Abstract
A set of 31 undecapeptides, incorporating 1 to 11 d-amino acids and derived from the antimicrobial peptide BP100 (KKLFKKILKYL-NH2), was designed and synthesized. This set was evaluated for inhibition of growth of the plant-pathogenic bacteria Erwinia amylovora, Pseudomonas syringae pv. syringae, and Xanthomonas axonopodis pv. vesicatoria, hemolysis, and protease degradation. Two derivatives were as active as BP100, and 10 peptides displayed improved activity, with the all-d isomer being the most active. Twenty-six peptides were less hemolytic than BP100, and all peptides were more stable against protease degradation. Plant extracts inhibited the activity of BP100 as well as that of the d-isomers. Ten derivatives incorporating one d-amino acid each were tested in an infectivity inhibition assay with the three plant-pathogenic bacteria by using detached pear and pepper leaves and pear fruits. All 10 peptides studied were active against E. amylovora, 6 displayed activity against P. syringae pv. syringae, and 2 displayed activity against X. axonopodis pv. vesicatoria. Peptides BP143 (KKLFKKILKYL-NH2) and BP145 (KKLFKKILKYL-NH2), containing one d-amino acid at positions 4 and 2 (underlined), respectively, were evaluated in whole-plant assays for the control of bacterial blight of pepper and pear and fire blight of pear. Peptide BP143 was as effective as streptomycin in the three pathosystems, was more effective than BP100 against bacterial blight of pepper and pear, and equally effective against fire blight of pear.
INTRODUCTION
Bacterial plant diseases are responsible for significant losses in agriculture and in natural resources, and their control is achieved mainly by treatments with copper compounds and antibiotics (1). Growing awareness of the negative effects caused by these bactericides in the environment and to consumer health as well as the development of resistance by plant-pathogenic bacteria (29, 33, 45, 46) have raised the need to study safer alternatives. In recent years, antimicrobial peptides, both endogenously produced by living organisms and de novo designed, have emerged as promising candidates (22, 27, 31, 35, 36).
Antimicrobial peptides exhibit a broad spectrum of activity mainly against bacteria and fungi but also against viruses, parasites, and tumor cells (2, 13, 15, 17, 26, 40, 53). Most of them are cationic and have the ability to adopt an amphipathic conformation, properties that govern their antibacterial activity. Their mechanism of action results from an electrostatic interaction with the negatively charged cytoplasmic membrane of bacteria. When a critical concentration is reached, peptides are inserted into the membrane, disturbing the bilayer integrity by either disruption or pore formation (9, 10, 12, 14, 22, 25, 30, 39, 50). This unique mode of action makes the selection of resistance in target pathogens difficult, because it requires dramatic changes in the cell membrane, mainly in phospholipid composition and/or organization (14, 43, 51). Apart from the classical mechanism of action based on membrane disruption, it has been described that certain antimicrobial peptides are immunomodulators in animals (49) or induce defense reactions in plants (16).
Antimicrobial peptides have been described to be effective against pathogen infections in plants, including postharvest products (27, 31, 35, 36). During our current research oriented to the development of new antimicrobial agents for use in plant protection, we designed linear undecapeptides (CECMEL11) using a combinatorial approach (5–7). The antimicrobial evaluation of the CECMEL11 library led to the identification of peptides with high activity against plant pathogenic bacteria, fungi, or both types of pathogens, including Erwinia amylovora, Xanthomonas axonopodis pv. vesicatoria, Pseudomonas syringae pv. syringae, and Penicillium expansum. The best bactericidal peptide, KKLFKKILKYL-NH2 (BP100), was effective in vivo to prevent infections of E. amylovora in detached apple and pear flowers. However, peptide concentrations required in vivo for an effective control of infections were 10 to 50 times higher than the corresponding in vitro MICs (2.5 to 7.5 μM) (5). This loss of activity could be attributed to the reaction or interaction of peptides with nontarget plant structures or compounds or to their enzymatic degradation by proteases from epiphytic microorganisms or by those intrinsic to the plant tissues (4, 19).
Peptide stability against protease hydrolysis can be increased by the development of synthetic analogues with similar structural features but containing nonproteinogenous amino acids. In particular, incorporation of d-amino acids is an approach used to protect peptides against enzymatic hydrolysis, since only a few enzymes are known to digest amide bonds involving d-configuration (32). This strategy has been used to improve the biological activity profiles of synthetic antimicrobial peptides, not only increasing the resistance to proteolytic enzymes but also reducing the hemolytic activity while maintaining the antimicrobial activity (23, 28, 42, 44, 47, 48, 54).
In the present study, we designed a set of BP100 analogues, incorporating d-amino acids, to find candidates with better biological profiles in planta against E. amylovora, X. axonopodis pv. vesicatoria, and P. syringae pv. syringae, as well as with low hemolytic activity and protease susceptibility.
MATERIALS AND METHODS
Peptide synthesis.
All peptides were synthesized manually by the solid-phase method using 9-fluorenylmethoxy carbonyl (Fmoc)-type chemistry, tert-butyloxycarbonyl side chain protection for Lys, and tert-butyl (tBu) for Tyr. Fmoc-Rink-4-methylbenzhydrylamine (MBHA) resin (0.64 mmol/g) was used as the solid support to obtain C-terminal peptide amides. Couplings of the Fmoc-amino acids (4 eq) were mediated by N-[1H-benzotriazol-1-yl) (dimethylamino)methylene]-N-methylmethanaminium hexafluorophosphate N-oxide (HBTU) (3.8 eq), 1-hydroxybenzotriazole (HOBt) (4 eq) and N,N-diisopropylethylamine (DIEA) (7.8 eq) in N,N-dimethylformamide (DMF) for 1 h and monitored by the ninhydrin test. The Fmoc group was removed by treating the resin with a mixture of piperidine-DMF (3:7 dilution ratio; two treatments of 2 and 10 min). Peptides were individually cleaved from the resin with trifluoroacetic acid (TFA)-H2O-triisopropylsilane (95:2.5:2.5; 2 h). Following TFA evaporation and diethyl ether extraction, the crude peptides were dissolved in H2O, lyophilized, and analyzed by high-performance liquid chromatography (HPLC; Dionex) performed at 1.0 ml/min using a Kromasil C18 reverse-phase column (4.6 by 40 mm; 3.5 μM particle size). Linear gradients of 0.1% aqueous TFA and 0.1% TFA in CH3CN were run from dilutions of 0.98:0.02 to 0:1 over 7 min, with UV detection at 220 nm. Peptides were obtained with 42 to 92% purity. Electrospray ionization mass spectrometry (ESI-MS; Bruker Daltonics) and matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF; Bruker) analysis were used to confirm peptide identity. Peptides were purified by a preparative HPLC equipped with an Ultrabase C18 ODS2 column (7.8 by 250 mm; 5 μm particle size). Different linear gradients of 0.1% aqueous TFA and 0.1% TFA in CH3CN were run at 3 ml/min. After the purification, peptides showed >95% HPLC purity.
Peptide BP163 was also prepared on a ChemMatrix aminomethyl resin (0.66 mmol/g). Until the ninth residue, couplings and Fmoc removal were carried out as described above. The deprotection of the ninth residue was performed by treatment with piperidine-DMF (3:7 dilution; once for 2 min and 3 times for 10 min). Fmoc removal of the tenth and eleventh residues required a treatment performed once for 2 min and 6 times for 10 min. For the coupling of the tenth and eleventh residues, two and three treatments for 24 h were necessary, respectively. This protocol led to BP163 at 86% HPLC purity, rather than the 48% purity obtained using the MBHA resin.
