Abstract
A DNA microarray (Enteroarray) was designed with probes targeting four species-specific taxonomic identifiers to discriminate among 18 different enterococcal species, while other probes were designed to identify 18 virulence factors and 174 antibiotic resistance genes. In total, 262 genes were utilized for rapid species identification of enterococcal isolates, while characterizing their virulence potential through the simultaneous identification of endogenous antibiotic resistance and virulence genes. Enterococcal isolates from broiler chicken farms were initially identified by using the API 20 Strep system, and the results were compared to those obtained with the taxonomic genes atpA, recA, pheS, and ddl represented on our microarray. Among the 171 isolates studied, five different enterococcal species were identified by using the API 20 Strep system: Enterococcus faecium, E. faecalis, E. durans, E. gallinarum, and E. avium. The Enteroarray detected the same species as API 20 Strep, as well as two more: E. casseliflavus and E. hirae. Species comparisons resulted in 15% (27 isolates) disagreement between the two methods among the five API 20 Strep identifiable species and 24% (42 isolates) disagreement when considering the seven Enteroarray identified species. The species specificity of key antibiotic and virulence genes identified by the Enteroarray were consistent with the literature adding further robustness to the redundant taxonomic probe data. Sequencing of the cpn60 gene further confirmed the complete accuracy of the microarray results. The new Enteroarray should prove to be a useful tool to accurately genotype strains of enterococci and assess their virulence potential.
INTRODUCTION
Enterococci are commensal bacteria found in the gastrointestinal tracts of humans, animals, and birds, as well as in soil and water. Sequenced Enterococcus faecalis and E. faecium genomes revealed the presence of virulence factors and mobile and/or exogenously acquired DNA giving some insight on how these bacteria made the transition from a commensal organism to a nosocomial pathogen (21, 46, 53). Enterococci can infect the endocardium, abdomen, urinary tract, burn or surgical wounds, and numerous other tissues, especially in immunocompromised patients (24, 59). E. faecalis causes the majority of infections, followed by E. faecium (53). Other enterococci such as E. avium, E. casseliflavus, E. durans, E. gallinarum, E. hirae, E. malodoratus, E. mundtii, E. raffinosus, and E. solitarius rarely cause infections but frequent misidentification by classical biochemical or microbiological methods suggests that their importance might be underestimated (14, 24, 35, 52, 55).
Resistance to antimicrobial agents can be intrinsic or acquired through several genetic mechanisms, including mutation, integration of foreign genetic material, or transfer of plasmids and transposons (21, 46, 52). While E. gallinarum and E. casseliflavus possess intrinsic low-level resistance to vancomycin, other enterococci have been shown to be resistant to aminoglycosides, ampicillin, and glycopeptides (30, 42, 55). Nosocomial infections involving vancomycin-resistant enterococci are difficult to treat with antimicrobial agents (2, 52). From an agricultural perspective, many antimicrobial agents have been approved for use in treating infections or to positively impact animal growth and feeding (15). Widespread use of antimicrobial agents in livestock and poultry has raised concerns that bacterial pathogens, having food animal reservoirs, could develop drug resistance and transfer these antimicrobial resistance (AMR) genes to humans through the consumption of retail meat (1, 49, 51). Consequently, there is an increasing interest in characterizing virulence factors and antibiotic resistance in enterococci causing nosocomial infections in hospitals or isolated from food products (17, 23, 25, 39, 43, 44, 47). Rapid and accurate identification of infection-related enterococcal species is crucial in choosing the appropriate treatment for patients (2).
Classical biochemical identification methods relying on enzymatic activity or sugar fermentation, such as the API 20 Strep, are time-consuming and only identify a limited number of species. More importantly, they have been shown to misidentify some species (5, 7, 22, 54, 57), underscoring the limitations of classical microbiological/biochemical methods. To circumvent these limitations, molecular identification and typing methods such as PCR, multilocus sequence typing, and pulsed-field gel electrophoresis have been developed (11, 26, 27, 33, 38, 40, 41, 45, 54). However, these methods usually only target a few strains or species and generally lack information on the presence of specific virulence factors and AMR genes.
The parallel processing power of DNA microarrays allows the rapid identification and virulence potential assessment of microbial pathogens (4, 18–20, 31, 39). Microarrays targeting enterococci have appeared in the literature to identify several species or to detect virulence factors in E. faecalis. In addition, independent microarrays have been developed for AMR gene detection (4, 18–20, 31, 39). Due to the importance of enterococci in nosocomial infections, increasing resistance to antimicrobials, and the undefined role of different enterococcal species in food-borne illnesses, a single tool to comprehensively identify and characterize the virulence potential of isolated enterococcal species would be useful in understanding their role in enterococcus-related disease.
In the present study, a DNA microarray (Enteroarray) was designed with three distinct modules: taxonomic identifiers, virulence factors, and antibiotic resistance genes, making it a powerful instrument for the rapid and comprehensive species identification of enterococcal isolates, while providing valuable information on their virulence potential. Identification of enterococcal farm isolates by the classical API 20 Strep method and the Enteroarray confirmed that the array was extremely accurate in species identification by correctly identifying isolates misidentified by API 20 Strep while simultaneously characterizing the virulence potential and antibiotic resistance phenotype of each isolate.
MATERIALS AND METHODS
Enterococcal isolation.
Enterococcal strains (n = 171) were isolated from various samples (fecal, cecal, and litter) from nine commercial and experimental broiler farms in the Fraser Valley of British Columbia. The commercial farms each raise, on average, 130,000 to 360,000 broilers annually, while approximately 3,600 broilers are produced annually on the experimental farms. The samples (5 to 6 g) in Carry-Blair medium (Quelab, Montreal, Quebec, Canada) were vigorously vortexed for 1 min, and then 10-fold serial dilutions were prepared in sterile saline. Presumptive Enterococcus populations were isolated by spreading 10-fold dilutions of individual samples from each farm onto KF streptococcal agar CM0701 (Oxoid, Nepean, Ontario, Canada) and incubating them at 37°C for 48 h as described elsewhere (9, 23). Six to eight typical presumptive colonies per sample were randomly selected, and a total of 171 isolates were confirmed as enterococci by Gram staining, catalase test, and API 20 Strep (bioMérieux, St-Laurent, Quebec, Canada). All 171 presumptive enterococcal isolates were subjected to the API 20 Strep (bioMérieux, St-Laurent, Quebec, Canada). Isolates from cecal samples were obtained at necropsy. Purified Enterococcus colonies were frozen in tryptic soy broth (TSB) containing 25% glycerol at −80°C for further analysis. Six Enterococcus species were obtained from the American Type Culture Collection (ATCC) as controls to validate the Enteroarray: E. avium ATCC 49463, E. durans ATCC 11576, E. faecium ATCC 35667, E. gallinarum ATCC 49608, E. faecalis ATCC 29212, and E. hirae ATCC 10541.
