Abstract
In order to better understand the ecological and virulence characteristics of the various clades of Escherichia, in vitro and in vivo experiments were undertaken. Members of the recently described cryptic clades of Escherichia (clades III, IV, and V) were found to have an enhanced ability to form biofilms compared to strains of Escherichia coli, E. fergusonii, or E. albertii. Members of the cryptic clades were also able to replicate at a lower temperature (5°C versus 11°C) than strains of the named species of Escherichia. Neither a strain's maximal growth rate nor its optimal temperature for growth varied with respect to the strain's phylogenetic affiliation. Escherichia strains not belonging to the species E. coli were positive for a mix of traits thought to enhance a strain's ability to cause either intestinal or extraintestinal disease. However, no non-E. coli Escherichia strain was virulent in a mouse model of extraintestinal infection. The frequency of resistance to antibiotics was low, and none of the strains tested harbored class 1, 2, or 3 integrons. The results of these experiments support the hypothesis that members of the cryptic Escherichia clades may be better able to persist in the external environment compared to E. coli, E. fergusonii, or E. albertii, isolates.
INTRODUCTION
The application of techniques such as multilocus sequence analysis and a growing interest in the ecology and evolution of bacteria from nonhuman sources has revealed a wealth of previously unrecognized diversity in even well-characterized groups such as the Enterobacteriaceae. This is nowhere more apparent than in the genus Escherichia. Escherichia coli is the type species for the genus and was first described in 1885 by Theodor Escherich (17). E. fergusonii was the second species to be recognized (9), and E. albertii was described in 2003 (12). Most recently, the existence of five novel clades of Escherichia has been reported (32). Unlike E. albertii and E. fergusonii, which are phenotypically distinct from E. coli, members of the novel clades cannot be distinguished from E. coli using traditional biochemical criteria and hence their designation as cryptic lineages (32).
Although we have a growing appreciation of the ecological and life-history characteristics of E. coli (30), we have a very poor understanding of these characteristics for most bacterial species. We can infer that E. albertii, E. fergusonii, and the cryptic clades of Escherichia have a cosmopolitan distribution, but there is little else we know about these clades (32). The data concerning the source of isolation suggests that the various members of the genus are nonrandomly distributed. E. coli is predominantly isolated from mammals (9), whereas data for Australian isolates suggest that E. albertii and E. fergusonii can only be detected as a dominant member of the aerobic microbial community in birds. Although members of the cryptic Escherichia clades can be isolated from birds and mammals, members of some clades appear to be more often detected in water and sediment samples (31, 32).
In order to better understand the ecological and life-history characteristics of the various clades of Escherichia, a series of in vitro and in vivo experiments were undertaken. Although E. albertii has been implicated as an intestinal pathogen of birds and humans (19) and E. fergusonii has been implicated as an intestinal and opportunistic extraintestinal pathogen (2, 8, 16), little is known of the virulence potential of the cryptic lineages of Escherichia. Consequently, the presence of genes associated with virulence was determined, and the potential of strains of these Escherichia lineages to cause extraintestinal infection was assessed in vivo.
MATERIALS AND METHODS
Strains examined.
The strains used in the present study included those described by Walk et al. (32), as well as Australian strains belonging to the cryptic lineages that were discovered subsequent to the Walk et al. study. The identity of these additional representatives of the cryptic clades was confirmed with multilocus sequence analysis using the seven-gene MLST scheme described at http://mlst.ucc.i.e. In addition, the characterization of a large collection of French and Gabonese strains using the Institute Pasteur MLST scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html) revealed additional members of the cryptic clades, and a subset of these strains was included in the present study. The French and Gabonese strains were from a collection of 450 wild and domestic animal (French and Gabon) and human (France) commensal strains (29) and from a collection of 1,100 human extraintestinal pathogenic strains from France (4, 21). The E. coli strains were selected from a collection of over 1,200 Escherichia isolates collected in an ad hoc manner from more than 350 native vertebrate species from throughout Australia (9, 10), from humans living in Australia (11), and from soil, sediments, and water samples (23).