Bacterial strains and growth conditions.
For the analysis of the in vitro activity of peptides, the following plant-pathogenic bacterial strains were used: E. amylovora PMV6076 (8), P. syringae pv. syringae EPS94 (37), and X. axonopodis pv. vesicatoria 2133-2 (20). For the infection assay, E. amylovora EPS101 (18) was used instead of strain PMV6076, a nonpathogenic mutant. Also, P. syringae pv. syringae EPS31 (Institut de Tecnologia Agroalimentària, Universitat de Girona, Spain) and X. axonopodis pv. vesicatoria 1779 (52) were used in the infection experiments due to their higher virulence than the strains used in vitro. All bacteria were stored in Luria-Bertani (LB) broth supplemented with glycerol (20%) and maintained at −80°C. All strains were scraped from LB agar plates incubated at 25°C after 24 h of growth, in the case of E. amylovora and P. syringae pv. syringae, or after 48 h of growth, in the case of X. axonopodis pv. vesicatoria. The cell material was suspended in sterile water to obtain a suspension of 108 CFU ml−1. This suspension was 10-fold serially diluted in sterile distilled water to obtain suitable concentrations when necessary.
Antibacterial activity.
Lyophilized peptides were solubilized in sterile distilled water to a concentration of 1,000 μM and filter sterilized through a 0.22-μm-pore-size filter. For MIC assessment, dilutions of the synthetic peptides were made to obtain final concentrations of 75, 50, 25, 12.5, and 6.25 μM. Twenty microliters of each dilution was mixed in a microtiter plate well with 20 μl of the corresponding suspension of the bacterial indicator at 108 CFU ml−1 and with 160 μl of Trypticase soy broth (TSB; bioMérieux, France) to a total volume of 200 μl. The final peptide concentrations assayed were 7.5, 5.0, 2.5, 1.25, and 0.62 μM. Three replicates for each strain, peptide, and concentration were used. Positive controls contained water instead of peptide, and negative controls contained peptide without bacterial suspension. Microbial growth was automatically determined by optical density measurement at 600 nm (Bioscreen C; Labsystems, Finland). Microplates were incubated at 25°C with 20 s of shaking before hourly absorbance measurement for 48 h. The experiment was repeated twice. The MIC was taken as the lowest peptide concentration with no growth at the end of the experiment.
Hemolytic activity.
The hemolytic activities of the peptides were evaluated by determining hemoglobin release from erythrocyte suspensions of fresh human blood (5%, vol/vol). Blood was aseptically collected using a BD Vacutainer K2E system with EDTA (Belliver Industrial State, Plymouth, United Kingdom) and stored for less than 2 h at 4°C. Blood was centrifuged at 6,000 × g for 5 min, washed three times with Tris buffer (10 mM Tris, 150 mM NaCl, pH 7.2), and diluted 10-fold in the same buffer. Peptides were solubilized in Tris buffer to a concentration of 500 μM. A total of 65 μl of human red blood cells was mixed with 65 μl of the peptide solution (final concentration of 250 μM) in a 96-well reaction plate and incubated under continuous shaking for 1 h at 37°C. Then, the plates were centrifuged at 3,500 × g for 10 min. Eighty-microliter aliquots of the supernatant were transferred to 100-well microplates and diluted with 80 μl of sterile distilled water. Three replicates for each peptide were used. Hemolysis was measured with a microplate reader as the absorbance at 540 nm. Complete hemolysis was determined in Tris buffer plus melittin at 200 μM (Sigma-Aldrich Corporation, Spain) as a positive control. The percentage of hemolysis (H) was calculated using the equation H = 100 × [(Op − Ob)/(Om − Ob)], where Op is the density for a given peptide concentration, Ob is that for the buffer, and Om is that for the melittin-positive control.
Susceptibility to protease degradation.
Digestion of the peptides was carried out by treating 50 μg/ml peptide with 1 μg/ml proteinase K (Sigma-Aldrich Corporation, Spain) in 100 mM Tris buffer, pH 7.6, at 25°C. The peptide cleavage after 5, 10, 15, 30, 45, and 60 min was monitored by HPLC or MALDI-TOF. Digestion of peptides BP138 to BP155 and BP157 to BP161 was monitored by HPLC using a Kromasil C18 reverse-phase column (4.6 by 40 mm; 3.5 μm particle size). Linear gradients of 0.1% aqueous TFA and 0.1% TFA in CH3CN were run from dilutions of 0.98:0.02 to 0:1 over 7 min, with UV detection at 220 nm. Digestion was estimated as the percentage of degraded peptide calculated from the decrease of the HPLC peak area of the native peptide. Digestion of peptides BP156 and BP162 to BP168 was monitored by freezing the samples after each reaction time, lyophilization, and analysis by MALDI-TOF.
Effect of plant extracts on the activity of peptides.
The activity levels of peptides BP100, BP143, and BP157 were studied in plant extracts prepared from young leaves of potted pear plants (Pyrus communis cv. Conference). Leaves were ground to a powder in liquid nitrogen, and the resulting material was extracted with chilled 100 mM Tris buffer, pH 7.6 (1 mg of material per 10 μl of buffer), at 4°C for 45 min with continuous stirring. The extract was diluted 5-fold in the same buffer, filtered through several layers of sterile cheesecloth, and sterilized by filtration through a 0.22-μm-pore-size filter. Then, the resulting extract was further diluted with buffer to obtain a final concentration of 25% or 50%. A total of 800 μl of the appropriate plant extract dilution was mixed with 100 μl of a 200 μM solution of the corresponding peptide and incubated for 30 min at 25°C. Controls containing buffer, leaf extract alone, or buffer with the corresponding peptide were used. After incubation, 100 μl of a bacterial suspension (107 CFU ml−1) of the same pathogen strains used for the in vitro assay was added. The number of surviving cells were determined after an additional incubation time at 25°C for 90 min (25% leaf extract) or 120 min (50% leaf extract). Then, samples were taken and spread onto LB agar plates for E. amylovora and P. syringae pv. syringae and onto King's B (KB) agar plates for X. axonopodis pv. vesicatoria using a spiral plater (Eddy Jet; IUL Instruments, Spain). Triplicates of each assay condition were performed. Colonies were counted after 48 h of incubation at 25°C with an automatic counter (Flash and Go; IUL Instruments, Spain) by incorporating the software CounterMat version 5.0.
Ex vivo assays.
