ExoI (lanes 4,6,8), digested with XhoI, and analyzed by nondenaturing in-gel hybridization with a C1–3A oligonucleotide probe (top); the same gel, denatured and rehybridized with the same oligonucleotide probe (bottom); lanes 1,2: pVL260 (with 80 bp of G1–3T telomeric repeat DNA) heat denatured or nondenatured, respectively. Size markers: λ HindIII-digested DNA. (B) DNA from a cdc13-5 strain, prepared from time points collected every 20 min after release from an α factor–induced G1 arrest, was examined by native in-gel hybridization with a C1–3A oligonucleotide probe (top, left), denaturation and rehybridization with the same probe (bottom, left), and FACS analysis (right); lanes 1,2: pVL260, denatured or nondenatured, respectively.
The elongated terminal extensions detected in a cdc13-5 strain could reflect a defect during S phase, when telomere synthesis by telomerase occurs (Marcand et al. 2000). The alternative possibility is that the altered G tails in the cdc13-5 mutant are the consequence of an alteration in end structure that persists throughout the cell cycle, such as that observed in yku70− and yku80− mutants (Gravel et al. 1998; Polotnianka et al. 1998), possibly as a consequence of exposure of telomeres to unregulated activities other than telomerase. To distinguish between these two possibilities, we determined whether extensive elongation of the G-rich strand could be detected at different periods of the cell cycle, using synchronized cultures of the cdc13-5 mutant strain. Cells were arrested with α factor in G1 and subsequently released, and the G-rich strand of the telomere was examined on native gels using a strand-specific probe (Fig. 2B). When cells were arrested in G1, no telomeric single strandedness could be detected. However, 40–60 min after release, when FACS analysis indicated that the majority of cells had entered S phase, the cdc13-5 mutant exhibited a peak of single-stranded G-rich telomeric DNA. This signal declined as the cells exited S phase and increased again during the next cell cycle, although the peak of the signal became less pronounced as the culture became more asynchronous. These results indicate that the single-stranded G tail is a substrate for extended synthesis by telomerase during S phase, resulting in the telomere length defect observed in the cdc13-5 mutant strain.
Overexpression of STN1 suppresses cdc13-5 and pol α telomere length defects
The similarity in phenotypes of the cdc13-5 and DNA polymerase α mutant strains suggests that the same step in telomere replication may be affected, which is also consistent with the recent demonstration that Cdc13 and DNA Pol α can be coimmunoprecipitated (Qi and Zakian 2000). To provide further support for a functional relationship between these two proteins, we asked whether alterations in the level of the Cdc13-interacting protein, Stn1, could influence the phenotypes of either pol1 or cdc13-5 mutants. Notably, increased expression of STN1 suppressed the telomere phenotypes of mutations in both genes, implicating the cdc13-5 defect in a step in telomere replication that depends on DNA polymerase α function. STN1 expression, driven by the GAL promoter and on a high-copy plasmid, reduced telomere length in a cdc13-5 strain to nearly wild-type length (Fig. 3A) whereas overexpression of STN1 had little or no effect on telomere length in a wild-type strain. In addition, single-stranded G-rich DNA was not detectable in asynchronous cultures of a cdc13-5 strain overexpressing STN1 (Fig. 3B), as would be predicted if the alterations in telomere length and the terminal G strand overhang are the result of the same defect. Increased expression of STN1 similarly influenced the telomere replication defect of DNA polymerase α mutants. The telomere elongation phenotype observed when pol1-12, pol1-16, and pol1-17 mutant strains were grown at semipermissive temperatures was substantially suppressed when STN1 was overexpressed (Fig. 3C; data not shown). Suppression was specific to the telomere replication phenotype of the pol1 mutant strains, as overexpression of STN1 did not suppress the temperature sensitivity of these pol1 mutant alleles (data not shown).
Figure 3.