Only two representatives of clade II are known and, consequently, clade II strains were only included in the antibiotic resistance and virulence assays. Clade I strains were not included in the ecological assays since there is no evidence to suggest that they are over-represented in soil, water, or sediment samples compared to vertebrate fecal samples.
Biofilm production.
The ability of an isolate to form biofilms was assessed using a method modified from Reisner et al. (25). The extent of biofilm formation was assessed on three occasions (repeats) for each strain. Strains were assigned at random to the wells of 96-well microtiter plate. The remaining wells on each plate acted as negative controls. Strains were propagated overnight in minimal glucose medium [7 g of K2HPO4, 2 g of KH2PO4, 0.5 g of Na3C3H5O(CO2)3, 0.1 g of MgSO4, 1 g of (NH4)2SO4, and 10 of glucose per liter of distilled water supplemented with 1.0 ml of 0.2% thiamine solution] before 10 μl of each overnight culture was inoculated into either 150 μl of minimal glucose medium or nutrient broth (Oxoid). Four plates were inoculated, two of the glucose-limited medium and two of nutrient broth. For each of the media, one plate was incubated for 48 h at 24°C, and the other plate was incubated at 37°C. After incubation, all media were removed from each well of the microtiter plates, and the wells were then rinsed twice with 200 μl of sterile saline solution (0.9%) in order to remove all nonadhering bacterial cells. The remaining adherent cells were stained by adding 160 μl of crystal violet (0.1% [wt/vol]). After 15 min of staining, excess crystal violet was removed from the wells, and the wells were then rinsed twice using 200 μl of a saline solution (0.9%). Ethanol (95%) was added to every well in order to destain the adherent bacterial cells. After 5 min of destaining, the amount of crystal violet in the ethanol was quantified by determining the absorbance at 590 nm using a Powerwave 10 scanning spectrophotometer (Bio-Tek Instruments, Inc.). The optical density data were log transformed (loge) prior to analysis. When the data were analyzed, the bacterial isolate represented an experimental unit, and the repeats of the experiment were considered a random effect in the model.
Maximum growth rate and optimal temperature for growth.
The manner in which the growth rate of all isolates changed in response to temperature was assessed in order to determine an isolate's maximum growth rate and optimal temperature for growth. Growth rate estimates for all isolates were made at 11 different temperatures: 27, 30, 32, 34, 36, 38, 40, 42, 44, 46, and 48°C. From the freezer cultures, all strains were inoculated into the wells of a microtiter plate containing 150 μl of LB broth, and the plates were incubated overnight at 37°C. A 10-μl aliquot of each culture was used to inoculate another microtiter plate containing 150 μl of LB broth. The plate was then incubated at one of the experimental temperatures, and during incubation optical density readings (630 nm) were taken every 5 min by using a Powerwave 10 microplate scanning spectrophotometer until each culture had reached stationary phase.
The peak growth rate of an isolate at a particular temperature was established in the following manner. For every 15-min interval, the rate of change in the optical density (ψ) was calculated as ψ = ln(ODt + 1)/ODt)/Δt, where t is the time interval in hours. The growth rate (ψ) of a strain was plotted against time, and the peak growth rate observed was taken to be the growth rate of the strain at the experimental temperature.
The relationship between an isolate's growth rate and temperature was described by using the function ψT = (B − CT)DT, where T is the temperature and B, C, and D are parameters describing the shape of the curve. This function was fitted to the data for every replicate of an isolate using a nonlinear curve fitting routine implemented in JMP 7.01 (SAS Institute). Using this function, the maximum growth rate of a strain (ψmax) is defined as ψmax = (C/lnD)exp[(BlnD/C) − 1], and the optimal growth temperature (Topt) is defined as Topt = B/C − 1/lnD.
Growth at low temperatures.
Overnight LB broth cultures of the isolates were prepared and incubated at 37°C. Aliquots (100 μl) of these overnight cultures were used to inoculate test tubes containing 5 ml of phenol red broth (10.0 g of pancreatic digest of casein, 5.0 g of NaCl, 5.0 g of glucose, 0.018 g of phenol red per liter of distilled H2O). Each isolate, together with a negative control, was cultured in phenol red broth and incubated at each of four temperatures: 2, 5, 8, and 11°C. After 11 days of incubation at each temperature, the cultures were examined to determine whether the medium had changed in color from red to yellow.