DNA extraction.
Bacterial lysates were prepared for each isolate. A single colony was grown overnight in 1 ml of TSB in a 15-ml snap-cap tube. The cells were centrifuged at 12,000 × g for 10 min, and the supernatant was discarded before resuspending the pellet in 200 μl of high-pressure liquid chromatography water. Cells were lysed by boiling for 15 min and then placed on ice for 1 min. The lysed preparation was centrifuged at 12,000 × g for 5 min to pellet cell debris, and the supernatant containing the DNA was transferred in a sterile 1.5-ml Eppendorf tube.
Enterococcal microarray design.
Our enterococcal microarray represents a composite of three separately designed modules: AMR genes, virulence genes, and taxonomic genes.
(i) Antibiotic resistance.
We had previously designed and validated antibiotic resistance microarrays for use with both Gram-negative and Gram-positive bacteria (4, 20, 37) using a 70-mer probe length to maximize the overall sensitivity of the microarray (32). A total of 173 probes were used, including 166 antibiotic resistance genes and a class 1 integron. Sixty-five oligonucleotide probes printed on the microarray were selected directly from a previous study (18).
(ii) Virulence factors.
OligoPicker software (56) was used to design 70-mer oligonucleotide probes targeting 15 virulence factors associated with E. faecalis and two associated with E. faecium (Table 1). The targeted virulence factors include hemolysins, gelatinases, surface adhesion proteins, aggregation substances, surface carbohydrates, collagen adhesins, and sex pheromones, which have been reported to be primarily found in isolates from foods of animal origin (13, 39).
Table 1.
Prevalence of virulence factors represented on the Enteroarray
| Virulence factor | Descriptiona | Presence (% total) in: |
||
|---|---|---|---|---|
| E. faecalis | E. faecium | E. hirae plus E. avium | ||
| ace | Collagen binding cell wall protein | 100 | 0 | 0 |
| agg | Aggregative adherence protein | 16 | 0 | 0 |
| agrBfs | AgrBfs protein in E. faecalis | 100 | 0 | 0 |
| cad1 | Pheromone cAD1 precursor lipoprotein | 100 | 0 | 100 |
| cAM373 gene | Sex pheromone cAM373 precursor | 100 | 0 | 0 |
| cCF10 gene | Pheromone cCF10 precursor lipoprotein | 100 | 0 | 0 |
| cob | Pheromone cOB1 precursor/lipoprotein | 100 | 0 | 0 |
| cpd1 | Pheromone cPD1 lipoprotein | 100 | 0 | 0 |
| cylA | Hemolysin | 6 | 0 | 0 |
| cylB | Hemolysin | 6 | 0 | 0 |
| cylL | Hemolysin | 3 | 0 | 0 |
| cylM | Hemolysin | 3 | 0 | 0 |
| efaAEfs | E. faecalis specific cell wall adhesin | 100 | 0 | 0 |
| efmAEfm | E. faecium specific cell wall adhesin | 0 | 90.1 | 0 |
| esp (E. faecalis) | E. faecalis enterococcal surface protein | 0 | 0 | 0 |
| esp (E. faecium) | E. faecium enterococcal surface protein | 0 | 0 | 0 |
| gelE | Gelatinase | 100 | 0 | 0 |
No virulence factors were found in E. casselflavus, E. gallinarum, and E. durans hybridized to the Enteroarray.
(iii) Taxonomic identifiers.
Taxonomic microarrays generally rely on 16S or 23S rRNA genes or the intervening sequence to identify bacterial species (31, 58). A previous enterococcus-specific array used 18-mer sequences to define species-specific probes. Because of the high similarities observed when the enterococcal 16S rRNA sequences were aligned, probes of >18 bases would lose their discriminatory ability if used on a microarray. Therefore, this approach could not be applied to the current virulence array when trying to match the length of the virulence factors and antibiotic resistance genes designed previously. Another more genetically diverse candidate gene, cpn60 (heat shock protein), also frequently used in bacterial taxonomy (10, 34) was also eliminated for the same reasons as the 16S rRNA sequence since 28-mers were the longest species-specific probes that could be designed for enterococci.
To replace 16S and cpn60, we chose four genes—ddl, pheS, atpA, and recA (12, 28, 41, 50)—based on the availability of their sequences in GenBank, and their high level of interspecies sequence variation. However, specific 70-mer probes could not be designed for all enterococcal species, therefore we opted for 50-mers. The Tm of the 50-mer probes was close enough to that of the 70-mer virulence factor and antibiotic resistance gene probes to allow their printing on the same microarray since, at those lengths, any intensity differences between the different size probes are minor and have no effect on positive gene scoring.
Eighteen Enterococcus species most commonly used for the development of new identification methods (28, 31) were targeted for the Enteroarray. Confirmation of selected enterococcal isolates was performed by sequencing a segment of the cpn60 gene amplified by the universal cpn60 primers H729 (5′-CGC CAG GGT TTT CCC AGT CAC GAC GAI III GCI GGI GAY GGI ACI ACI AC-3′) and H730 (5′-AGC GGA TAA CAA TTT CAC ACA GGA YKI YKI TCI CCR AAI CCI GGI GCY TT-3′), including the M13F and M13R sequences, respectively (underlined) (34).
The universal eubacterial probe EUB338 was used as a positive control, and two types of negative controls were present, a shuffled version of the EUB338 probe and 50% dimethyl sulfoxide buffer with no oligonucleotide probe sequence. The microarrays were printed as previously described (3, 4).
Enterococcus DNA labeling.