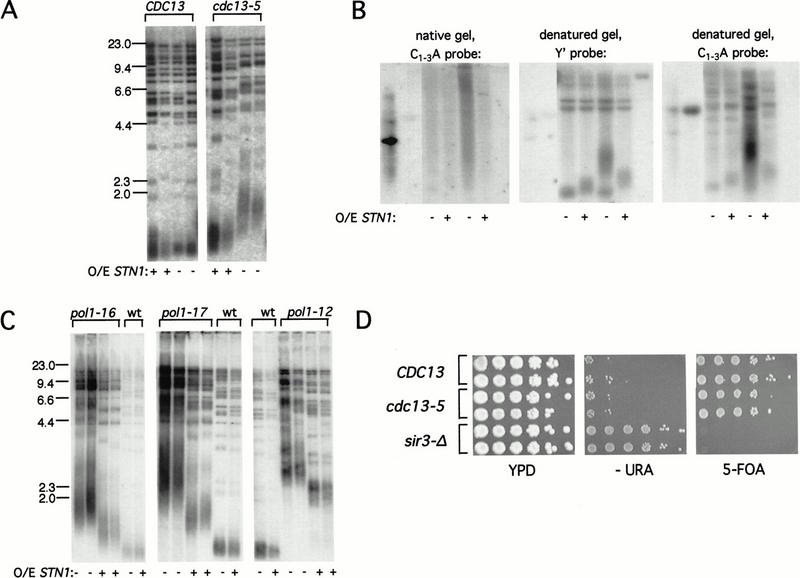
Overexpression of STN1 suppresses the telomere elongation phenotypes of cdc13-5 and pol1 mutant strains. (A) Telomere length of a cdc13-Δ strain covered by a CDC13+ plasmid (pVL440; lanes 1–4) or a cdc13-5 plasmid (pVL1033; lanes 5–8) plus a plasmid overexpressing STN1 (pVL1035) or a control vector (pVL1036). (B) DNA prepared from a cdc13-Δ strain covered by a CDC13+ plasmid (pVL440; lanes 3,4) or a cdc13-5 plasmid (pVL1033; lanes 5,6) plus a plasmid overexpressing STN1 (pVL1035; lanes 4,6) or a control vector (pVL1036; lanes 3,5) was analyzed by nondenaturing in-gel hybridization with a C1–3A oligonucleotide probe (left); denatured and rehybridized with a Y′ probe (middle); and denatured and rehybridized with a C1–3A oligonucleotide probe (right); lanes 1,2: pVL260 heat denatured or nondenatured, respectively; lane 7: pJH354, containing a region of Y′ sequence. (C) Telomere length of the indicated pol1 mutant strains or the isogenic POL1+ strain (indicated as wt) transformed with either a plasmid overexpressing STN1 (pVL1034) or vector (YEplac112). (D) CDC13 (UCC3505, Singer and Gottschling 1994), cdc13-5 (TVL422) and sir3-Δ (TV318), each containing a telomere-proximal URA3 gene, were grown in rich media, and 10-fold serial dilutions were plated on rich media, on media lacking uracil, and on media containing 5-FOA and incubated for 48 h at 30°C.
Duplex telomeric chromatin is not altered in the cdc13-5 strain
One mechanism for telomere length control proposes that telomeric chromatin equilibrates between an open conformation that permits telomerase access and a higher-order closed conformation that inhibits access of the telomerase enzyme through sequestration of the chromosome terminus (for review, see Evans and Lundblad 2000). This length-determining conformational switch is determined by binding of negative regulatory proteins to duplex telomeric DNA. Therefore, the increased telomere length of pol1 strains could be due to reduced assembly of inhibitory proteins on duplex telomeric DNA, possibly as a consequence of altered rates of lagging strand synthesis (Diede and Gottschling 1999; Adams Martin et al. 2000). Alterations in telomeric chromatin are often assessed by monitoring the metastable repression of transcription of reporter genes placed adjacent to the telomeric G1–3T telomeric tract, referred to as telomeric silencing (Gottschling et al. 1990). In a pol1-17 mutant strain, telomeric silencing is greatly reduced, even under growth conditions where telomeres have not yet undergone extensive elongation (Adams Martin et al. 2000). In contrast, telomeric silencing is unaltered in the cdc13-5 strain (Fig. 3D), indicating that the structure of duplex telomeric chromatin is unaffected in this mutant. This suggests that the telomere elongation observed in the cdc13-5 mutant strain is a consequence of events that are restricted to the very terminus.
Stn1 interaction with Cdc13 is abolished by the cdc13-2 mutation
Increased expression of STN1 suppressed the telomere length phenotypes of the cdc13-5 strain (Fig. 3) but did not suppress the cdc13-2 defect; in fact, the senescence phenotype of a cdc13-2 strain was enhanced when the negative regulator STN1 was overexpressed (data not shown). In contrast, the cdc13-2 phenotype is partially suppressed when the positive regulator EST1 is overexpressed (Nugent et al. 1996), which is also consistent with recent work indicating that Est1 binds to a site on Cdc13 that is abolished by the cdc13-2 mutation (Pennock et al., 2001). Since two-hybrid analysis had previously shown that Stn1 and Cdc13 also interact (Grandin et al. 1997), we tested whether Stn1 performs its negative regulatory role by binding to a similar domain on Cdc13 as is bound by Est1. Figure 4 shows that Stn1 and Cdc13 exhibit a strong two-hybrid interaction, as previously observed. Strikingly, the cdc13-2 mutation abrogates this Stn1-Cdc13 interaction. This effect is specific to this mutation, as association between Stn1 and Cdc13 is still observed when either the cdc13-1 or cdc13-3 mutations are introduced (Fig. 4; data not shown).
Figure 4.
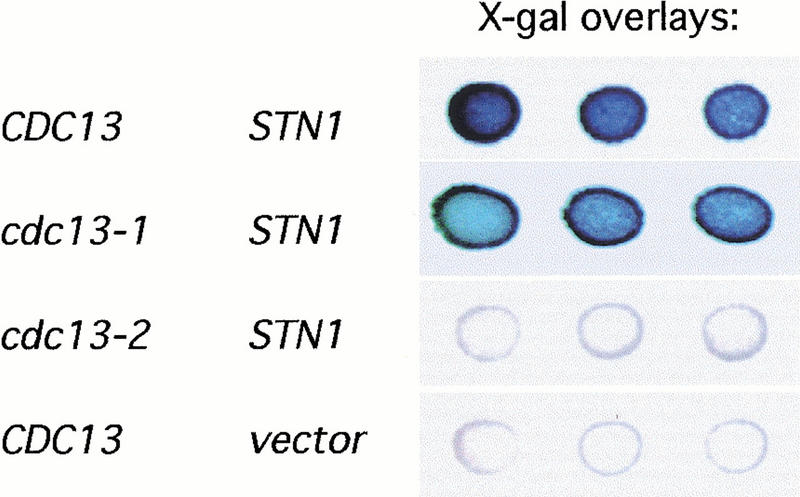
Cdc13, but not Cdc13-2, interacts with Stn1, by a two-hybrid test. Activation of a lacZ two-hybrid reporter gene was evaluated using X-gal overlay assays to detect β-galactosidase activity. Results are shown for three independent transformants each of CDC13 and STN1 (pVL705 and pVL859), cdc13-1 and STN1 (pVL667 and pVL859), cdc13-2 and STN1 (pVL854 and pVL859), and CDC13 and vector (pVL705 and pACT2). The ADE2 and HIS3 reporter genes present in this strain (pJ69-4A; James et al. 1996) gave results similar to the lacZ reporter (data not shown).