Low temperature survival in the absence of nutrients.
Overnight LB broth cultures of the strains were prepared and incubated at 37°C. The overnight cultures were then serially diluted using sterile saline (0.9%) in order to achieve a density of 105 cells ml−1. Cells from this dilution were then added to test tubes containing 12.5 ml of sterile water such that the final cell density was ∼103 cells ml−1. One tube containing cells suspended in water was prepared for each isolate and incubated at 5°C. The change in viable cell density over time was determined at regular intervals by plating a 50-μl aliquot of the water in the experimental tube onto LB agar plates. The plates were incubated overnight at 37°C, and the number of colonies on each plate was determined. The average life span of each isolate at each temperature was determined by fitting an exponential decay function to the number of cells alive at time t days, that is Nt = Noe−μt, where No is the initial number of cells and μ is the mortality rate (1/μ = the average life span of a cell in days). Although life span estimates are typically obtained using initial cell densities (No) in the range of 108 to 109 cells ml−1, such high initial densities are not required. The value of No is only important in that in determines, in part, how many samples can be taken prior to the number of cells in a culture declining below detection frequency. Provided there are cell density estimates for at least four time points, an adequate estimate of μ can be obtained. Indeed, high initial cell densities can lead to increased experimental error because high cell densities initially require the experimental cultures to be serially diluted one or more times prior to obtaining an estimate of cell density. Each serial dilution step adds error, and these errors are compounded with each dilution step (15). Since the number of dilutions required generally declines as cells die, the number of dilution steps required also declines, and this leads to a situation where the amount of experimental error associated with each cell density estimate changes through time. The model of exponential decay was fitted to the data for each of the isolates using the nonlinear curve fitting routine implemented in JMP 7.01 (SAS Institute). Survival estimates were log transformed prior to analysis.
Virulence factor screening.
Escherichia strains not belonging to the E. coli lineage were PCR screened for the presence of virulence factors involved in intestinal (elt [LT-A], astA, eaeA, and cdtA) and extraintestinal virulence (kpsE, sfa, iroN, aer/iutA, papC, papG, hlyD, cnf1, ompT, ibeA, and fyuA) as described by Gordon et al. (11) and Diard et al. (6).
Mouse virulence assay.
A mouse model of systemic infection was used to assess the intrinsic virulence of the strains (21). For each strain, 10 outbred female Swiss OF1 mice (3 to 4 weeks old, 14 to 16 g) were challenged subcutaneously in the abdomen with a standardized bacterial inoculum (109 CFU of log-phase bacteria/ml in 0.2 ml of Ringer solution). Mortality was assessed over 7 days postchallenge. The urosepsis strain CFT073 was used as a positive control (killing 10 mice out of 10), whereas the feces-derived strain K12-MG1655 was used as a negative control (killing no mouse out of 10) (13).
Antibiotic resistance assays.
Antimicrobial susceptibilities for the strains sourced from France and Gabon were determined by the method of disc diffusion, according to the guidelines of the French Society of Microbiology Antibiogram Committee (CASFM; www.sfm.asso.fr). The resistance profiles of all other strains were determined using the BBL Sensi-Disk antimicrobial susceptibility test disks according to the manufacturer's protocol. All strains were tested for sensitivity to chloramphenicol, kanamycin, streptomycin, tetracycline, and nalidixic acid, and the strains sourced from France and Gabon were also screened for amoxicillin and sulfamethoxazole resistance. Carriage of class 1, 2, and 3 integrons were determined using the methods described by Skurnik et al. (28).
RESULTS
The in vitro experiments were undertaken using the strains reported in Walk et al. (32), as well as additional isolates from Australia. For each of the experiments, the number of strains of each of the Escherichia clades is reported in Tables 1, 2, and 3.
Table 1.
Differences in the extent of biofilm formation among Escherichia cladesa
| Escherichia clade | No. of strains | Predicted meanb |
|---|---|---|
| Clade IV | 5 | 0.610A |
| Clade III | 6 | 0.592A |
| Clade V | 22 | 0.118B |
| E. fergusonii | 6 | −0.185C |
| E. albertii | 13 | −0.390C |
| E. coli | 39 | −0.415C |
Predicted means (loge optical density) after adjusting for the effects of culture media and temperature.