The efficacy of the peptides was determined using infection assays in detached plant organs. Immature pear fruits (Pyrus communis cv. Passe Crassane) were used for E. amylovora, detached pear leaves (P. communis cv. Conference) for P. syringae pv. syringae, and detached pepper leaves (Capsicum annuum cv. Dulce Italiano) for X. axonopodis pv. vesicatoria. Immature pear fruits obtained from commercial orchards were collected in early June at the 6-weeks stage from the fruit set and kept in the dark at 0 to 4°C. Fruits were used before 1 month of storage to avoid significant physiological changes which could affect the assay. Before inoculation, fruits were surface disinfected by immersion for 1 min in a diluted solution of sodium hypochlorite (1% active chlorine), washed twice in distilled water, and left under airflow in a sterile cabinet to remove excess water. Each fruit was wounded four times in opposite sides with a cork borer (approximately 2 mm diameter and 5 mm in depth). Fruits were placed in polystyrene tray packs and put into boxes. The leaves of pears or peppers were obtained from potted plants cultivated in the greenhouse. The youngest leaves were selected and surface disinfected by following the same procedure described above for immature fruits. Leaves were wounded approximately in the middle by a double transverse incision (∼1 mm) to the midrib and placed in plastic boxes over a humidified paper towel. Then, 10 μl of a 200 μM solution of the corresponding peptide was delivered onto the wounds of leaves or immature fruits, and the treated materials were left at room temperature for 1 h. The treated materials were inoculated with 10 μl of a suspension of E. amylovora EPS101 (107 CFU ml−1), P. syringae. pv. syringae EPS31 (107 CFU ml−1), or X. axonopodis pv. vesicatoria 1779 (108 CFU ml−1). The inoculated plant material was incubated at 23°C with high relative humidity for 5 days. The experimental design consisted of three replicates of eight immature fruits or nine leaves per treatment. A nontreated control inoculated only with the corresponding pathogen and a control treated with 100 mg/liter of streptomycin were included. Controls for the phytotoxicity assessment consisted of uninoculated immature fruits or leaves, treated only with the peptides. Infection severity levels were determined for each fruit using the following scale of severity from 0 to 3, according to the symptoms observed: 0, no symptoms; 1, local necrosis around the wound or the presence of exudates; 2, necrosis progression around the wound, with a diameter between 3 to 5 mm; and 3, necrosis progression around the wound, with a diameter higher than 5 mm. Infection severity levels were determined for each leaf using the following scale of severity from 0 to 3, according to the symptoms observed: 0, no symptoms; 1, leaf necrosis localized around the wound; 2, necrosis progression far from the wound; and 3, necrosis of the whole leaf. The mean severity for each replicate was calculated. Two independent experiments were done.
In planta assays.
The efficacy of the peptides was determined using whole-plant infection assays with pear plants (P. communis cv. Conference) inoculated with E. amylovora or P. syringae pv. syringae and with pepper plants (C. annuum cv. Dulce Italiano) inoculated with X. axonopodis pv. vesicatoria. Two-to-three-year-old pear plants grown in 20-cm-diameter plastic pots in the greenhouse were used. Plants were pruned to leave 3 to 4 shoots per plant and were forced to bud in the greenhouse. Plants were fertilized once a week with a 200-ppm N-P-K solution (20:10:20) and were used when the shoots were about 3 to 4 cm in length and had 5 to 6 young leaves per shoot. Pepper plants were obtained from seeds grown in 10-cm-diameter plastic pots in the greenhouse, fertilized as described for pear plants, and used when they achieved 25 to 30 cm in height. Standard insecticide and miticide sprays were applied to pear and pepper plants. Fungicides or bactericides were not applied to prevent interference with the assays. For the assays, the three youngest expanded leaves of each shoot were wounded approximately in the middle of the leaf by a double transverse incision (∼1 mm) to the midrib. Then, 10 μl of a 200 μM solution of the corresponding peptide was delivered onto the wounds, and the plants were left at room temperature for 1 h. Each leaf wound of the treated plants was inoculated with 10 μl of a suspension of the corresponding bacteria, as described for the ex vivo assays. Plants were incubated in a controlled environment greenhouse for 9 days for pear plants and 12 days for pepper plants at 23 ± 2°C during the day and 15 ± 2°C at night. The photoperiod used was 16 h of light and 8 h of dark. The experimental design consisted of three replicates of three plants per treatment. Appropriate nontreated, noninoculated, and reference controls were used as described for the ex vivo assays. Activity was assessed according to the incidence of infection, i.e., appearance of necrosis at the inoculation point. The mean incidence was calculated for each replicate. Two independent experiments were performed.
Statistical analysis.
The effect of the peptides on plant material infection was determined using analysis of variance (ANOVA) with the general linear model (GLM) procedure of the software Statistical Analysis System (SAS) (version 8.2; SAS Institute Inc., NC). The means were separated using Tukey's test (P < 0.05).
RESULTS
Design and synthesis of peptides.
Peptides were designed and synthesized in order to study the influence of selected single or multiple substitutions of l-amino acids by the corresponding d-isomers on the biological activity of BP100 (KKLFKKILKYL-NH2) (Table 1). A total of 31 sequences were prepared containing the following: (i) one d-amino acid (BP138 to BP148); (ii) two or three d-amino acids at consecutive and nonconsecutive positions (BP149 to BP152, BP154); (iii) 4 to 10 d-amino acids at the C terminus (BP155 to BP161); (iv) 4 to 10 d-amino acids at the N terminus (BP162 to BP168); and (v) all d-amino acids (BP153). The synthesis followed a standard Fmoc/tBu protocol, and peptides were obtained in >95% purity after purification by preparative HPLC. Their structure was confirmed by mass spectrometry (data not shown).
Table 1.
Antibacterial activity (MIC) against three plant pathogenic bacteria, cytotoxicity, and stability against protease degradation of linear undecapeptides with d-amino acids
| Peptide | Sequencea | MIC (μM)c |
Hemolysis (%)b | Digestion (%)d | ||
|---|---|---|---|---|---|---|
| Xav | Pss | Ea | ||||
| BP100 | KKLFKKILKYL | 5–7.5 | 2.5–5 | 2.5–5 | 54 ± 0.1 | 75 |
| BP138 | KKLFKKILKYL | >7.5 | >7.5 | >7.5 | 7 ± 0.1 | 53 |
| BP139 | KKLFKKILKYL | >7.5 | >7.5 | >7.5 | 23 ± 0.9 | 6 |
| BP140 | KKLFKKILKYL | >7.5 | 5–7.5 | >7.5 | 0 ± 0 | 1 |
| BP141 | KKLFKKILKYL | >7.5 | 2.5–5 | 2.5–5 | 4 ± 0.3 | 1 |
| BP142 | KKLFKKILKYL | >7.5 | 2.5–5 | 5–7.5 | 3 ± 0.6 | 35 |
| BP143 | KKLFKKILKYL | 5–7.5 | 2.5–5 | 2.5–5 | 5 ± 0.8 | 18 |
| BP144 | KKLFKKILKYL | >7.5 | 2.5–5 | >7.5 | 7 ± 0.4 | 50 |
| BP145 | KKLFKKILKYL | 5–7.5 | 2.5–5 | 2.5–5 | 51 ± 0.5 | 61 |
| BP146 | KKLFKKILKYL | >7.5 | 2.5–5 | >7.5 | 53 ± 1.1 | 24 |
| BP147 | KKLFKKILKYL | 2.5–5 | 2.5–5 | 2.5–5 | 71 ± 0.4 | 47 |
| BP148 | KKLFKKILKYL | >7.5 | >7.5 | >7.5 | 0 ± 0 | 0 |
| BP149 | KKLFKKILKYL | >7.5 | >7.5 | >7.5 | 0 ± 0 | 6 |
| BP150 | KKLFKKILKYL | 2.5–5 | >7.5 | >7.5 | 1 ± 0.1 | 0 |
| BP151 | KKLFKKILKYL | 5–7.5 | 5–7.5 | >7.5 | 1 ± 0.3 | 0 |
| BP152 | KKLFKKILKYL | 5–7.5 | >7.5 | >7.5 | 5 ± 1.6 | 1 |
| BP154 | KKLFKKILKYL | 5–7.5 | 2.5–5 | 5–7.5 | 41 ± 6.2 | 37 |
| BP155 | KKLFKKILKYL | >7.5 | >7.5 | >7.5 | 3 ± 0.1 | 6 |
| BP156 | KKLFKKILKYL | 5–7.5 | 2.5–5 | >7.5 | 49 ± 1.2 | —e |
| BP157 | KKLFKKILKYL | 1.25–2.5 | 2.5–5 | 1.25–2.5 | 3 ± 1.0 | 0 |
| BP158 | KKLFKKILKYL | 2.5–5 | 1.25–2.5 | 2.5–5 | 28 ± 2.1 | 0 |
| BP159 | KKLFKKILKYL | 2.5–5 | 1.25–2.5 | 2.5–5 | 58 ± 5.7 | 12 |
| BP160 | KKLFKKILKYL | 1.25–2.5 | 1.25–2.5 | 2.5–5 | 65 ± 1.9 | 0 |
| BP161 | KKLFKKILKYL | 1.25–2.5 | 1.25–2.5 | 1.25–2.5 | 66 ± 2.9 | 1 |
| BP162 | KKLFKKILKYL | 2.5–5 | 1.25–2.5 | 1.25–2.5 | 17 ± 0.7 | — |
| BP163 | KKLFKKILKYL | 5–7.5 | >7.5 | 2.5–5 | 8 ± 5.0 | — |
| BP164 | KKLFKKILKYL | >7.5 | 2.5–5 | >7.5 | 0 ± 0 | — |
| BP165 | KKLFKKILKYL | >7.5 | 1.25–2.5 | >7.5 | 6 ± 0.2 | — |
| BP166 | KKLFKKILKYL | >7.5 | 2.5–5 | >7.5 | 1 ± 0.2 | — |
| BP167 | KKLFKKILKYL | 2.5–5 | 2.5–5 | 2.5–5 | 21 ± 7.4 | — |
| BP168 | KKLFKKILKYL | 5–7.5 | 1.25–2.5 | 2.5–5 | 3 ± 2.7 | — |
| BP153 | KKLFKKILKYL | 0.62–1.25 | 1.25–2.5 | 1.25–2.5 | 50 ± 2.1 | 5 |
Underlined amino acids are d-enantiomers. All peptides are C-terminal amides.