The increased single-stranded telomeric DNA present in a cdc13-5 strain does not invoke a DNA damage response
Cdc13 has an essential function in protecting the chromosome termini from degradation, as loss of CDC13 function (using the thermolabile cdc13-1 allele characterized previously) results in substantial resection of the C strand of the telomere and a resulting RAD9-mediated G2/M arrest (Weinert and Hartwell 1993; Garvik et al. 1995; Diede and Gottschling 1999). This indicates that Cdc13 has an essential function in protecting the chromosome termini from degradation (Garvik et al. 1995; Nugent et al. 1996). The cdc13-5 strain, in contrast, does not appear to be defective for this essential function of CDC13 because it does not exhibit either growth defects or a DNA damage response. The growth of a cdc13-5 strain was comparable to that of a wild-type strain at both 30°C and 36°C, temperatures that are not permissive for growth of the thermolabile cdc13-1 strain (Fig. 5A). Both FACS data (Fig. 2B) and bud index analysis (data not shown) showed that the cdc13-5 strain exhibited the same cell cycle distribution as a wild-type strain at 30°C. In addition, the kinetics of microcolony formation for a cdc13-5 strain was comparable to that of wild type, whereas a cdc13-1 strain arrested as large budded cells at 30°C (Fig. 5B). Thus, the cdc13-5 mutant does not show a noticeable growth defect or an alteration in cell cycle progression, showing that the activity of CDC13 that is responsible for negative regulation of telomere length is distinct from the essential end protection function of CDC13. Furthermore, expression of the RNR genes, which are normally induced in response to DNA damage or replication blocks (Elledge et al. 1993), was comparable in the cdc13-5 and CDC13 strains, in contrast to the robust induction of RNR expression that was observed either in a cdc13-1 strain at a nonpermissive temperature (30°C) or in a CDC13 strain treated with HU (Fig. 5C). In addition, genome-wide expression analysis of a cdc13-3 strain did not reveal any characteristic expression patterns suggesting a response to DNA damage, clearly distinguishing it from cdc13-1 (T.R. Hughes et al., in prep.). Therefore, the substantial increase in the amount of single-stranded DNA at the telomere that is observed in cdc13-3 and cdc13-5 strains is not sufficient to invoke a DNA damage response.
Figure 5.
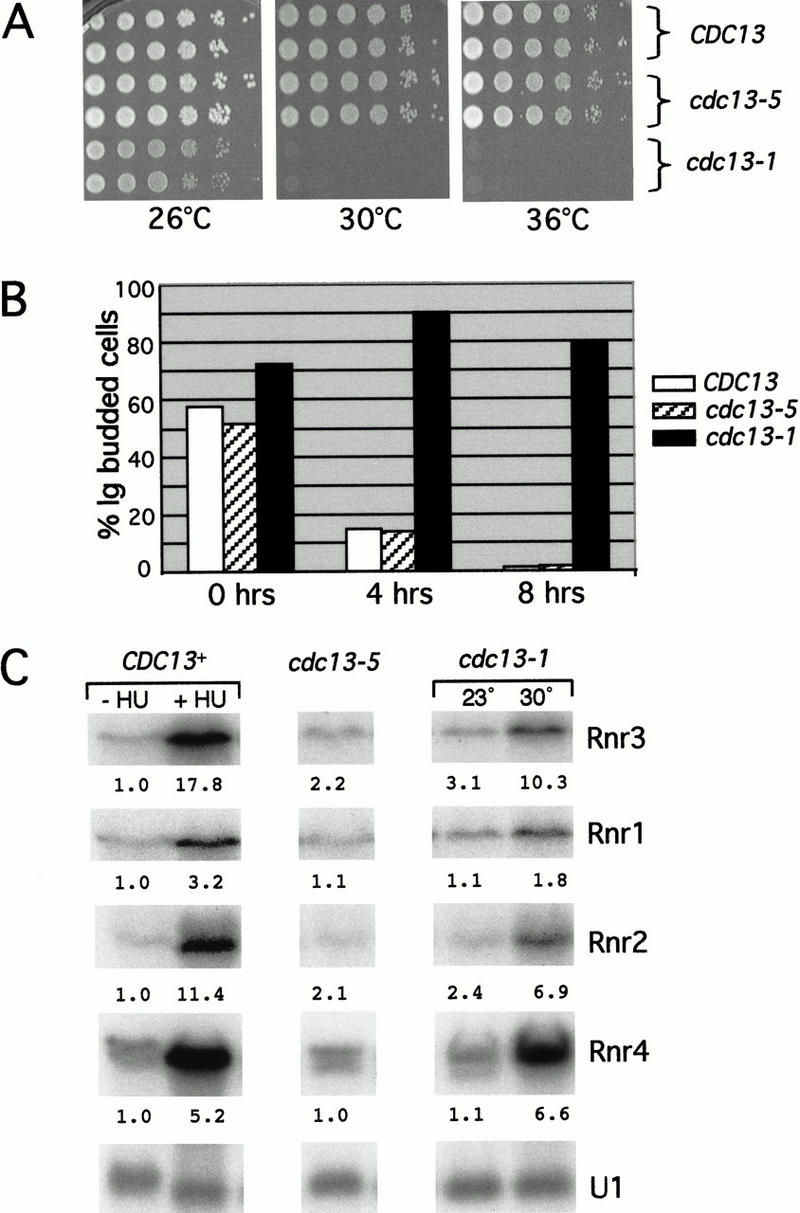