Bacterial DNA was labeled using a Bioprime DNA labeling system (Invitrogen Life Technologies, Burlington, Ontario, Canada). A 10-μl portion of bacterial lysate was added to 10 μl of a random primer solution (750 μg of oligodeoxyribonucleotide primers [random octamers]/ml in 125 mM Tris-HCl [pH 6.8], 12.5 mM MgCl2, and 25 mM 2-mercaptoethanol) and denatured at 95°C for 5 min. After a 5-min incubation on ice, the reaction mixture was completed by adding 5 μl of a deoxyribonucleotide triphosphate mix (1.2 mM dATP, 1.2 mM dGTP, 1.2 mM dTTP, and 0.6 mM dCTP in 10 mM Tris [pH 8.0] and 1 mM disodium EDTA), 2 μl of 1 mM Cy5-dCTP and 0.5 μl of high-concentration DNA polymerase (Klenow fragment, 40 U/μl) for a total reaction volume of 27.5 μl. Labeling reactions were performed in the dark at 37°C for 3.5 h and stopped by the addition of 2.5 μl of 5 M disodium EDTA (pH 8.0). The labeled samples were then purified with a Pure-Link PCR purification kit (Invitrogen Life Technologies) according to the manufacturer's protocol. The amount of incorporated fluorescent Cy5 dye was then quantified by scanning the DNA sample with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) from 200 to 700 nm. The data were analyzed by using an online percent incorporation calculator (http://www.pangloss.com/seidel/Protocols/percent_inc.html).
Hybridization of labeled DNA.
Microarrays were prehybridized at 50°C for 1 h under a Hybri Slip coverslip (22 by 60 mm; Sigma Chemical Co., St. Louis, MO) in a Slide Booster hybridization work station (model SB800; Advalytix, Germany) with 50 μl of DIG Easy Hyb buffer (Roche Diagnostics, Laval, Quebec, Canada) containing 5% (vol/vol) bovine serum albumin (1 mg/ml; New England BioLabs, Inc., Beverly, MA). The slide was then dipped in 0.1× SSC (15 mM NaCl plus 1.5 mM trisodium citrate [pH 7.0]) to remove the coverslip and air dried. Before hybridization, 1 μg of labeled DNA was dried by using a SpeedVac (DNA SpeedVac DNA110; Savant Scientific) and resuspended in 18 μl of DIG Easy Hyb buffer (Roche Diagnostics). The samples were denatured by heating 5 min at 95°C and pipetted under a Lifter cover slip (18 by 18 mm; Erie Scientific, Portsmouth, NH). Microarrays were then hybridized overnight at 50°C in the slide booster. The coverslips were removed by dipping the slide in 0.1× SSC–0.1% (wt/vol) sodium dodecyl sulfate (pH 7.2) and four stringency washes (three in 0.1× SSC–0.1% [wt/vol] sodium dodecyl sulfate and one in 0.1× SSC) were performed at 37°C for 5 min with agitation. The slide was then air dried and scanned at a resolution of 10 μm at 90% laser power with a ScanArray Lite fluorescent microarray analysis system (Perkin-Elmer, Mississauga, Ontario, Canada). Acquisition of fluorescent spots and quantification of fluorescent spot intensities were performed using the ScanArray Express software, version 2.1 (Perkin-Elmer, Foster City, CA). The data was normalized by subtracting local background intensity from the recorded spot intensities from one subarray. For each subarray, the median value for each set of triplicate-spotted oligonucleotides was compared to the median value for the whole subarray. Oligonucleotides with a signal-to-noise fluorescence ratio above 2 were considered positive. To identify an isolate down to the species level, a minimum of three of the four taxonomic probes designed for a particular species must show a positive signal before it was considered positively identified.
RESULTS
Taxonomy.
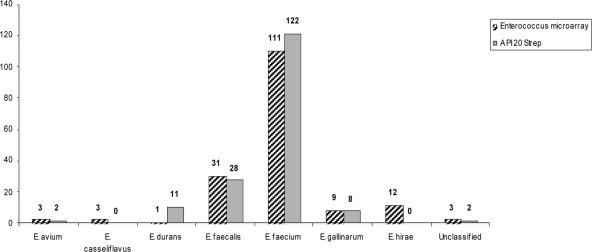
A total of 171 presumptive enterococcal isolates were subjected to the API 20 Strep commercial identification system. Of the 171 isolates, all were confirmed to be enterococci (Fig. 1) and were distributed among five different enterococcal species as follows: E. faecium (n = 122), E. faecalis (n = 28), E. durans (n = 11), E. gallinarum (n = 8), and E. avium (n = 2).
Fig. 1.
Identification of enterococcal isolates by the Enteroarray and API 20 Strep tests. The results of the Enteroarray are presented in the hatched columns, while the gray columns represent the results of the API 20 Strep biochemical method. The biochemical method failed to identify all of the E. casseliflavus and E. hirae isolates.
The same 171 isolates were labeled and hybridized on the Enteroarray. Similar to the API 20 Strep results, all isolates were confirmed to be enterococci; however, they were distributed among seven different enterococcal species as follows: E. faecium (n = 111), E. faecalis (n = 31), E. hirae (n = 12), E. gallinarum (n = 9), E. avium (n = 3), E. casseliflavus (n = 3), and E. durans (n = 1). One isolate (isolate 2941) hybridized with probes representing both E. faecium and E. faecalis, suggesting that this sample was not obtained from a pure colony (Table 2). The same isolate was identified by API 20 Strep as being only E. faecium. All five ATCC strains were identified correctly on the Enteroarray. Therefore, between the two methodologies, an identification discrepancy was observed among 42 of the confirmed 171 enterococcal isolates. Unlike the 18 species that the Enteroarray was designed to identify, API 20 Strep only recognizes 5 and does not include E. hirae and E. casseliflavus. If the latter are removed from the pool of isolates, the discrepancies in species identification between the methods is ca. 15%. However, rather than indicating unknown species, API 20 Strep misclassified all three E. casseliflavus isolates as E. faecium at 99.9% certainty and all of the E. hirae as either E. durans or E. faecium at 91 to 99.9% certainty (Table 2). Therefore, a more conservative estimate of divergence between the methods is >24%.
Table 2.