Percent hemolysis at 250 μM plus confidence interval (α = 0.05).
Xav, Xanthomonas axonopodis pv. vesicatoria; Pss, Pseudomonas syringae pv. syringae; Ea, Erwinia amylovora.
Percentage of degraded peptide, as calculated by HPLC.
—, degradation was monitored by MALDI-TOF analysis, and the peptide was detected after 1 h.
Antibacterial activity.
Peptides were tested for in vitro growth inhibition of E. amylovora, P. syringae pv. syringae, and X. axonopodis pv. vesicatoria at 0.62, 1.25, 2.5, 5.0, and 7.5 μM and compared to that of BP100 (Table 1). Two derivatives were as active as BP100, and 10 peptides displayed improved activity, with the all-d isomer being the most active. Among the 31 isomers, the activity levels were increased or maintained for 18 peptides against X. axonopodis pv. vesicatoria, for 14 peptides against E. amylovora, and for 21 peptides against P. syringae pv. syringae.
Single d-amino acid replacement had a pronounced effect on the peptide activity against X. axonopodis pv. vesicatoria. Most of the peptides were less active than BP100, and only analogs BP143 (d-F4), BP145 (d-K2), and BP147 (d-K1) displayed MIC values below 7.5 μM. A decrease in activity against E. amylovora was also observed, with only the above-mentioned peptides together with BP141 (d-L8) being as active as BP100 (MIC of 2.5 to 5.0 μM). In contrast, in the case of P. syringae pv. syringae, the following seven sequences retained the same activity as that of BP100 (MIC of 2.5 to 5.0 μM): BP141 (d-L8), BP142 (d-K5), BP143 (d-F4), BP144 (d-L3), BP145 (d-K2), BP146 (d-K6), and BP147 (d-K1). Therefore, peptides BP143 (d-F4), BP145 (d-K2), and BP147 (d-K1) were the most active against the three bacteria, with MIC values ranging from 2.5 to 7.5 μM.
Double or triple d-amino acid replacement led to peptides BP149 (d-L3, d-K9), BP150 (d-L8, d-K9), BP151 (d-L3, d-K6, d-K9), and BP152 (d-I7, d-L8, d-K9), which were less active than BP100, with the exception of BP150 against X. axonopodis pv. vesicatoria (MIC of 2.5 to 5.0 μM). In contrast, the analogue BP154 (d-K1, d-K2, d-F4) had activity similar to that of BP100 (MIC of 2.5 to 7.5 μM).
For peptides incorporating 4 to 10 d-amino acids at the C terminus (BP155 to BP161), activity increased with the number of d-residues. Peptides containing four (BP155) and five (BP156) d-amino acids were less active, whereas the incorporation of six to nine d-amino acids resulted in peptides BP157 to BP160, displaying higher antibacterial activities than BP100 against two pathogens (MIC of 1.25 to 5.0 μM). Accordingly, BP161, with 10 d-amino acids, and the all-d isomer BP153 were more active than the parent peptide against the three bacteria (MIC of 0.62 to 2.5 μM). When 4 to 10 d-amino acids were incorporated at the N terminus (BP162 to BP168), a decrease of activity was generally observed. However, three sequences (BP162, BP167, and BP168) were more active than BP100, with MIC values ranging from 1.25 to 7.5 μM.
Hemolytic activity.
Since the mechanism of action of AMPs consists of cell membrane disruption, toxicity to animal or plant cells (cytotoxicity) may be a problem. This cytotoxicity can be assessed with animal or plant cell model systems, with erythrocytes being more frequently used due to the easier comparison with cells used in existing reports (5–7, 34). Thus, the toxicity of the peptides to eukaryotic cells was determined as the ability to lyse erythrocytes in comparison to the toxicity of melittin (Table 1). A total of 18 of 31 peptides displayed less than 10% hemolysis at 250 μM. The analysis of the influence of a single d-amino acid replacement pointed out that peptides BP138 (d-Y10), BP140 (d-K9), BP141 (d-L8), BP142 (d-K5), BP143 (d-F4), BP144 (d-L3), and BP148 (d-I7) were the least hemolytic (≤7%). Increased hemolysis was observed for peptides BP139 (d-L11), B145 (d-K2), and BP146 (d-K6), ranging from 23 to 53%, and BP147 (d-K1) was the most hemolytic (71%).
Double or triple d-amino acid replacement resulted in analogs BP149 to BP152, which exhibited low hemolysis (≤5%), while BP154 displayed higher cytotoxicity (41%). Peptides incorporating 4 to 10 d-amino acids at the N terminus were less hemolytic (0 to 21%) than those containing d-isomers at the C terminus. The latter showed significant hemolysis (28 to 66%), except for BP155 and BP157 (3%). In addition, the all-d peptide BP153 was as hemolytic as BP100.
Susceptibility to protease degradation.
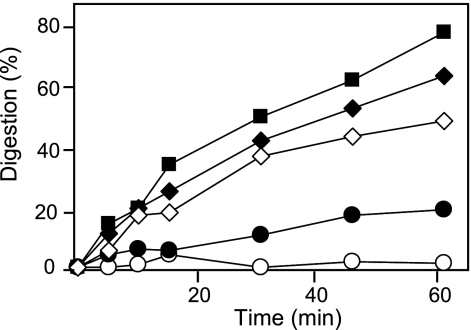
The susceptibility of peptides BP138 to BP155 and BP157 to BP161 to proteolysis was studied by exposure to proteinase K, and degradation was monitored by reverse-phase HPLC over time (Table 1). As expected, all peptides were more stable than BP100 (Fig. 1). Sixteen sequences displayed degradation below 20% after 1 h of incubation. The least stable peptides were BP138 (d-Y10), BP142 (d-K5), BP144 (d-L3), BP145 (d-K2), BP146 (d-K6), BP147 (d-K1), and BP154 (d-K1, d-K2, d-F4), with 24 to 61% degradation. The degradation of peptides BP156 and BP162 to BP168 could not be monitored by HPLC and was analyzed by MALDI-TOF analysis. They were detected after 1 h of incubation with protease, which is indicative that these peptides were not completely degraded.