Means with the same superscript capital letter are not significantly different (Student t test, α = 0.05).
Table 2.
Differences among Escherichia clades in the growth rate of isolatesa
| Escherichia clade | No. of strains | Maximum growth rate (h−1) | Optimal temp (°C) |
|---|---|---|---|
| E. fergusonii | 6 | 0.95A | 41.1A |
| Clade V | 22 | 0.94A | 41.7A |
| E. coli | 38 | 0.93A | 41.7A |
| E. albertii | 13 | 0.92A | 41.9A |
| Clade IV | 5 | 0.91A | 41.5A |
| Clade III | 6 | 0.88A | 41.0A |
Means with the same superscript capital letter are not significantly different (Student t test, α = 0.05).
Table 3.
Differences among Escherichia clades in the average life span (days) of isolates at 5°C in the absence of nutrients
| Escherichia clade | No. of strains | Mean life span (days)a |
|---|---|---|
| E. coli | 15 | 31.2A |
| Clade IV | 5 | 18.6A,B |
| Clade V | 15 | 15.6A,B |
| Clade III | 6 | 15.6A,B |
| E. albertii | 13 | 7.0B |
| E. fergusonii | 6 | 4.6B |
Means connected by the same superscript capital letter are not significantly different (Student t test, α = 0.05).
Biofilm formation.
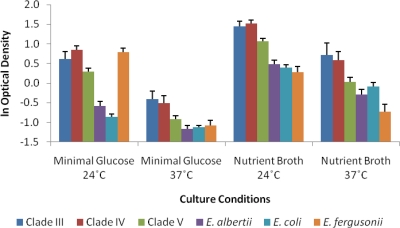
Escherichia clade, temperature, and media all accounted for a significant amount of the observed variation in the degree of biofilm formation. Although all interaction terms were significant, the effects were small relative to the main effects and cumulatively accounted for <10% of the observed variation (results not presented). In general, there was greater biofilm formation at 24°C than at 37°C and greater biofilm formation when the strains were grown in nutrient broth compared to minimal glucose medium (Fig. 1). The one exception to these outcomes was for E. fergusonii, where the greatest degree of biofilm formation was observed when the isolates were grown at 24°C in minimal glucose medium (Fig. 1). This outcome (high E. fergusonii biofilm formation at 24°C on minimal glucose medium) accounted for the majority of the observed interaction effects. Overall, isolates from clades III, IV, and V showed enhanced levels of biofilm formation relative to isolates of E. coli, E. fergusonii, and E. albertii (Table 1).
Fig. 1.
Biofilm formation by the six Escherichia clades in rich (L broth) and poor (minimal glucose) media at both 37 and 24°C. The mean optical density readings for each of the Escherichia clades are plotted on a log scale. The optical density readings were used to quantify biofilm formation by measuring the absorbance values for each bacterial culture at 590 nm obtained by extracting the cell bound crystal violet.
Thermal “niche.”
The Escherichia clade membership of an isolate did not predict its maximum growth rate (analysis of variance [ANOVA]: F(5, 83) = 0.598 P = 0.701) or the temperature at which the maximum growth rate occurred (ANOVA: F(5, 83) = 0.70, P = 0.624). Growth rates ranged between 0.88 and 0.95 h−1, while the optimal temperatures for growth varied by less than 1°C among Escherichia clades (Table 2).
Isolates were tested for their ability to replicate at low temperatures. This experiment revealed that Escherichia clade membership of an isolate was a significant predictor of an isolate's lower growth limit (Kruskal-Wallis test, χ2(5) = 49.58, P < 0.0001). None of the strains tested had the ability to replicate at 2°C. All strains belonging to clades III, IV, and V had the ability to replicate at 5°C. No isolates of E. coli, E. fergusonii, or E. albertii could grow at 5°C. Some isolates of E. coli (5 of 15 strains) and E. fergusonii (2 of 6 strains) could grow at 8°C, but most could only replicate at 11°C, and no E. albertii isolate could grow at less than 11°C.