Differential isolate identification by API 20 Strep, Enteroarray, and cpn60 sequencing
| Isolate | Phenotypic identification API 20 Strep |
Genotypic identification (Enteroarray) |
cpn60 sequencing BLAST resulta |
|||
|---|---|---|---|---|---|---|
| Species | % Identity | Species | % Identity | Accession no. | ||
| 2954 | E. faecalis | 97.0 | E. avium | ND | ||
| 2911 | E. faecium | 99.8 | E. avium | E. avium | 98 | AF417583 |
| 2803 | E. faecium | 99.9 | E. casseliflavus | E. casseliflavus | 98 | AF417584 |
| 2876 | E. faecium | 99.9 | E. casseliflavus | E. casseliflavus | 98 | AF417584 |
| 2982 | E. faecium | 99.9 | E. casseliflavus | E. casseliflavus | 100 | AF417584 |
| 2902 | E. durans | 91.9 | E. faecalis | E. faecalis | 98 | DQ074968 |
| 1737 | E. faecium | 92.0 | E. faecalis | E. faecalis | 99 | AF335185 |
| 2814 | E. faecium | 99.8 | E. faecalis | E. faecalis | 99 | DQ074968 |
| 2934 | E. faecium | 69.3 | E. faecalis | E. faecalis | 99 | DQ074968 |
| 2989 | E. faecium | 69.3 | E. faecalis | E. faecalis | 99 | DQ074968 |
| 2990 | E. faecium | 69.3 | E. faecalis | E. faecalis | 99 | DQ074968 |
| 2917 | E. durans | 90.1 | E. faecalis | E. faecium | 100 | AF417582 |
| 1695 | E. faecalis | 99.9 | E. faecium | E. faecium | 98 | AF417582 |
| 2804 | E. faecalis | 99.7 | E. faecium | E. faecium | 98 | AF417582 |
| 1746 | E. gallinarum | 66.0 | E. faecium | E. faecium | 99 | AF417582 |
| 1740 | E. gallinarum | 66.0 | E. faecium | E. faecium | 99 | AF417582 |
| 2019 | E. gallinarum | 77.4 | E. faecium | E. faecium | 99 | AF417582 |
| 2026 | E. gallinarum | 77.4 | E. faecium | ND | ||
| 2033 | E. gallinarum | 80.6 | E. faecium | E. faecium | 98 | AF417582 |
| 2020 | E. gallinarum | 93.8 | E. faecium | E. faecium | 99 | AF417582 |
| 1734 | E. gallinarum | 99.6 | E. faecium | E. faecium | 99 | AF417582 |
| 1745 | E. avium | 97.0 | E. gallinarum | E. gallinarum | 97 | AF417587 |
| 2828 | E. faecium | 93.8 | E. gallinarum | E. gallinarum | 95 | AF417587 |
| 2799 | E. faecium | 99.8 | E. gallinarum | E. gallinarum | 99 | AF417587 |
| 2802 | E. faecium | 99.8 | E. gallinarum | E. gallinarum | 95 | AF417587 |
| 2821 | E. faecium | 99.8 | E. gallinarum | E. gallinarum | 95 | AF417587 |
| 2823 | E. faecium | 99.8 | E. gallinarum | E. gallinarum | 98 | AF417587 |
| 2839 | E. faecium | 99.8 | E. gallinarum | E. gallinarum | 95 | AF417587 |
| 2853 | E. faecium | 99.8 | E. gallinarum | E. gallinarum | 99 | AF417587 |
| 2866 | E. durans | 91.7 | E. hirae | E. hirae | 97 | AF417586 |
| 2906 | E. durans | 91.7 | E. hirae | E. hirae | 100 | AF417586 |
| 1687 | E. durans | 95.0 | E. hirae | E. hirae | 98 | AF417586 |
| 1696 | E. durans | 95.0 | E. hirae | E. hirae | 98 | AF417586 |
| 1713 | E. durans | 95.0 | E. hirae | E. hirae | 99 | AF417586 |
| 1722 | E. durans | 95.0 | E. hirae | E. hirae | 98 | AF417586 |
| 1732 | E. durans | 95.0 | E. hirae | E. hirae | 99 | AF417586 |
| 2029 | E. durans | 96.1 | E. hirae | E. hirae | 98 | AF417586 |
| 1731 | E. faecium | 91.0 | E. hirae | E. hirae | 98 | AF417586 |
| 1739 | E. faecium | 97.0 | E. hirae | E. hirae | 98 | AF417586 |
| 1721 | E. faecium | 99.0 | E. hirae | E. hirae | 98 | AF417586 |
| 2819 | E. faecium | 99.9 | E. hirae | ND | ||
| 2998 | Aerococcus viridans | 99.3 | No taxo probe | ND | ||
| 2941 | E. faecium | 99.8 | E. faecium and E. faecalis | ND | ||
| 2985 | Not valid | Not valid | One E. faecalis probe | ND | ||
cpn60 was amplified with universal primers (see Materials and Methods). ND, not done.
Although all four taxonomic probes printed on the Enteroarray for each species always gave signals well above background, hybridization results from seven isolates gave signals for only three of the four positive probes. However, three isolates were identified as E. casseliflavus whose identification relies on only three taxonomic probes since the recA sequence was not available in GenBank. The built in redundancy of the multiple independent taxonomic probes allowed clear identification of the remaining four isolates based on the three positive signals. To further validate the accuracy of the Enteroarray, the identities of 38 isolates covering all combinations of species differences were confirmed by sequencing their cpn60 gene, 34 of which were from misidentified isolates and 4 isolates that were found to be the same species by the API 20 Strep and the Enteroarray. The top single species cpn60 BLAST matches ranged between 95 and 100% identity with the second nearest species matching only at 89% or less. In all cases, sequencing of the cpn60 gene agreed with the Enteroarray identification, including the four isolates that were in agreement by both methods (Table 2).
Virulence factors.
All 31 isolates identified as E. faecalis by the Enteroarray also scored positively for a core of nine virulence factors: ace, agrBEfs (the agrB gene of E. faecalis) cad1, the cAM373 gene, the cCF10 gene, cob, cpd1, efaAEfs (the efaA gene of E. faecalis), and gelE. In addition to these nine virulence factors, five isolates were also positive for agg, one of which also possessed two hemolysins, cylA and cylB, and another with all four hemolysins (cylA, cylB, cylL, and cylM) for a total of 14 virulence factors in the same isolate (isolate 2968). Interestingly, the API 20 Strep system identified three fewer E. faecalis isolates than the Enteroarray. The three isolates were identified as an E. avium and two E. faecium strains. The possession of the same nine core virulence factors found in all of the Enteroarray identified isolates provides indirect support for their classification as E. faecalis strains as shown by the Enteroarray. None of the Enteroarray-identified isolates possessed the specific enterococcal surface protein (esp) from either E. faecalis or E. faecium. Esps are cell wall-associated proteins shown to be linked to urinary tract infections and are more frequently found in clinical isolates than commensal isolates (21).