The cdc13-5 mutation is not compromised for the essential role of CDC13. (A) 10-fold serial dilutions of CDC13+, cdc13-5, and cdc13-1 strains were plated on rich media and incubated at 26°C, 30°C, and 36°C. (B) Progression of individual cells to microcolonies, for CDC13, cdc13-1, and cdc13-5 strains, measured as described by Weinert and Hartwell 1993. Strains were grown to early log growth phase at 23°C and plated for individual cells on prewarmed plates at 30°C (a nonpermissive temperature for the cdc13-1 strain); for each genotype, ≥100 cells were monitored at 0, 4, and 8 h for the proportion of large budded cells. (C) Northern analysis monitoring the expression level of RNR genes. CDC13+ and cdc13-5 strains were grown to an OD600 of 0.4 at 30°C, and an aliquot of CDC13+ cells was incubated in the presence of 200 mM hydroxyurea for 3 h; the cdc13-1 strain was grown at 23°C and an aliquot shifted to 30°C for 3 h. Relative RNR transcript levels are shown, normalized to the fold difference between the level of the RNR transcript relative to the U1 transcript levels in the wild-type strain.
This suggests that the DNA damage response of the cdc13-1 mutant strain may be the combined consequence of the simultaneous increase in single-stranded DNA and the loss of the unstable Cdc13-1 protein from chromosome ends. The lack of a DNA damage response evoked by the cdc13-5 mutant could be explained if the extended single-stranded G tails are transient and/or are still bound (and hence protected) by the Cdc13 protein. The latter suggestion is also compatible with the previous demonstration that a single-stranded telomeric oligomer is capable of binding multiple Cdc13 molecules (Hughes et al. 2000a). Consistent with such a model, the binding affinity of the truncated 80-kD version of the Cdc13-3 protein to a 24-base oligonucleotide was almost identical to that of wild-type Cdc13 protein (Fig. 6; data not shown). We therefore propose that the extended single-stranded DNA present at telomeres in cdc13-3 and cdc13-5 strains is still bound by Cdc13 (and presumably other factors) and is incapable of sending a DNA damage signal.
Figure 6.
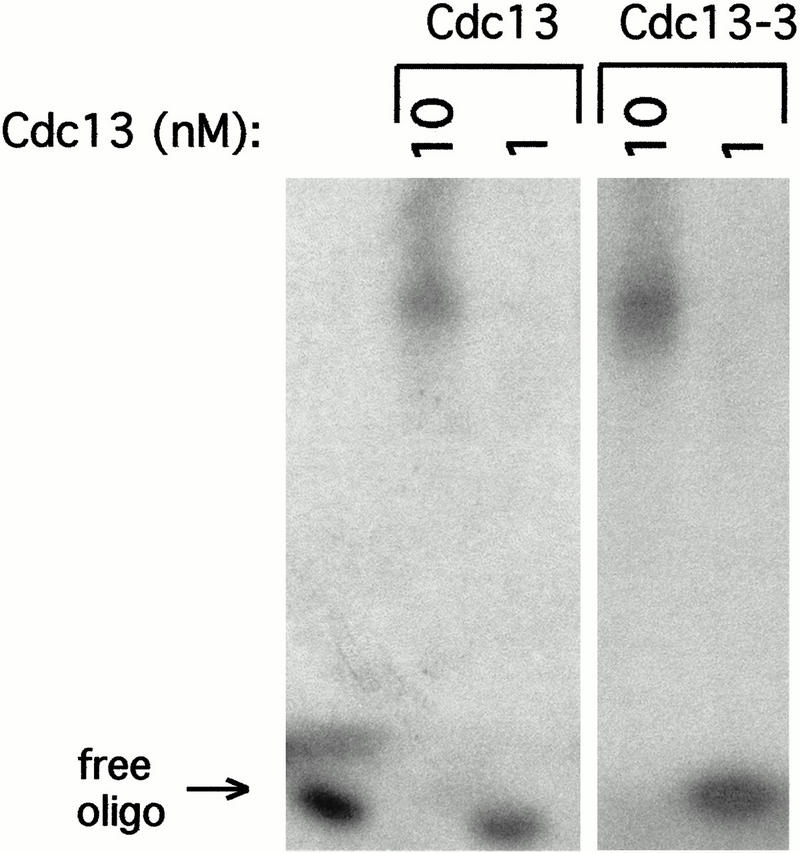
The 80-kD truncated Cdc13 protein retains DNA binding activity. DNA binding ability of wild-type Cdc13 (lanes 2,3) and the 80-kD truncated derivative of Cdc13-3 (lanes 4,5), as assessed by DNA gel shift assays with 5′ end-labeled d(TGT GTGGG)3 oligomer at 1 nM and protein concentration at 10 or 1 nM, as indicated; lane 1, no protein added.