Fig. 1.
Digestion kinetics of peptides BP100 (▪), BP141 (○), BP143 (•), BP145 (♦), and BP147 (⋄), as shown by results with proteinase K.
Activity of peptides in the presence of plant extracts.
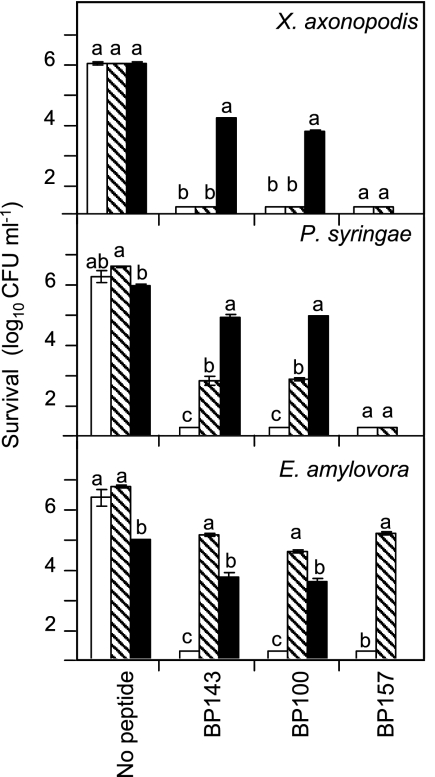
The effects of pear leaf extracts on the antibacterial activities of peptide BP100 and the two d-diastereoisomers BP143 and BP157 were studied (Fig. 2). These two peptides were chosen for a more detailed assay because they are highly antibacterial in vitro and have low hemolytic activity and susceptibility to protease degradation (Table 1) but differ in their numbers of d-amino acid substitutions (one in BP143 and six in BP157). A peptide assay was performed in buffer or in pear leaf extracts diluted at 25% or at 50%. In the presence of buffer, the three peptides were highly inhibitory. In the absence of peptide, the survival of the three bacterial pathogens in 25% pear leaf extract did not differed from that in buffer, but in 50% pear leaf extract, survival slightly decreased in the case of E. amylovora and P. syringae pv. syringae. Peptide activity decreased in the presence of leaf extract. At the 50% pear leaf extract concentration, the antibacterial activity of BP100 and BP143 decreased significantly against the three pathogens. At the 25% pear leaf extract concentration, an important decrease in the activity of all three peptides against E. amylovora was observed. The activity of BP100 and BP143 decreased moderately against P. syringae pv. syringae, while that of BP157 was not affected. In contrast, against X. axonopodis pv. vesicatoria, the activity of the three peptides remained unaffected.
Fig. 2.
Influence of leaf extract on the antibacterial activity levels of peptides BP100, BP143, and BP157. The effect was measured as the survival of X. axonopodis pv. vesicatoria, P. syringae pv. syringae, and E. amylovora after exposure to a 20 μM peptide concentration. The peptide assay was carried out in buffer (white bars) or in pear leaf extracts diluted at 25% (shaded bars) and 50% (black bars). Data represent the mean values of the two assays performed. The confidence intervals for the means are indicated on top of the bars. Letters over the bars indicate the significance of the difference between treatment extracts (P ≤ 0.05), according to Tukey's test.
Ex vivo infection assays.
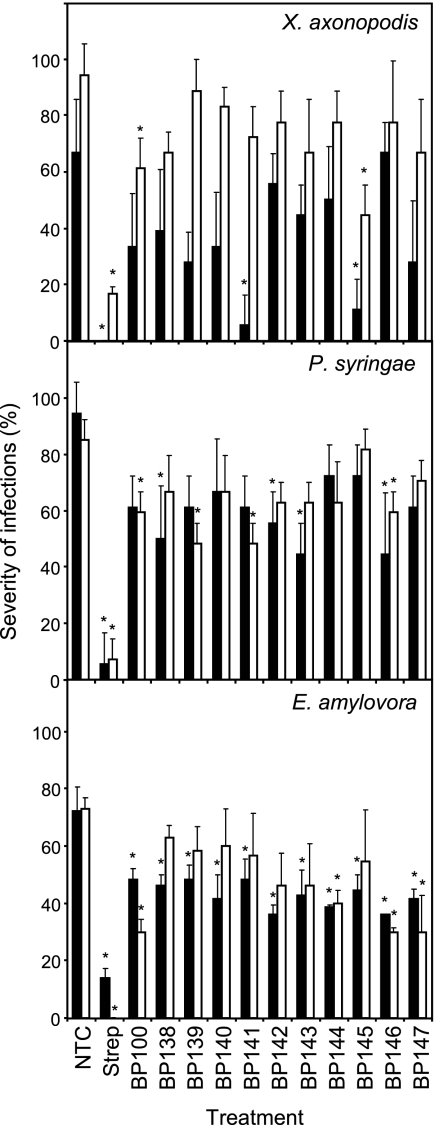
The activities of peptides BP138 to BP147, which contain one d-amino acid, were compared to that of the parent peptide BP100 in a detached plant organ assay using a preventative application (Fig. 3). The disease intensities in the nontreated controls for the three plant-pathogenic bacteria were high and ranged from 65 to 95% (necrosis progressed around the wound, with a mean diameter of >3 mm). For the three pathogens in the two experiments performed, streptomycin-treated wounds were not infected or infections remained at the site of inoculation. In pepper leaves inoculated with X. axonopodis pv. vesicatoria, peptides BP100 and BP141 were effective in one of the two trials performed, whereas peptide BP145 was effective in both trials, decreasing infection levels from 65 to 10% (assay 1) and from 95 to 42% (assay 2). Besides, peptides BP141 and BP145 were as effective as streptomycin in assay 1. In pear leaves inoculated with P. syringae pv. syringae, 6 out of the 11 peptides tested (BP100, BP138, BP139, BP141, BP142, and BP143) were active in one of the two experiments performed, whereas peptide BP146 was effective in both experiments, decreasing infection levels from 95 to 43% (assay 1) and from 85 to 60% (assay 2). In immature pear fruits inoculated with E. amylovora, 7 of the 11 peptides tested (BP138, BP139, BP140, BP141, BP142, BP143, and BP145) were effective in one of the two experiments, whereas peptides BP100, BP144, BP146, and BP147 were effective in both assays, decreasing infection levels from 73% to around 35%. Thus, peptide BP145 was the most effective one in the ex vivo assay for X. axonopodis pv. vesicatoria, BP146 for P. syringae pv. syringae, and BP100, BP144, BP146, and BP147 for E. amylovora.
Fig. 3.
Effect of the preventative application of linear undecapeptides in control of infections in detached pear (P. syringae pv. syringae) and pepper (X. axonopodis pv. vesicatoria) leaves and in immature pear fruits (E. amylovora). Two independent experiments, assay 1 (black bars) and assay 2 (white bars), were performed. A nontreated control (NTC) and a reference treatment with streptomycin (Strep) were used. The confidence intervals for the means are indicated on top of the bars. An asterisk over a bar indicates a significant difference from the nontreated control for a given experiment, according to Tukey's test (P < 0.05).
Efficacy in whole-plant assays.