Although all 31 E. faecalis isolates were positive for the cell wall adhesin (efaAEfs), 100 of the 111 E. faecium isolates were positive for the E. faecium-specific cell wall adhesin efmAEfm (the efmA gene of E. faecium). Two other species scored positive signals for virulence factors. All E. avium and E. hirae isolates were positive for the cad1 gene. This was the only virulence factor found in these two species, and no other virulence factors were found in species other than E. faecalis and E. faecium.
Antibiotic resistance.
Many antibiotic resistance genes were identified among all of the enterococcal isolates. These genes are involved in resistance against aminoglycosides, kanamycin, neomycin, gentamicin, erythromycin, lincosamide, streptothricin, streptogramin, tetracycline, and bacitracin, and well as vancomycin. We also identified some genes associated with macrolide efflux pumps and transposons.
The E. faecium isolates possessed antibiotic resistance genes associated with 10 different antimicrobial agent classes, more than the six other species identified by the Enteroarray, where it ranged from two to eight classes (Table 3). Transposon-associated genes were found in all seven species and mostly in E. casseliflavus, E. durans, E. faecium, and E. hirae. Genes associated with tetracycline resistance were found in six species and were prevalent in 67 to 100% of the isolates for each species. Genes associated with aminoglycosides and bacitracin resistance were found in five species and in 100% of E. durans and E. hirae isolates for bacitracin and in 98% of the E. faecium isolates for aminoglycosides. Erythromycin-associated genes were found in four species, mostly in E. faecalis and E. faecium, and in ca. 50% of the E. gallinarum and E. hirae isolates. Vancomycin resistance genes were only present in E. casseliflavus and E. gallinarum, with all E. gallinarum and E. casseliflavus isolates carrying the vanC1 and vanC3 genes, respectively. The other antimicrobial agent classes were present in one to three species and in <45% of the isolates.
Table 3.
Distribution of antimicrobial agents in enterococcal isolates
| Species (no. of strains) | No. of strains (%) | Antibiotic resistance gene class(es)a | % Isolates with indicated ABR gene(s) |
|---|---|---|---|
| E. durans (1) | 1 (100) | TE, B | TetO, 100; BcrR, 100 |
| E. avium (3) | 1 (33) | TE | Tetracycline, 67 |
| 1 (33) | A, TE | tetL, 33 | |
| 1 (33) | No ABR gene | tetM, 67 | |
| Aac(6), 33 | |||
| E. casseliflavus (3) | 3 (100) | V | VanC3, 100 |
| 1 (10) | VanC1, 100 | ||
| 5 (50) | V | Tetracycline, 90 | |
| E. gallinarum (10) | 1 (10) | TE, V | tetM, 60 |
| 1 (10) | E, TE, V | tetS, 30 | |
| 1 (10) | E, TE, V, B | tetL, 20 | |
| 1 (10) | A, E, TE, V | Erythromycin, 40 | |
| A, E, TE, V, B | ermAM, 30 | ||
| ermB, 30 | |||
| ermA, 10 | |||
| BcrR, 20 | |||
| Aminoglycosides, 20 | |||
| aac(6′)-aph(2′), 10 | |||
| ant(9′)-Ia, 10 | |||
| 1 (8) | BcrR, 100 | ||
| 2 (17) | A, B | Tetracycline, 83 | |
| 1 (8) | TE, B | tetM, 58 | |
| 2 (17) | E, B | tetL, 50 | |
| E. hirae (12) | 1 (8) | E, TE, B | tetO, 42 |
| 1 (8) | A, L, TE, B | Aminoglycosides, 50 | |
| 1 (8) | A, E, TE, B | aac(6), 25 | |
| 1 (8) | TE, STA, B | aac(6′)-aph(2′), 17 | |
| 1 (8) | A, E, TE, STA, B | ant(9)-Ia, 17 | |
| 1 (8) | A, L, TE, B, STA | aph(3′)-IIIa, 8 | |
| A, E, STC, TE, STA, B | Erythromycin, 50 | ||
| ermAM, 33 | |||
| ermB, 33 | |||
| ermA, 17 | |||
| SatG-VatE8, 33 | |||
| LinB, 17 | |||
| Sat4, 8 | |||
| 3 (10) | E, B | Erythromycin, 90 | |
| 1 (3) | TE, B | ermB, 81 | |
| 1 (3) | E, TE | ermAM, 87 | |
| E. faecalis (31) | 2 (6) | A, TE | Tetracycline, 87 |
| 14 (45) | E, TE, B | tetK, 3 | |
| 1 (3) | A, E, STC, B | tetL, 71 | |
| 1 (3) | A, E, STC, TE | tetM, 84 | |
| 8 (26) | A, E, STC, TE, B | BcrR, 84 | |
| Aminoglycosides, 39 | |||
| aac(6), 29 | |||
| aac(6′)-aph(2′), 6 | |||
| aph(3′)-IIIa, 29 | |||
| Sat4, 29 | |||
| 3 (3) | A | Aminoglycosides, 99 | |
| 1 (1) | TE, B | aac(6′)-Ii, 99 | |
| 1 (1) | A, B | aac(6), 55 | |
| 9 (8) | A, EES | aac(6′)-aph(2′), 1 | |
| 2 (2) | A, L, TE | ant(9)-Ia, 5 | |
| 2 (2) | A, TE, B | ant(2″)-Ia, 1 | |
| 1 (1) | A, E, EES | aph(3′)-IIIa, 33 | |
| 2 (2) | A, STA, EES | Macrolide efflux system, 89 | |
| 6 (5) | A, B, EES | mefA-mefE, 16 | |
| 1 (1) | A, TE, EES | msrC, 85 | |
| 9 (8) | A, TE, B, EES | Tetracycline, 80 | |
| 8 (7) | A, E, TE, EES | tetL, 65 | |
| 1 (1) | A, TE, STA, EES | tetM, 78 | |
| 7 (6) | A, TE, STA, B, EES | tetO, 2 | |
| 5 (4) | A, E, TE, B, EES | tetS, 1 | |
| E. faecium (111) | 1 (1) | A, L, TE, STC, B | BcrR, 74 |
| 2 (2) | A, E, TE, STA, B | Erythromycin, 54 | |
| 3 (3) | A, L, TE, B, EES | ermA, 5 | |
| 2 (2) | A, L, TE, STA, B, EES | ermB, 42 | |
| 2 (2) | A, E, L, TE, B, EES | ermAM, 52 | |
| 2 (2) | A, E, TE, STA, B, EES | LinB, 43 | |
| 2 (2) | A, E, TE, STC, B, EES | SatG-VatE, 40 | |
| 1 (1) | A, E, L, TE, STA, EES | Sat4, 28 | |
| 1 (1) | A, E, TE, STC, STA, B, EES | ||
| 5 (4) | A, E, L, TE, STA, B, EES | ||
| 1 (1) | A, L, TE, STC, STA, B, EES | ||
| 5 (4) | A, E, L, TE, STA, B, EES | ||
| 1 (1) | A, E, L, TE, STC, STA, EES | ||
| 11 (10) | A, E, L, TE, STC, B, EES | ||
| 14 (13) | A, E, L, TE, STC, STA, B, EES |
A, aminoglycosides; TE, tetracycline; B, bacitracin; L, lincosamide; STA, streptogramin A; V, vancomycin; E, erythromycin; STC, streptothricin; EES, erythromycin efflux system.