Discussion
Several previous studies, in both ciliates and budding yeast, indicate that coordinated synthesis of the two strands of the telomere is required for telomere length maintenance. Disruption of the activity of the lagging strand DNA replication machinery, by either mutation or pharmacological intervention, leads to unregulated elongation of the G-rich strand, accompanied by an impaired inability to regulate synthesis of the C-rich strand. However, the specific mechanism that coordinates G-strand synthesis by telomerase with C-strand synthesis by the lagging strand synthesis machinery has not been elucidated.
The work presented here shows that Cdc13 negatively regulates elongation of the G strand and that this activity is a key component in the coordination between G-strand and C-strand synthesis. A model to explain this negative regulatory role must also take into account the fact that Cdc13 positively regulates telomere replication as well. One simple proposal, presented in Figure 7, is derived from a model presented by Fan and Price (1997), based on their studies of de novo telomere replication in the ciliate Euplotes crassus. These authors proposed that following elongation of the G strand by telomerase, fill-in synthesis of the C strand could limit further addition of telomeric repeats, possibly by displacement or inhibition of telomerase by the pol α/primase complex. In Figure 7, we expand their model by proposing that the two regulatory activities of Cdc13, defined by the cdc13-2 and cdc13-5 defects, correspond to two distinct steps in telomere replication that coordinate and regulate synthesis of the two strands of the telomere. Binding of Cdc13 to the transiently exposed single-strand G tails present in S phase initiates telomere replication by recruiting telomerase, which can then extend the 3′ terminus of the G-rich strand. Subsequent fill-in synthesis of the C strand generates a terminus that no longer exhibits an extended single-stranded end structure. We propose that synthesis of the C strand by lagging strand synthesis machinery acts to limit extension of the G strand by telomerase, and it is this inhibitory second regulatory step that is defective in the cdc13-5 mutant. Thus, the first step (telomerase recruitment) occurs normally in the cdc13-5 strain, but the subsequent response to the initiation of C-strand DNA replication is impaired or delayed, allowing telomerase to continue unchecked in its synthesis of the G strand.
Figure 7.
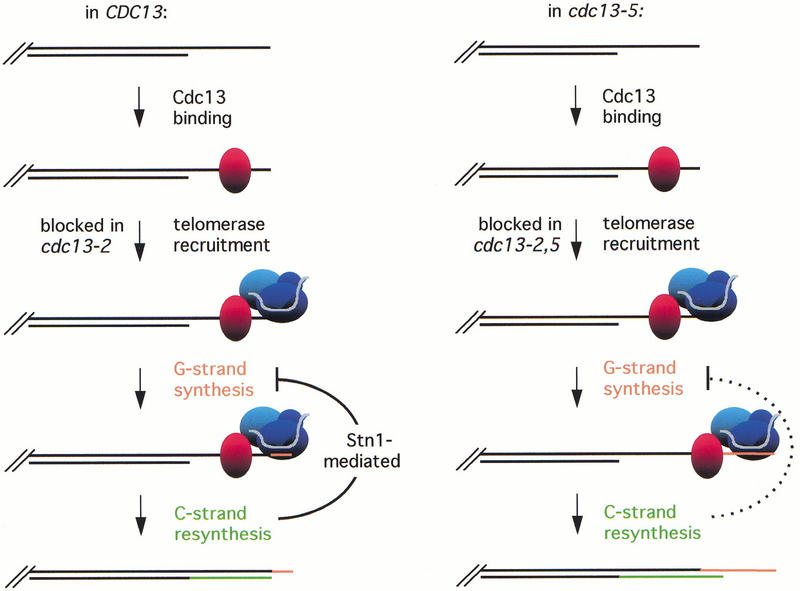
Cdc13 plays a dual role in regulation of G-strand synthesis by telomerase. The extended G strand, which is generated by a telomerase-independent mechanism (Wellinger et al. 1996), provides a substrate for binding by Cdc13. Once bound to the telomere, Cdc13 initiates synthesis of the G strand by recruiting telomerase through an interaction with the Est1 telomerase subunit. Binding of Est1 to Cdc13, and hence enzyme recruitment, is blocked by the cdc13-2 mutation. Elongation of the G strand by telomerase is subsequently followed by fill-in synthesis of the C strand by lagging strand synthesis machinery, which inhibits further synthesis of the G strand by telomerase. This inhibitory step is defective in the cdc13-5 mutant, allowing telomerase to further extend the G strand. We propose that binding of Stn1 to Cdc13, thereby displacing telomerase, is the mechanism by which unchecked telomerase synthesis is inhibited.