Although several d-diastereomeric peptides with multiple d-amino acid substitutions displayed good in vitro activity profiles, their high production costs limit their application as plant protection products. Therefore, in the present study, only isomers containing single d-amino acid substitutions were considered candidates for the whole-plant assays. In addition, the selection of peptides BP143 and BP145 for the whole-plant assays was based primarily on the results from the ex vivo infection assays and secondly on the values of the in vitro activity. In the ex vivo assays, peptides BP138, BP139, BP141, BP142, BP143, BP145, and BP146 were active against at least two pathogens. From this set, peptides BP143 and BP145 were selected because they displayed the highest in vitro activity levels, being as active as BP100. Moreover, peptide BP143 was almost 4-fold less susceptible to protease degradation and 10-fold less hemolytic than BP100. Peptide BP145 showed a hemolytic and protease degradation profile similar to that of BP100.
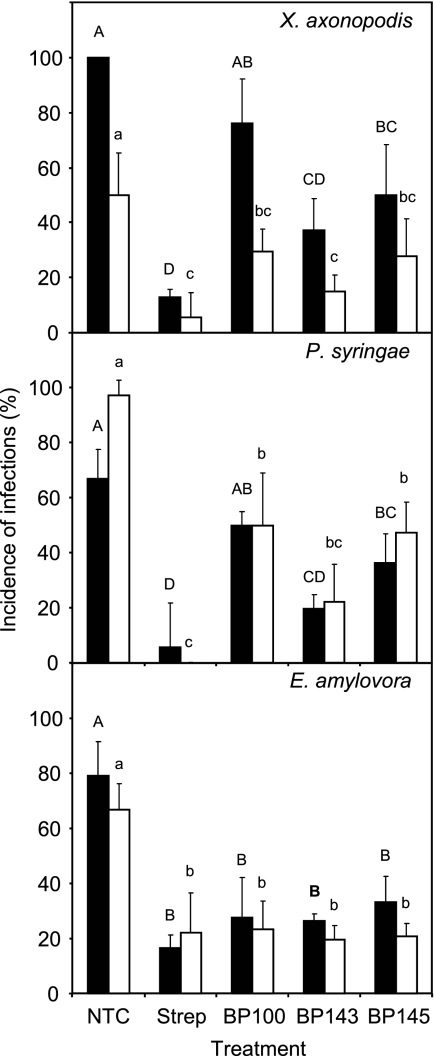
The activity levels of peptides BP143 and BP145 containing one d-amino acid were studied and compared to those of the parent peptide BP100 and streptomycin in whole-pear (E. amylovora and P. syringae pv. syringae) and -pepper (X. axonopodis pv. vesicatoria) plant assays (Fig. 4).
Fig. 4.
Effect of the preventative application of linear undecapeptides in the control of bacterial blight of pepper (X. axonopodis pv. vesicatoria), bacterial blight of pear (P. syringae pv. syringae), and fire blight of pear (E. amylovora) under controlled greenhouse conditions. Two independent experiments, assay 1 (black bars) and assay 2 (white bars), were performed. A nontreated control (NTC) and a reference treatment with streptomycin (Strep) were used. The confidence intervals for the means are indicated on top of the bars. Letters over the bars indicate the significance of the difference for assay 1 (capital letters) and assay 2 (lowercase letters) (P ≤ 0.05), according to Tukey's test.
The three peptides effectively controlled the three pathogens in the corresponding plant host, except for BP100 against P. syringae pv. syringae and X. axonopodis pv. vesicatoria in one experiment (Fig. 4). Streptomycin controlled the disease caused by the three pathogens in the two experiments performed. Peptide BP100 did not differ from streptomycin in the two experiments against E. amylovora on the pear and in one experiment against X. axonopodis pv. vesicatoria on the pepper. Peptide BP145 was as effective as BP100 in the three pathosystems. Peptide BP143 was more effective than BP100 in one experiment against X. axonopodis pv. vesicatoria on the pepper and in one experiment against P. syringae pv. syringae on the pear and did not differ significantly against E. amylovora on the pear. Interestingly, peptide BP143 was as effective as streptomycin in all three pathosystems. No symptoms of phytotoxicity in leaf wounds of plants treated with the peptide solutions were observed.
DISCUSSION
The use of antimicrobial peptides as pesticides for plant disease control is hampered by their susceptibility to proteases that are present in plant tissues, limiting the in vivo efficacy. Our main objective in this study was to improve the activity of the antimicrobial peptide KKLFKKILKYL-NH2 (BP100) under in vivo conditions (5) by introducing d-amino acids into the sequence. Besides testing the influence of replacing all-l amino acids on activity, the influence of incorporating 1 d-amino acid, 2 or 3 d-amino acids at adjacent or nonconsecutive positions, and 4 to 10 d-amino acids at either the N or the C terminus was also investigated.
d-Amino acid incorporation has been previously applied to other antimicrobial peptides, such as pardaxin, melittin, or magainin, leading to d-analogues with similar antimicrobial activities, lower hemolytic activity levels, and higher stability levels against protease degradation (11, 28). In the present study, the antibacterial activity of the BP100 isomers depended on the pathogen, the number of d-amino acids introduced, and the residue that was replaced. Regarding the pathogen, in general, peptides did not display a clear pattern of activity. Among the 31 isomers studied, there were more peptides as active as or more active than BP100 against P. syringae pv. syringae and X. axonopodis pv. vesicatoria than against E. amylovora. On the other hand, a differential susceptibility of bacteria to a given peptide was observed. This has been attributed to differences in the membrane components of the target microorganism, e.g., charge and lipid composition that would influence rates of binding of cationic peptides to the membranes (24).
d-Diastereoisomers exhibited complicated activity patterns and did not display a simple dependence on the polarity of the residue. However, general trends on the positions that influence the activity were observed. Analysis of the results obtained for single d-amino acid substitutions reflected that replacement of residues I7, K9, Y10, and L11 decreased the antibacterial activity against the three pathogens, indicating that these amino acids are crucial for activity and cannot be replaced. In contrast, residues F4, K2, and K1 can be substituted without weakening the activity against the three bacteria, leading to peptides BP143, BP145, and BP147, respectively (MIC = 2.5 to 7.5 μM). These results confirm previous data indicating that subtle changes in a peptide sequence influence the antimicrobial and hemolytic activities (5, 6, 34). Consistently, the previous tendency for peptides containing two and three d-amino acids was also observed. Thus, sequences containing d-K9 or d-I7 and d-K9 were less active than that of BP100, whereas BP154, which incorporates d-K1, d-K2, and d-F4, showed activity similar to that of the parent peptide. On the other hand, the introduction of d-amino acids at the N or C terminus resulted in distinctly improved activity. Peptides containing d-amino acids at the C terminus were more active, and their activity increased with the number of d-amino acids, leading to peptides with MIC values of 1.25 to 5.0 μM. The all-d amino acid isomer BP153 was also analyzed and resulted in being the most active sequence.
The hemolytic activity of BP100 isomers depended on the residue that was replaced but followed a different pattern than the one followed by antibacterial activity. Single d-amino acid substitution of residues L3, F4, K5, I7, L8, K9, and Y10 led to peptides with low hemolytic activity (≤7%). Replacement of the other amino acids resulted in a substantial increase in the hemolysis (23 to 71%). Notably, BP143, which incorporates d-F4, combined both high antibacterial activity (MIC of 2.5 to 7.5 μM) and low hemolysis (5%). Consistently, when two or three of the above-described residues were replaced with the corresponding enantiomer, the resulting peptides BP149 to BP152 displayed low hemolytic activity (<5%). In contrast, BP154 containing d-K1, d-K2, and d-F4 showed a higher cytotoxicity level (41%). Hemolysis of peptides incorporating 4 to 10 d-amino acids at the N or C terminus did not depend on the number of d-amino acids incorporated. However, peptides incorporating d-amino acids at the N terminus were less hemolytic (0 to 21%). Moreover, even though attempts to use enantiomeric peptides solely composed of d-amino acids to enhance peptide cell selectivity have been made (32), in the present study, the all-d peptide BP153 was as cytotoxic as BP100.