DISCUSSION
The custom-designed Enteroarray, possessing a taxonomic module in addition to virulence and antibiotic resistance gene modules, was able to accurately identify all isolates to the species level, unlike a standard methodology such as API 20 Strep, which had a 24% misidentification rate in our study.
Identification accuracy with the Enteroarray was high due to the built-in redundancy of using four ubiquitous enterococcal taxonomic probes per species. Validation of the Enteroarray with confirmed ATCC species, and later confirmation by cpn60 sequence analysis of selected isolates showing disagreement between the two methods (Table 2), showed that the probes are highly specific since only one probe for E. hermanniensis showed cross-hybridization with all of the E. hirae isolates (data not shown). Although any one gene would have been sufficient to correctly identify most of the 18 enterococcal species targeted in the present study, the built-in redundancy helped bypass cases where genetic drift would be sufficient to reduce or eliminate hybridization of one of the genes. Indeed, there were 9 isolates among the 171 examined where one of the four genes did not result in a positive signal. Among those nine, only two taxonomic probes resulted in a false-negative result, ddl (three E. gallinarum isolates) and recA (five E. faecalis and one E. faecium isolate). As noted earlier, E. casseliflavus had only three probes designed for its identification.
In the present study, of the five species that API 20 Strep claims to identify, this methodology misidentified 15% of them. This underestimates the real rate (∼24% in this study) since this method does not give a negative or “unknown” response to those enterococcal species not included here. For example, E. casseliflavus and E. hirae isolates were identified as E. faecium or E. durans to a high degree of certainty.
In addition to validating the taxonomic identification section of the Enteroarray using five ATCC Enterococcus species recognized by the API 20 Strep kit, we also picked other isolates from each of the seven species identified in the present study to independently confirm species identification by cpn60 sequencing. Species identification of all sequences was confirmed by BLAST analysis and in all cases was in agreement with the Enteroarray results. It is especially important to note that of five API 20 Strep-identified E. faecium isolates at high levels of certainty, only one was correct, and the other four were misidentified as four separate species (E. casseliflavus, E. faecalis, E. gallinarum, and E. avium). Therefore, we can conclude that the Enterococcus microarray is a more powerful tool for the identification of enterococci to the species level than the API 20 Strep system that relies on biochemical tests. Although the cost of the microarray is higher than the API 20 Strep, the poor accuracy of the API 20 leaves the validity of any conclusions related to species identified by this methodology in doubt. One other publication has used a 16S/23S oligonucleotide DNA microarray to identify a range of enterococcal species similar to our study (31). Successful species discrimination was achieved by hybridizing specific PCR amplicons derived from either whole strains or E. faecium/E. faecalis-spiked milk. By utilizing four different housekeeping genes rather than rRNA gene regions, we were able to increase the number of different specific probes per species to allow for a more robust assay. Moreover, with longer oligonucleotides (50 to 70 versus 18), the Enteroarray has increased target signal intensity. This also permits the addition of a wealth of different gene probes to extract a wide range of genomic information per hybridization with regard to virulence and antibiotic resistance gene content, thus representing a large improvement over PCR-based assays.
Infections caused by E. faecium are becoming more frequent, presumably due to increasing resistance to many antimicrobial agents commonly used in patient treatment regimes. E. faecium is equally responsible, along with E. faecalis, for enterococcus-related nosocomial infections in hospitals through two different mechanisms: antimicrobial agent resistance and virulence (53). According to the literature, no virulence factors have been characterized in enterococcal species other than E. faecalis and E. faecium (44). However, Fontana et al. (16) found cad1 in E. casseliflavus but since this isolate did not display a vanC resistance phenotype, a hallmark of this species, its identity remains unclear. With the Enteroarray, the cad1 gene probe was positive in all E. faecalis, E. hirae, and E. avium isolates, as well as in the ATCC strains used as positive controls for these species. It was not found in E. casseliflavus, E. durans, or E. faecium. Surprisingly, subsequent verification by PCR with primers directed against the E. faecalis gene sequence showed that only E. faecalis isolates produced an amplicon. To validate the genomic target sequence in either E. hirae and E. avium, we performed asymmetric genome walking to amplify the target for sequencing (48). Interestingly, sequence analysis from cad1 primed amplicons from either E. hirae or E. avium produced a similar hypothetical protein. The lack of a positive cad1 probe in E. casseliflavus, E. durans, or E. faecium indicates that this hypothetical protein is not universal, nor is it a variant of cad1, suggesting that our cad1 probe needs to be redesigned.
There is a growing interest for the characterization of enterococci isolated from food since microbial AMR genes present in retail meat or dairies could possibly be transferred to the consumer (17, 23, 39, 43, 47, 49). Some AMR genes have been described in the literature as species characteristic and consequently have been used for their identification (5, 6). For example, Costa et al. (6) indicated that the presence of the aac(6′)-Ii gene, causing resistance to aminoglycosides was specific for E. faecium. With 110 of the 111 E. faecium isolates being positive for this gene, our data clearly support that observation. This gene was not found in any other isolate belonging to six other species. Therefore, the aac(6′)-Ii gene appears to be a good marker to identify E. faecium but only when present, as some E. faecium isolates can be found lacking this gene. This observation has also been observed by Kobayashi et al. (29), who detected aac(6′)-Ii exclusively in E. faecium but not in every isolate. Indirectly, the aac(6′)-Ii probe on our Enteroarray further confirms and supports the accuracy of the four taxonomic probes in properly identifying E. faecium isolates misidentified by the API 20 Strep method. The intrinsic low-level vancomycin resistance observed in E. gallinarum and E. casseliflavus conferred by the vanC1 and vanC3 genes, respectively, has become a species-specific characteristic and is commonly used as a molecular marker for their identification (5, 22, 54). All isolates in our study identified as E. gallinarum and E. casseliflavus by the Enteroarray, including the two ATCC strains, possessed either the vanC1 or the vanC3 gene. None of the isolates distributed among the other five species possessed either of these genes. This is consistent with the literature and also indirectly confirmed the identity of these isolates. The relationship between phenotypic antibiotic resistance and the genotype identified by the Enteroarray was presented elsewhere (8).