This model is supported by several observations. First, the telomere elongation defect of the cdc13-5 mutant is dependent on an intact telomerase complex, as well as the recruitment activity of Cdc13. In addition, the enhanced single-strand extension of the G strand observed in the cdc13-5 mutant is also restricted to the same period of the cell cycle when telomeres are normally elongated (Marcand et al. 2000). Two experiments indicate that regulation of C-strand synthesis is also an important component of the phenotype of the cdc13-5 mutant. The telomere length defects of both cdc13-5 and pol1 mutant strains are suppressed by overexpression of the Stn1 protein, indicating that Stn1 helps coordinate the action of the lagging strand replication complex with the regulatory activity of CDC13. Furthermore, the cdc13-5 strain displays an extreme enhancement of the telomere defect in response to altered dosage of one subunit of the lagging strand DNA replication machinery, arguing that the cdc13-5 defect is correlated with the ability to synthesize the C strand (data not shown). Finally, the lack of a silencing defect in the cdc13-5 strain argues that the telomere elongation phenotype is not a secondary consequence of a more general disruption of duplex telomeric chromatin structure.
Several lines of evidence indicate that both Stn1 and Est1 bind to the Cdc13 protein. Therefore, we suggest that the two steps in regulation of G-strand synthesis may be the consequence of the binding of two successive complexes to Cdc13. Work presented elsewhere demonstrates that the telomerase-associated Est1 protein promotes the positive regulatory step in telomere replication by binding to the recruitment domain of Cdc13, an interaction that is eliminated by the cdc13-2 mutation (Nugent et al. 1996; Qi and Zakian 2000; Pennock et al., 2001). In this work, we show that interaction of the negative regulator Stn1 with Cdc13 is also abolished by the same mutation. Thus, binding of Stn1 to Cdc13, occurring in response to C-strand synthesis, could limit the extent of G-strand synthesis by displacing telomerase from Cdc13. This proposed mechanism predicts that Est1 and Stn1 should compete for binding to overlapping binding sites on Cdc13 (defined by the cdc13-2 missense mutation), a possibility that is currently under investigation.
The cdc13-5 mutation, which is caused by the loss of the 230 terminal amino acids of Cdc13, still retains the site defined by the cdc13-2 mutation. Based on the above model, loss of the C-terminal region presumably impairs the ability of the Cdc13-5 protein to interact with Stn1. This is consistent with rescue of the cdc13-5 defect by overexpression of STN1, indicating that increased Stn1 protein levels can restore an interaction between Stn1 and the truncated Cdc13-5 protein. Therefore, the presence of the C-terminal 230 amino acids of Cdc13, although not essential for interaction with Stn1, may help stabilize or facilitate the Stn1–Cdc13 association. It is possible that the C terminus of Cdc13 interacts with yet another protein that could modulate the proposed competition for binding between Est1 and Stn1.
We have previously described another means by which the limit on telomere length homeostasis can be overcome, by fusing a subunit of telomerase to Cdc13 or to the DNA binding domain of Cdc13, DBDCdc13 (Evans and Lundblad 1999; Hughes et al. 2000b). These telomerase fusions result in greatly elongated telomeres, presumably because the first step in telomere length regulation—the recruitment step—has been greatly enhanced. This contrasts with the proposal, above, that telomere elongation caused by the cdc13-5 mutation is due to a defect subsequent to the recruitment step. Comparison of the properties of these two perturbations of telomere length control supports the idea that two different regulatory steps in telomere replication are altered. First, telomere elongation by DBDCdc13-telomerase or Cdc13-telomerase fusions is the result of a gain-of-function property, whereas the cdc13-5 defect is a recessive mutation. Second, elongation by the Cdc13-telomerase fusions is not dependent on the activity defined by the cdc13-2 mutation (Evans and Lundblad 1999; Hughes et al. 2000b), whereas telomerase elongation by the cdc13-5 mutation cannot occur in the presence of the recruitment-defective cdc13-2 allele. Finally, the Cdc13-5 mutant protein does not exhibit a detectable association with telomerase (S.K. Evans, A. Chandra, and V. Lundblad, unpubl.), whereas the Cdc13–Est1 and Cdc13–Est2 fusions both retain tight association with an active enzyme complex (Evans and Lundblad 1999).
One potentially surprising consequence of this study is the observation that the extensive single-stranded regions present during S phase in a cdc13-5 strain can be tolerated without obvious effects on cell cycle progression or genomic stability. A cdc13-5 strain does not exhibit a DNA damage response (Fig. 5), nor is chromosome loss elevated in this strain (data not shown). This contrasts with previous studies of the cdc13-1 mutant strain, which exhibits increased mitotic recombination and chromosome loss at semipermissive temperatures (Carson and Hartwell 1985; Hartwell and Smith 1985) and a severe DNA damage response and extensive loss of C-strand DNA under nonpermissive conditions (Weinert and Hartwell 1993; Garvik et al. 1995). Diede and Gottschling (1999) have proposed that tight coupling between telomerase synthesis and the activity of DNA polymerases α and δ is necessary to prevent inappropriately elongated G-tails, which could elicit the same detrimental consequences similar to those observed in a cdc13-1 mutant. One explanation for the behavior of the cdc13-5 strain that would still be consistent with their proposal is that the cdc13-5 mutation may not be a complete coupling defect, as the elongated G tails are only observed in S phase in this strain. Because telomeres appear to be primarily duplex at other stages of the cell cycle in the cdc13-5 strain, most of the C strand must be eventually synthesized during each cell cycle in this mutant. This indicates that there is a delay, rather than a complete block, in coupling between synthesis of the two strands. However, if the proposed coupling function is essential for cell cycle progression, it is notable that the cdc13-5 strain displays no apparent cell cycle delay. This suggests that tight coupling may not be essential and that the cell may be able to maintain genomic integrity even in the presence of elongated single-stranded tracts at the telomere (at least during one period of the cell cycle), as long as these single-stranded regions are bound and protected by Cdc13.