The Escherichia clades were tested for their ability to survive in the absence of nutrients. Escherichia clade membership was a significant predictor of an isolate's life span at 5°C (ANOVA: F(5, 54) = 4.41, P = 0.0019). On average, E. coli cells survived the longest, followed by strains of clades III, IV, and V, whereas E. albertii and E. fergusonii isolates had the shortest life spans (Table 3).
Virulence characteristics.
Escherichia strains not belonging to the E. coli lineage were screened for a variety of traits thought to enhance a strain's ability to cause intestinal or extraintestinal disease. Many of the traits were detected and in some cases, such as astA, kpsE, ibeA, and ompT, at high frequencies (Table 4). The traits sfa, cnf1, papC, and papG were not detected. A number of traits were also detected at higher frequencies in some lineages compared to others, and these traits included astA, cdtA, elt, aer/iutA, fyuA, and ibeA (Table 4).
Table 4.
Frequency of intestinal and extraintestinal virulence traits in Escherichia strains not belonging to the E. coli lineage
| Trait | Frequency (%)a |
|||||
|---|---|---|---|---|---|---|
| Clade I (n = 19) | Clade III (n = 14) | Clade IV (n = 6) | Clade V (n = 52) | E. albertii (n = 13) | E. fergusonii (n = 6) | |
| astA | 21 | 50 | 0 | 67 | 23 | 0 |
| eaeA | 5 | 7 | 0 | 0 | 100 | 0 |
| cdtA | 0 | 0 | 0 | 33 | 38 | 0 |
| hylD | 5 | 0 | 0 | 0 | 0 | 0 |
| elt (LT-A) | 32 | 0 | 0 | 0 | 0 | 0 |
| aer/iutA | 16 | 7 | 0 | 32 | 15 | 17 |
| iroN | 0 | 0 | 0 | 4 | 8 | 0 |
| fyuA | 5 | 14 | 0 | 10 | 8 | 17 |
| kpsE | 74 | 100 | 67 | 88 | 85 | 100 |
| ibeA | 5 | 14 | 17 | 44 | 62 | 0 |
| ompT | 47 | 79 | 33 | 77 | 23 | 0 |
n, number of strains screened.
Representatives of each of the Escherichia lineages originating from both Australian and French/Gabonese collections and isolated from feces or extraintestinal sites (two urinary tract and one septicemia isolate) were tested in a mouse model of septicemia: four clade I, two clade II, three clade III, two clade IV, and five clade V strains, as well as the type strain of E. albertii and E. fergusonii. All of the strains were avirulent in this model as none of the 10 mice tested per strain were killed.
Antibiotic resistance assays.
The frequency of antibiotic resistance was low, and most strains were susceptible to most of the antibiotics tested (Table 5). In most cases, the frequency of resistance and sample sizes were too low for meaningful statistical analysis. However, no resistance to tetracycline was observed among clade V isolates from Australia, while 57% of the clade V isolates from France were resistant, a difference that was statistically significant (contingency analysis: likelihood ratio χ2 = 18.52, P < 0.001). No integron was detected in the strains from France or Gabon.
Table 5.
Frequency of antibiotic resistance in Escherichia strains not belonging to the E. coli lineage
| Antibiotic | Frequency (%)a |
|||||
|---|---|---|---|---|---|---|
| Clade I | Clade III | Clade IV | Clade V | E. albertii | E. fergusonii | |
| Nalidixic acid | 20 (5) | 0 (6) | 0 (6) | 2 (52) | 8 (13) | 0 (6) |
| Chloramphenicol | 33 (3) | 0 (13) | 0 (6) | 0 (50) | 0 (13) | 0 (6) |
| Kanamycin | 0 (6) | 0 (13) | 0 (5) | 0 (45) | 8 (13) | 0 (6) |
| Streptomycin | 0 (6) | 0 (13) | 0 (5) | 0 (44) | 0 (13) | 17 (6) |
| Tetracycline | 67 (3) | 8 (12) | 0 (4) | 37 (43) | 15 (13) | 50 (6) |
| Amoxicillin | 17 (6) | 0 (8) | ND | 0 (30) | ND | ND |
| Sulfamethoxazole | 33 (6) | 0 (8) | ND | 0 (30) | ND | ND |
The number of strains screened is indicated in parentheses. ND, not determined.