In addition to their use as therapeutic agents for animals, antimicrobial agents are also used to accelerate animal growth (15). The increase in the occurrence of antibiotic resistance genes in food is possibly a consequence of the use of antimicrobial agents in agriculture. In the present study, the treatment with antimicrobial agents of the broiler chickens, from which the enterococci were isolated, may have influenced the occurrence and type of AMR genes associated with the different antimicrobial agent classes (8). Bacitracin was used as a growth promoter by all of the farms from which the chicken enterococcal samples were isolated. The bcrR gene, associated with bacitracin resistance, was present in most of the isolates but in none of the ATCC strains, which suggests that it could be related to the bacitracin used as in the study farm's feed supplement. This type of dissemination has also been observed in another study (36).
The presence of virulence factors and antibiotic resistance genes in the present study does not mean that these isolates can lead to difficulties in treating infections but rather that these genes are a good indicator of their potential as human pathogens. Consequently, their presence should not be underestimated. The Enteroarray is a rapid and robust tool for the identification and characterization of Enterococcus isolates to the species level. This microarray can be very useful in a clinical setting where enterococci causing nosocomial infections need to be characterized to direct subsequent patient treatment as well as in food safety and environmental assessment studies. In addition, it can be useful in agricultural settings to measure the impact of different feed or treatment methods on Enterococcus populations.
ACKNOWLEDGMENTS
This study was supported by Agriculture and Agri-Food Canada and the National Research Council of Canada.
We acknowledge the technical assistance of J. Takizawa, the British Columbia Chicken Marketing Board, and participating farmers from the Fraser Valley. We also thank E. Yergeau for help with the statistical analyses.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Angulo F. J., Baker N. L., Olsen S. J., Anderson A., Barrett T. J. 2004. Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans. Semin. Pediatr. Infect. Dis. 15:78–85 [DOI] [PubMed] [Google Scholar]
- 2. Arias C. A., Contreras G. A., Murray B. E. 2010. Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 16:555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bekal S., et al. 2003. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. J. Clin. Microbiol. 41:2113–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruant G., et al. 2006. Development and validation of an oligonucleotide microarray for detection of multiple virulence and antimicrobial resistance genes in Escherichia coli. Appl. Environ. Microbiol. 72:3780–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clark N. C., Teixeira L. M., Facklam R. R., Tenover F. C. 1998. Detection and differentiation of vanC-1, vanC-2, and vanC-3 glycopeptide resistance genes in enterococci. J. Clin. Microbiol. 36:2294–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costa Y., Galimand M., Leclercq R., Duval J., Courvalin P. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37:1896–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Devriese L. A., Pot B., Van Damme L., Kersters K., Haesebrouck F. 1995. Identification of Enterococcus species isolated from foods of animal origin. Int. J. Food Microbiol. 26:187–197 [DOI] [PubMed] [Google Scholar]
- 8. Diarra M. S., et al. 2010. Distribution of antimicrobial resistance and virulence genes in Enterococcus spp. and characterization of isolates from broiler chickens. Appl. Environ. Microbiol. 76:8033–8043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diarra M. S., et al. 2007. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and Enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolates. Appl. Environ. Microbiol. 73:6566–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubois J. W., et al. 2004. The development of a DNA microarray-based assay for the characterization of commercially formulated microbial products. J. Microbiol. Methods 58:251–262 [DOI] [PubMed] [Google Scholar]
- 11. Dutka-Malen S., Evers S., Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dybvig K., et al. 1992. Degenerate oligonucleotide primers for enzymatic amplification of recA sequences from gram-positive bacteria and mycoplasmas. J. Bacteriol. 174:2729–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eaton T. J., Gasson M. J. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67:1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Facklam R. R., Collins M. D. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Falkow S., Kennedy D. 2001. Antibiotics, animals, and people—again! Science 291:397. [DOI] [PubMed] [Google Scholar]
- 16. Fontana C., Gazzola S., Cocconcelli P. S., Vignolo G. 2009. Population structure and safety aspects of Enterococcus strains isolated from artisanal dry fermented sausages produced in Argentina. Lett. Appl. Microbiol. 49:411–414 [DOI] [PubMed] [Google Scholar]
- 17. Fracalanzza S. A., Scheidegger E. M., Santos P. F., Leite P. C., Teixeira L. M. 2007. Antimicrobial resistance profiles of enterococci isolated from poultry meat and pasteurized milk in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 102:853–859 [DOI] [PubMed] [Google Scholar]
- 18. Frye J. G., et al. 2006. DNA microarray detection of antimicrobial resistance genes in diverse bacteria. Int. J. Antimicrob. Agents 27:138–151 [DOI] [PubMed] [Google Scholar]
- 19. Frye J. G., et al. Development of a DNA microarray to detect antimicrobial resistance genes identified in the National Center for Biotechnology Information database. Microb. Drug Resist. 16:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garneau P., et al. 2010. Diagnostic microarray for antimicrobial resistance bacterial gene (ABG) identification. Zoonoses Public Health 57:94–99 [DOI] [PubMed] [Google Scholar]
- 21. Giridhara Upadhyaya P., Ravikumar K., Umapathy B. 2009. Review of virulence factors of Enterococcus: an emerging nosocomial pathogen. Indian J. Med. Microbiol. 27:301–305 [DOI] [PubMed] [Google Scholar]
- 22. Gomes B. C., et al. 2007. Correlation between API 20 STREP and multiplex PCR for identification of Enterococcus spp. isolated from Brazilian foods. Braz. J. Microbiol. 38:617–619 [Google Scholar]