In this article, we have described a role for Cdc13 in the negative regulation of telomere length by coordinating G-strand and C-strand synthesis. This mechanism appears to be distinct from that promoted by duplex telomere DNA binding proteins, which mediate a conformational switch that affects the accessibility of the chromosome end to telomerase (for review, see Evans and Lundblad 2000). Notably, like CDC13, the mammalian telomere binding protein TRF2 is required for both negative regulation of telomere length (Smogorzewska et al. 2000) and an essential role in chromosome end protection (van Steensel et al. 1998). Both end protection and length regulation of human telomeres may rely on the regulated formation of a unique DNA structure, dubbed the t-loop, in which the G-rich 3′ overhang invades the duplex telomeric tract (Griffith et al. 1999). Whether such a structure forms at yeast telomeres, and is regulated by the Cdc13 protein, is an interesting experimental question.
Materials and methods
Strains and plasmids
All strains used in this study are isogenic, derived from previously published strains by introduction of the relevant deletion mutations or cdc13 mutations by standard molecular techniques, with the exception of the strains used in Figure 3C and 3D. DVL233 (MATa/α ura3-52/ura3-52 ade2-101/ade2-101 trp1Δ-1/trp1Δ-1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 TLC1/tlc1Δ::LEU2 RAD52/rad52-Δ::LEU2 CDC13/cdc13-3 CF-SUP11-TRP1) and DVL326 (MATa/α ura3-52/ura3-52 ade2-101/ade2-101 trp1Δ-1/trp1Δ-1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 TLC1/tlc1-Δ::LEU2 CDC13/cdc13-5 CF-SUP11-TRP1) were derived from DVL131 as described (Lendvay et al. 1996), DVL328 (MATa/α ura3-52/ura3-52 ade2-101/ade2-101, trp1Δ-1/trp1Δ-1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 EST1/est1-Δ3::HIS3 RAD52/rad52-Δ::LEU2 CDC13/cdc13-5 CF-SUP11-TRP1) was derived from TVL140 as described previously (Lundblad and Blackburn 1993), and DVL162 (MATa/α ura3-52/ura3-52 ade2-101/ade2-101, trp1Δ-1/trp1Δ-1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 CDC13/cdc13-Δ::LYS2 CF-SUP11-TRP1/pVL 438) from YPH275 (Lundblad and Szostak 1989). Haploid cdc13-1, cdc13-3, cdc13-5, or cdc13-2,5 strains were derived by either dissecting the relevant diploid or by introducing the relevant plasmids into a cdc13-Δ/pVL438 haploid strain (derived from dissection of DVL162), followed by eviction of pVL438 on selective media containing 5-FOA.
All CDC13 plasmids used in this study were derived from pVL438 (CEN URA3 CDC13; YCplac33-based) or pVL440 (CEN LEU2 CDC13; YCplac111-based); both contained the same 4.5-kb genomic insert with the wild-type CDC13 gene. pVL690 (CEN LEU2 cdc13-2), pVL762 (CEN LEU2 cdc13-1), and pVL1360 (CEN LEU2 cdc13-2,5) were derived from pVL440 by introducing the relevant CDC13 mutant alleles. The cdc13-3 mutation (K695A, R697A, D698A, E699A) was constructed by site-directed mutagenesis in the E. coli expression plasmid pVL427 (Hughes et al. 2000a) and subsequently subcloned to generate pVL821 (CEN LEU2 cdc13-3). The cdc13-5 mutation was constructed by PCR to introduce a stop codon following amino acid 694 to generate pVL1033 (CEN LEU2 cdc13-5). The cdc13-3 and cdc13-5 mutations were integrated into the genome of appropriate strains using pVL780 (YIp URA3 cdc13-3) or pVL1215 (YIp URA3 cdc13-5), using a pop-in/pop-out strategy. pVL841 and pVL903, which encode HA3–Cdc13p and HA3–Cdc13-5p, respectively, are derived from pVL440; the HA3 epitope tag introduced into the N terminus of Cdc13 was described previously (pVL842; Hughes et al. 2000b).