DISCUSSION
Bacteria that form biofilms are protected from conditions found in the secondary habitat, such as predation and UV radiation (3). This phenotype has been linked to persistence and regrowth of E. coli in drinking water distribution systems (18), and the most significant phenotypic characteristic identified in three E. coli strains associated with bacterial bloom events in two Australian lakes was the formation of a thick extracellular matrix, or capsule (23). Escherichia strains of the cryptic clades III, IV and V exhibited a greater degree of biofilm formation than did strains of E. albertii, E. coli or E. fergusonii, so it appears that this phenotype has been conserved among these distantly related lineages.
The thermal tolerance exhibited by species is a key indicator of a species' ecological niche. In the Enterobacteriaceae there is a distinct separation in the thermal profiles of host-associated species compared to free-living species (14, 20). Free-living bacteria, such as Buttiauxella agrestis and Rahnella aquatilis, are frequently isolated from a variety of water sources (14). These organisms are able to grow at temperatures as low as 1°C and have an optimal temperature for growth of 25°C, and, as a result, these species are classed as psychrotrophs (14, 24). In contrast, predominantly host-associated species, such as E. coli and Citrobacter freundii, have thermal limits ranging from 10°C to 45°C (20). These mesophilic species have evolved to temperatures that do not frequently occur in the secondary habitat, but span the range of body temperatures of birds and eutherian mammals. Members of clades III, IV, and V are able to replicate at lower temperatures than strains of the other Escherichia spp. However, members of these cryptic clades have retained their ability to grow at higher temperatures (Table 2). Thus, clades III, IV, and V break the general trend within the Enterobacteriaceae since it appears that their thermal tolerance enables them to cope with a wider range of temperatures, like those found in both the primary and secondary habitats.
No significant differences between the optimal growth temperatures or maximum growth temperatures of the various Escherichia clades were detected. Consequently, it appears that Escherichia clades III, IV, and V have extended their thermal niche without sacrificing their fitness at higher temperatures. The lack of any apparent trade-off between growth at high and low temperatures contrasts with the results of long-term evolution experiments that have shown such a trade-off to occur in E. coli laboratory populations (1, 22). Bacteria grown for 2,000 generations at lower temperatures demonstrated a loss of fitness in terms of growth rate at high temperatures relative to strains that had been maintained at higher temperatures.
There are a number of explanations for the lack of any observed trade-off in thermal tolerance. It is likely that the cryptic clades are still dependent on a host population, meaning that compensatory mutations occurred elsewhere in the genomes of these strains so that any fitness costs due to low temperature adaptation were overcome. Finally, it may be that the cryptic clades represent the ancestral Escherichia phenotype and that by adapting to warmer host environments, contemporary E. coli, E. fergusonii, and E. albertii strains lost the ability to grow at lower temperatures. The most appropriate explanation is likely to become clear as more data are collected from representative strains of known and yet-to-be-discovered Escherichia clades.
The enhanced ability of the three cryptic Escherichia clades to form biofilms and to replicate at lower temperatures relative to the other named Escherichia species supports the hypothesis that they are better adapted to secondary habitats that strains of E. coli. However, these strains did not have an increased ability to survive in the complete absence of nutrients (Table 3), suggesting that this type of selective pressure was not important during the evolution of these lineages. It could be that although nutrient concentrations are often lower in the secondary habitat compared to the primary habitat, the amount of available nutrients may be sufficient to allow cell division to replace the losses due to cell death. Future studies focused on nutrient scavenging and not persistence when nutrients are absent may shed light on this hypothesis.