- 23. Hayes J. R., et al. 2003. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 69:7153–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jett B. D., Huycke M. M., Gilmore M. S. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kayaoglu G., Orstavik D. 2004. Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 15:308–320 [DOI] [PubMed] [Google Scholar]
- 26. Ke D., et al. 1999. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 37:3497–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S. Y., et al. 2010. Characterization of Enterococcus spp. from human and animal feces using 16S rRNA sequences, the esp gene, and PFGE for microbial source tracking in Korea. Environ. Sci. Technol. 44:3423–3428 [DOI] [PubMed] [Google Scholar]
- 28. Knijff E., Dellaglio F., Lombardi A., Andrighetto C., Torriani S. 2001. Rapid identification of Enterococcus durans and Enterococcus hirae by PCR with primers targeted to the ddl genes. J. Microbiol. Methods 47:35–40 [DOI] [PubMed] [Google Scholar]
- 29. Kobayashi N., et al. 2001. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 126:197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leclercq R., Derlot E., Duval J., Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161 [DOI] [PubMed] [Google Scholar]
- 31. Lehner A., et al. 2005. Oligonucleotide microarray for identification of Enterococcus species. FEMS Microbiol. Lett. 246:133–142 [DOI] [PubMed] [Google Scholar]
- 32. Letowski J., Brousseau R., Masson L. 2004. Designing better probes: effect of probe size, mismatch position and number on hybridization in DNA oligonucleotide microarrays. J. Microbiol. Methods 57:269–278 [DOI] [PubMed] [Google Scholar]
- 33. Martin B., Garriga M., Aymerich T. 2008. Identification of Enterococcus species by melting curve analysis of restriction fragments. J. Microbiol. Methods 75:145–147 [DOI] [PubMed] [Google Scholar]
- 34. Masson L., et al. 2006. Identification of pathogenic Helicobacter species by chaperonin-60 differentiation on plastic DNA arrays. Genomics 87:104–112 [DOI] [PubMed] [Google Scholar]
- 35. Mastroianni A. 2009. Enterococcus raffinosus endocarditis. First case and literature review. Infez. Med. 17:14–20 [PubMed] [Google Scholar]
- 36. Matos R., Pinto V. V., Ruivo M., Lopes Mde F. 2009. Study on the dissemination of the bcrABDR cluster in Enterococcus spp. reveals that the BcrAB transporter is sufficient to confer high-level bacitracin resistance. Int. J. Antimicrob. Agents 34:142–147 [DOI] [PubMed] [Google Scholar]
- 37. Maynard C., et al. 2003. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 47:3214–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McBride S. M., et al. 2009. Genetic variation and evolution of the pathogenicity island of Enterococcus faecalis. J. Bacteriol. 191:3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGowan-Spicer L. L., et al. 2008. Antimicrobial resistance and virulence of Enterococcus faecalis isolated from retail food. J. Food Prot. 71:760–769 [DOI] [PubMed] [Google Scholar]
- 40. Naser S. M., et al. 2005. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151:2141–2150 [DOI] [PubMed] [Google Scholar]
- 41. Naser S. M., et al. 2005. Enterococcus canintestini sp. nov., from faecal samples of healthy dogs. Int. J. Syst. Evol. Microbiol. 55:2177–2182 [DOI] [PubMed] [Google Scholar]
- 42. Navarro F., Courvalin P. 1994. Analysis of genes encoding d-alanine-d-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob. Agents Chemother. 38:1788–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Novais C., et al. 2005. High occurrence and persistence of antibiotic-resistant enterococci in poultry food samples in Portugal. J. Antimicrob. Chemother. 56:1139–1143 [DOI] [PubMed] [Google Scholar]
- 44. Ogier J. C., Serror P. 2008. Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126:291–301 [DOI] [PubMed] [Google Scholar]
- 45. Pangallo D., et al. 2008. Evaluation of different PCR-based approaches for the identification and typing of environmental enterococci. Antonie Van Leeuwenhoek 93:193–203 [DOI] [PubMed] [Google Scholar]
- 46. Paulsen I. T., et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 [DOI] [PubMed] [Google Scholar]
- 47. Peters J., Mac K., Wichmann-Schauer H., Klein G., Ellerbroek L. 2003. Species distribution and antibiotic resistance patterns of enterococci isolated from food of animal origin in Germany. Int. J. Food Microbiol. 88:311–314 [DOI] [PubMed] [Google Scholar]
- 48. Pilhofer M., et al. 2007. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res. 35:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Presi P., et al. 2009. Risk scoring for setting priorities in a monitoring of antimicrobial resistance in meat and meat products. Int. J. Food Microbiol. 130:94–100 [DOI] [PubMed] [Google Scholar]
- 50. Rodriguez H., de Las Rivas B., Munoz R. 2007. Efficacy of recA gene sequence analysis in the identification and discrimination of Lactobacillus hilgardii strains isolated from stuck wine fermentations. Int. J. Food Microbiol. 115:70–78 [DOI] [PubMed] [Google Scholar]
- 51. Shea K. M. 2003. Antibiotic resistance: what is the impact of agricultural uses of antibiotics on children's health? Pediatrics 112:253–258 [PubMed] [Google Scholar]
- 52. Sood S., Malhotra M., Das B. K., Kapil A. 2008. Enterococcal infections and antimicrobial resistance. Indian J. Med. Res. 128:111–121 [PubMed] [Google Scholar]
- 53. van Schaik W., Willems R. J. 2010. Genome-based insights into the evolution of enterococci. Clin. Microbiol. Infect. 16:527–532 [DOI] [PubMed] [Google Scholar]
- 54. Velasco D., et al. 2004. Lack of correlation between phenotypic techniques and PCR-based genotypic methods for identification of Enterococcus spp. Diagn. Microbiol. Infect. Dis. 49:151–156 [DOI] [PubMed] [Google Scholar]
- 55. Vincent S., Knight R. G., Green M., Sahm D. F., Shlaes D. M. 1991. Vancomycin susceptibility and identification of motile enterococci. J. Clin. Microbiol. 29:2335–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X., Seed B. 2003. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics 19:796–802 [DOI] [PubMed] [Google Scholar]
- 57. Winston L. G., et al. 2004. API 20 Strep identification system may incorrectly speciate enterococci with low level resistance to vancomycin. Diagn. Microbiol. Infect. Dis. 48:287–288 [DOI] [PubMed] [Google Scholar]
- 58. Yoo S. M., et al. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol. Cell Probes 24:44–52 [DOI] [PubMed] [Google Scholar]
- 59. Zehnder M., Guggenheim B. 2009. The mysterious appearance of enterococci in filled root canals. Int. Endod. J. 42:277–287 [DOI] [PubMed] [Google Scholar]