Plant extracts inhibited the antibacterial activity of BP100 as well as that of peptides containing d-amino acids. The reduction of antimicrobial peptide activity by use of leaf extracts of different plants for cecropin B and SB-37 (38), Pep11 and Pep20 (4), and Pep3 (19) has been previously reported. Moreover, it has been described that the degradative activity of proteases from the leaf intercellular fluids can reduce the activity of cecropin B analogues expressed in transgenic plants (41). As exemplified by BP157, which was stable to proteinase K degradation and was only slightly affected by the pear leaf extract, the loss of activity of our linear undecapeptides in plant material could also be attributed to degradation by plant proteases. However, BP143 and BP100 were similarly inhibited by leaf extracts, even though they differed by 4-fold in susceptibility to proteinase K. This is in agreement with previous reports that describe that other factors besides plant proteases present in the plant extract, such as phenolic compounds, could be implicated in the loss of peptide activity (3, 21).
Several d-diastereomeric peptides displayed good in vitro activity profiles; however, their high production costs impose severe limitations to their application as plant protection products. Therefore, in the present study, only isomers containing single d-amino acid substitutions were evaluated in plant material and whole-plant assays. Globally, several BP100 derivatives with single d-amino acid substitutions were active against the plant-pathogenic bacteria in an infectivity inhibition assay performed on detached leaves and fruits. More peptides were found to be active against P. syringae pv. syringae and E. amylovora on pear leaves or fruits than against X. axonopodis pv. vesicatoria on pepper leaves. This finding is in agreement with the results of the in vitro activity reported here. Also, a considerable specificity of the peptides for the pathogen-host system was confirmed, in comparison to streptomycin, which was consistently effective in the three pathosystems used. For example, the best peptide against X. axonopodis pv. vesicatoria on pepper leaves was BP145, whereas BP146 was the best peptide against P. syringae pv. syringae on pear leaves, and BP144, BP146, and BP147 were the most effective peptides against E. amylovora on immature pear fruits.
The selection of peptides BP143 and BP145 for the whole-plant assays was because these peptides were active against at least two pathogens in the ex vivo infection assays and displayed the highest in vitro activity levels, being as active as BP100, and because they showed equal or more favorable protease degradation and hemolytic profiles to those of BP100. In these whole-plant assays, peptide BP143 (KKLFKKILKYL-NH2) was more effective than BP100 against X. axonopodis pv. vesicatoria on the pepper and P. syringae pv. syringae on the pear. Despite this higher in planta activity, BP143 did not differ from BP100 in its in vitro activity against the three plant-pathogenic bacteria nor in its susceptibility to the plant leaf extract, but it was more stable to proteinase K. Interestingly, BP145 (KKLFKKILKYL-NH2), which was as effective as BP100 in planta, also displayed in vitro activity and susceptibility to protease degradation similar to those of BP100. Thus, these results clearly suggest that subtle changes in the structures of antimicrobial peptides affect not only in vitro antibacterial and hemolytic activities but also other biological properties like in planta activity and protease degradation susceptibility.
Notably, BP143 was as effective as streptomycin for the control of fire blight and bacterial blight on pear plants and bacterial blight on pepper plants. Antibiotics like streptomycin, tetracycline, and kasugamycin are used in certain countries to control plant bacterial diseases, at doses that are only slightly lower than those used with this antimicrobial peptide in the present study (1, 33, 46).
In addition, probably due to the inhibition of peptides by plant extracts, it was necessary to apply peptide doses that were 25 to 80 times higher than the in vitro MIC values to be effective in plant wound treatment to prevent infection. The need for higher doses in in vivo assays has been reported for other antimicrobial peptides as well as for most antifungal and antibacterial compounds used to control plant diseases (1, 33, 36).
Finally, the present study demonstrates how the activity of a linear undecapeptide against plant-pathogenic bacteria can be improved through d-amino acid substitution. It also shows that screening based on in vitro and ex vivo procedures can deliver effective compounds to control plant infections in model plant-pathogenic bacteria. The fact that peptide BP143 showed efficacy comparable to that of the reference antibiotic streptomycin against three bacterial diseases of plants confirms the potential of antimicrobial peptides as plant protection products. Future studies will involve field tests under variable agroclimatic conditions and improvement of the peptide synthesis process to reduce production costs.
ACKNOWLEDGMENTS
Imma Güell was the recipient of a predoctoral fellowship from the Generalitat de Catalunya. This work was financed by grants AGL2009-13255-C02-02/AGR, AGL-2009-13255-C02-01/AGR, and PPT-060000-2008-2 from MICINN of Spain. The LIPPSO and CIDSAV groups are recognized as 2009SGR182 and 2009SGR812 by the Catalonian Government.
We also acknowledge the Serveis Tècnics de Recerca of the University of Girona for the ESI-MS analysis and Josep Pereda for greenhouse assistance.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Agrios G. N. 2005. Plant pathology, 5th ed Academic Press, San Diego, CA [Google Scholar]
- 2. Ajesh K., Sreejith K. 2009. Peptide antibiotics: an alternative and effective antimicrobial strategy to circumvent fungal infections. Peptides 30:999–1006 [DOI] [PubMed] [Google Scholar]
- 3. Alan A. R., Earle E. 2002. Sensitivity of bacterial and fungal plant pathogens to the lytic peptides, MSI-99, magainin II, and cecropin B. Mol. Plant Microbe Interact. 15:701–708 [DOI] [PubMed] [Google Scholar]
- 4. Ali G. S., Reddy A. S. N. 2000. Inhibition of fungal and bacterial plant pathogens by synthetic peptides: in vitro growth inhibition, interaction between peptides, and inhibition of disease progression. Mol. Plant Microbe Interact. 13:847–859 [DOI] [PubMed] [Google Scholar]
- 5. Badosa E., et al. 2007. A library of linear undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 28:2276–2285 [DOI] [PubMed] [Google Scholar]
- 6. Badosa E., et al. 2009. Sporicidal activity of synthetic antifungal undecapeptides and control of Penicillium rot of apples. Appl. Environ. Microbiol. 75:5563–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bardají E., et al. April 2006. Antimicrobial linear peptides. Patent WO/2007/125142 A1. [Google Scholar]
- 8. Barny M. A., et al. 1990. Cloning of a large gene cluster involved in Erwinia amylovora CFBP1430 virulence. Mol. Microbiol. 4:777–786 [DOI] [PubMed] [Google Scholar]
- 9. Bechinger B. 2004. Structure and function of membrane-lytic peptides. Crit. Rev. Plant Sci. 23:271–292 [Google Scholar]
- 10. Bechinger B., Lohner K. 2006. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochim. Biophys. Acta 1758:1529–1539 [DOI] [PubMed] [Google Scholar]
- 11. Bessalle R., Kapitkovsky A., Gorea A., Shalit I., Fridkin M. 1990. All-d-magainin: chirality, antimicrobial activity and proteolytic resistance. FEBS Lett. 274:151–155 [DOI] [PubMed] [Google Scholar]