Plasmids used for high copy expression of STN1 were derived from YEplac112. pVL1034 (2 μ TRP1 STN1, with expression of STN1 driven by its native promoter) contains a 2.9-kb genomic insert that includes the STN1 gene. pVL1035 (2 μ TRP1 GAL-STN1) was derived from pVL1036 (YEplac112 containing the ∼0.8-kb GAL1 promoter) by insertion of a PCR-amplified copy of STN1 behind the GAL1 promoter. The plasmids used for two-hybrid analysis were derived from pAS1 and pACT1. pVL705 was constructed by insertion of a full-length fragment of CDC13 bearing an in-frame deletion of amino acids 585–677 (within the DNA binding domain of Cdc13; Hughes et al. 2000a) into the NdeI site of pAS1 to generate a fusion to the Gal4 DNA-binding domain; deletion of a portion of the DNA-binding domain was necessary to prevent overexpression lethality (C. Nugent, unpubl.). pVL854 is identical to pVL705 except for the presence of the cdc13-2 missense mutation. pVL667 contains the full-length cdc13-1 gene inserted into pAS1. pVL859 was recovered from a library screen as a Cdc13-interacting clone and contains amino acids 5–495 of the Stn1 protein fused in frame with the Gal4 activation domain of pACT1. pVL260 contains 80 bp of G1–3T DNA cloned into the polylinker of pDS67 (a pBluescript plasmid that also contains ARG4).
Molecular methods
Standard denaturing Southern gel conditions were used for determining telomere length (Lendvay et al. 1996). Single-strand chromosome termini were analyzed by nondenaturing in-gel hybridization with a 22-mer CA-rich oligonucleotide as described previously (Dionne and Wellinger 1996). To assay chromosome end structure during the cell cycle, the cdc13-5 strain was grown in YPD to OD600 0.2, followed by the addition of 3 μM α factor. Additional α factor (at a concentration of 3 μM) was added every hour until ∼90% of the cells appeared unbudded (for ∼3–4 h). The α factor was removed by centrifugation and cells released into YPD at OD600 0.4. Cells for FACS analysis were fixed in 70% ethanol, washed in 50 mM Na citrate (pH 7.0), digested with RNase overnight, washed again and stained with propidium iodide (15 μg/mL), and analyzed by flow cytometry.
For Northern blot analysis, total RNA was isolated by a phenol-freeze method (Schmitt et al. 1990), resolved on formaldehyde-1% agarose, and probed with the 1.7-kb BglII–EcoRI RNR1 fragment from pSE738, the 1.1-kb HindIII RNR2 fragment from pSE310, the 1-kb BamHI–EcoRI RNR3 fragment from pSE734, the 1.4-kb StuI–SwaI RNR4 fragment from pMH120, or a 600-bp fragment of the U1 gene (recovered by PCR of yeast genomic DNA).
Biochemical methods
For detection of the HA-tagged Cdc13 and Cdc13-5 proteins, cdc13-Δ/pVL440 (Cdc13), cdc13-Δ/pVL841 (HA3–Cdc13), and cdc13-Δ/pVL903 (HA3–Cdc13-5) were grown in selective media to an OD600 0.6. Extract preparation and Western analyses were done as described in Hughes et al. (2000b), except that TMG 300+ (10 mM Tris-HCl at pH 8.0, 1 mM MgCl2, 5% glycerol, 1 mM PMSF, 1 mM DTT, 300 mM NaCl) was used for immunoprecipitations and washes.
His6-tagged proteins (Cdc13 or Cdc13-3) were expressed in SF9 cells as described previously (Hughes et al. 2000a). For protein production, cells were pelleted, washed, and resuspended in SB 300 10% + (50 mM Na phosphate at pH 8.0, 300 mM NaCl, 10% glycerol, 0.5% Tween-20, 10 mM imidazole, 1 mM PMSF), and sonicated for 3–4 repetitions of 10–30 sec; the degree of sonication required was monitored by visual inspection.Extracts were clarified by centrifugation for 10 min, and His6-tagged proteins were purified on Ni-NTA agarose at 4° (QIAGEN) by batch method, according to the manufacturer's instructions. Protein concentration was determined by comparison to BSA standards on Coomassie-stained SDS-PAGE gels. Binding reactions were performed as described in Nugent et al. (1996).
Acknowledgments
We thank the members of the Lundblad laboratory for many helpful discussions and also thank the Flow Cytometry Core Lab, Texas Children's Hospital. We gratefully acknowledge the generous gifts of strains or plasmids from Connie Holm, Oscar Aparicio, Stephen Bell, Judith Campbell, Steve Elledge, Tim Formosa, and Peter Burgers. This work was supported by NIH grant GM55867 (to V.L.) and a postdoctoral fellowship from the U.S. Army and Materiel Command (to C.N.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lundblad@bcm.tmc.edu; FAX (713) 798-5931.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.861001.
References
- Adams AK, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams Martin A, Dionne I, Wellinger RJ, Holm C. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol Cell Biol. 2000;20:786–796. doi: 10.1128/mcb.20.3.786-796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Carson M, Hartwell L. CDC17: An essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and β. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Dionne I, Wellinger RJ. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- ————— Positive and negative regulation of telomerase access to the telomere. J Cell Sci. 2000;113:3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- Fan X, Price CM. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol Biol Cell. 1997;8:2145–2155. doi: 10.1091/mbc.8.11.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes & Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol Cell Biol. 2000;20:8397–8408. doi: 10.1128/mcb.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–745. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Ann Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Hartwell L, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR. “Biochemical characterization of telomere replication proteins in Saccharomyces cerevisiae.” Ph.D. thesis. Baylor College of Medicine; 1998. [Google Scholar]
- Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc Natl Acad Sci. 2000a;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000b;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, II, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: No longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E. Cell cycle restriction of telomere elongation. Curr Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- Price CM. Synthesis of the telomeric C-strand: A review. Biochemistry. 1997;62:1216–1223. [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes & Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]