- 12. Boman H. G. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197–215 [DOI] [PubMed] [Google Scholar]
- 13. Broekaert W. F., et al. 1997. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 16:297–323 [Google Scholar]
- 14. Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 15. Brogden K. A., Ackermann M., McCray P. B., Jr., Tack B. F. 2003. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 22:465–478 [DOI] [PubMed] [Google Scholar]
- 16. Brotman Y., Makovitzki A., Shai Y., Chet I., Viterbo A. 2009. Synthetic ultrashort cationic lipopeptides induce systemic plant defense responses against bacterial and fungal pathogens. Appl. Environ. Microbiol. 75:5373–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bulet P., Stöcklin R., Menin L. 2004. Antimicrobial peptides: from invertebrates to vertebrates. Immunol. Rev. 198:169–184 [DOI] [PubMed] [Google Scholar]
- 18. Cabrefiga J., Montesinos E. 2005. Analysis of aggressiveness of Erwinia amylovora using disease-dose and time relationships. Phytopathology 95:1430–1437 [DOI] [PubMed] [Google Scholar]
- 19. Cavallarin L., Andreu D., San Segundo B. 1998. Cecropin A-derived peptides are potent inhibitors of fungal plant pathogens. Mol. Plant Microbe Interact. 11:218–227 [DOI] [PubMed] [Google Scholar]
- 20. Ferre R., et al. 2006. Inhibition of plant-pathogenic bacteria by short synthetic cecropin A-melittin hybrid peptides. Appl. Environ. Microbiol. 72:3302–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Florack D., et al. 1995. Expression of giant silkmoth cecropin B genes in tobacco. Transgenic Res. 4:132–141 [DOI] [PubMed] [Google Scholar]
- 22. Hancock R. E. W., Sahl H. G. 2006. Antimicrobial and host defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557 [DOI] [PubMed] [Google Scholar]
- 23. Hong S. Y., Oh J. E., Lee K.-H. 1999. Effect of d-amino acid substitution on the stability, the secondary structure, and the activity of membrane-active peptide. Biochem. Pharmacol. 58:1775–1780 [DOI] [PubMed] [Google Scholar]
- 24. Huang H. W. 2000. Action of antimicrobial peptides: two-state model. Biochemistry 39:8347–8352 [DOI] [PubMed] [Google Scholar]
- 25. Huang H. W. 2006. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim. Biophys. Acta 1758:1292–1302 [DOI] [PubMed] [Google Scholar]
- 26. Jenssen H., Hamill P., Hancock R. E. W. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keymanesh K., Soltani S., Sardari S. 2009. Application of antimicrobial peptides in agriculture and food industry. World J. Microbiol. Biotechnol. 25:933–944 [Google Scholar]
- 28. Li X., Li Y., Han H., Miller D. W., Wang G. 2006. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J. Am. Chem. Soc. 128:5776–5785 [DOI] [PubMed] [Google Scholar]
- 29. Loper J. E., et al. 1991. Evaluation of streptomycin, oxytetracycline and copper resistance of Erwinia amylovora isolated from pear orchards in Washington state. Plant Dis. 75:287–290 [Google Scholar]
- 30. Marcos J. F., Gandía M. 2009. Antimicrobial peptides: to membranes and beyond. Expert Opin. Drug Discov. 4:659–671 [DOI] [PubMed] [Google Scholar]
- 31. Marcos J. F., Muñoz A., Pérez-Payá E., Misra S., López-García B. 2008. Identification and rational design of novel antimicrobial peptides for plant protection. Annu. Rev. Phytopathol. 46:271–301 [DOI] [PubMed] [Google Scholar]
- 32. Matsuzaki K. 2009. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 1788:1687–1692 [DOI] [PubMed] [Google Scholar]
- 33. McManus P. S., Stockwell V. O., Sundin V. O., Jones A. L. 2002. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40:443–465 [DOI] [PubMed] [Google Scholar]
- 34. Monroc S., et al. 2006. Improvement of cyclic decapeptides against plant pathogenic bacteria using a combinatorial chemistry approach. Peptides 27:2575–2584 [DOI] [PubMed] [Google Scholar]
- 35. Montesinos E. 2007. Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett. 270:1–11 [DOI] [PubMed] [Google Scholar]
- 36. Montesinos E., Bardají E. 2008. Synthetic antimicrobial peptides as agricultural pesticides for plant-disease control. Chem. Biodivers. 5:1225–1237 [DOI] [PubMed] [Google Scholar]
- 37. Moragrega C., Manceau C., Montesinos E. 1998. Evaluation of drench treatments with phosphonate derivatives against Pseudomonas syringae pv. syringae on pear under controlled environment conditions. Eur. J. Plant Pathol. 104:171–180 [Google Scholar]
- 38. Mourgues F., Brisset M., Chevreau E. 1998. Activity of different antibacterial peptides on Erwinia amylovora growth, and evaluation of the phytotoxicity and stability of cecropins. Plant Sci. 139:83–91 [Google Scholar]
- 39. Nicolas P. 2009. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 276:6483–6496 [DOI] [PubMed] [Google Scholar]
- 40. Otvos L., Jr 2000. Antibacterial peptides isolated from insects. J. Pept. Sci. 6:497–511 [DOI] [PubMed] [Google Scholar]
- 41. Owens L. D., Heutte T. M. 1997. A single amino acid substitution in the antimicrobial defense protein cecropin B is associated with diminished degradation by leaf intercellular fluid. Mol. Plant Microbe Interact. 10:525–528 [DOI] [PubMed] [Google Scholar]
- 42. Papo N., Oren Z., Pag U., Sahl H.-G., Shai Y. 2002. The consequence of sequence alteration of an amphipathic α-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 277:33913–33921 [DOI] [PubMed] [Google Scholar]
- 43. Peschel A., Sahl H. G. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529–536 [DOI] [PubMed] [Google Scholar]
- 44. Sharma R. K., Sundriyal S., Wangoo N., Tegge W., Jain R. 2010. New antimicrobial hexapeptides: synthesis, antimicrobial activities, cytotoxicity, and mechanistic studies. Chem. Med. Chem. 5:86–95 [DOI] [PubMed] [Google Scholar]
- 45. Sundin G. W., Bender C. L. 1993. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 59:1018–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vidaver A. K. 2002. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 34:S107–110 [DOI] [PubMed] [Google Scholar]
- 47. Wei G.-X., Bobek L. A. 2005. Human salivary MUC7 12-mer-L and 12-mer-d peptides: antifungal activity in saliva, enhancement of activity with protease inhibitor cocktail or EDTA, and cytotoxicity to human cells. Antimicrob. Agents Chemother. 49:2336–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wessolowski A., Bienert M., Dathe M. 2004. Antimicrobial activity of arginine- and tryptophan-rich hexapeptides: the effects of aromatic clusters, d-amino acid substitution and cyclization. J. Peptide Res. 64:159–169 [DOI] [PubMed] [Google Scholar]
- 49. Wieczorek M., et al. 2010. Structural studies of a peptide with immune modulating and direct antimicrobial activity. Chem. Biol. 17:970–980 [DOI] [PubMed] [Google Scholar]
- 50. Yeaman M. R., Yount N. Y. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27–55 [DOI] [PubMed] [Google Scholar]
- 51. Yount N. Y., Yeaman M. R. 2005. Immunocontinuum: perspectives in antimicrobial peptide mechanisms of action and resistance. Protein Pept. Lett. 12:49–67 [DOI] [PubMed] [Google Scholar]
- 52. Zaccardelli M., Spasiano A., Bassi C., Merighi M. 2005. Identification and in planta detection of Pseudomonas syringae pv. tomato using PCR amplification of hrpZPst. Eur. J. Plant Pathol. 111:85–90 [Google Scholar]
- 53. Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 54. Zhu W. L., Nan Y. H., Hahm K.-S., Shin S. Y. 2007. Cell selectivity of an antimicrobial peptide melittin diastereomer with d-amino acid in the leucine zipper sequence. J. Biochem. Mol. Biol. 40:1090–1094 [DOI] [PubMed] [Google Scholar]