Strains of the cryptic lineages possess an unusual mix of virulence traits, and the virulence profiles are neither typical of extraintestinal pathogens, such as those found in phylogroup B2, nor similar to those found in intestinal pathogens, such as those belonging to phylogroup E. Their avirulent phenotype in a mouse model of septicemia suggests that they have little potential to cause extraintestinal infection, despite the presence of important extraintestinal virulence determinants such as the capsule and the high pathogenicity island (screened with the fuyA gene). This observation accords with the fact that strains belonging to the cryptic clades are rarely isolated from extraintestinal sites in humans and, when isolated, the patients are usually immunocompromised patients (data not shown). These strains can thus be considered opportunistic pathogens. Interestingly, E. albertii and E. fergusonii are also not virulent in mice. This suggests that extraintestinal virulence has emerged relatively recently in the history of the genus and is largely restricted to E. coli strains, especially phylogroup B2 strains (21), where a fine-tuning between the genetic background and the virulence determinants allows the expression of virulence (7). There is a low frequency of antibiotic resistance in the cryptic lineages, and this also accords with the observation that strains belonging to the cryptic lineages are more likely to be isolated from birds and nonhuman mammals than from humans, the host group more likely to be exposed to antibiotics (29). E. coli isolated from native Australian vertebrates has a low frequency of resistance to antibiotics compared to humans living in Australia (27), and this pattern holds for strains belonging to other members of the genus Escherichia. The high frequency of tetracycline resistance among clade V strains from France probably reflects, compared to Australia, France's greater human and domestic animal population densities, and the widespread use of tetracycline in veterinary medicine.
All of the available evidence indicates that members of the cryptic Escherichia clades are rarely isolated from vertebrate hosts (32). Less than 3% of hosts examined (humans and animals) using conventional isolation techniques carried strains of these lineages (O. Clermont, D. M. Gordon, S. Brisse, S. T. Walk, and E. Denamur, unpublished data). Although we cannot discount the possibility that members of these clades are abundant in some rarely examined host group (e.g., molluscs or insects), they are certainly uncommon among vertebrate hosts. Like the cryptic clades, little is known concerning the distribution of E. albertii and E. fergusonii. These species have never been detected as a dominant member of the aerobic community of fish, frogs, reptiles, or mammals living in Australia, and they are rarely isolated from most Australian bird species (19). However, both species can be quite common in certain host species and, for example, E. albertii has been isolated from one in five clinically normal chickens (19), whereas E. fergusonii can represent one in six clinical isolates from humans (16). In stark contrast to the other Escherichia clades, E. coli can be detected in most individuals of most mammalian species (9).
There is some evidence to suggest that E. coli is competitively dominant to other members of the Enterobacteriaceae in the lower intestinal tract of mammals (10). The rarity of the cryptic Escherichia clades in vertebrate hosts, particularly humans, suggests that these strains are competitively inferior to E. coli in the gastrointestinal tracts of many mammals. However, biofilm production and growth at lower temperatures may increase the persistence of these organisms in the secondary habitat and allow for better transmission between hosts compared to E. coli. Consequently, E. coli may dominate the primary habitat (26), but this dominance may not translate to the secondary habitat (26). In such situations, mathematical models describing the population dynamics of competing bacterial strains have shown that there is a range of conditions under which strains with the characteristics of E. coli and the cryptic clades can stably coexist (5).
The results of the present study have revealed that strains of different clades of Escherichia vary in their ecological characteristics, specifically their ability to form biofilms and replicate at low temperatures, but are sensitive to most antibiotics and avirulent in a mouse model of extraintestinal infection. Further studies are needed to quantify other ecologically relevant traits in order to further characterize potentially important persistence phenotypes in the secondary habitat.
ACKNOWLEDGMENTS
We are grateful to Louis Garry for his help in the mouse virulence assay.
This study has been partially funded by the Alliance for the Prudent Use of Antibiotics in the frame of the Reservoirs of Antibiotic Resistance projects 2006-2007 obtained by E.D. and D.M.G.
Footnotes
Published ahead of print on 18 February 2011.
REFERENCES
- 1. Bennett A. F., Lenski R. E. 2002. Evolutionary adaptation to temperature. 2. Thermal niches of experimental lines of Escherichia coli. Evolution 47:1–12 [DOI] [PubMed] [Google Scholar]
- 2. Chaudhury A., Nath G., Tikoo A., Sanyal S. C. 1999. Enteropathogenicity and antimicrobial susceptibility of new Escherichia spp. J. Diarrhoeal Dis. Res. 17:85–87 [PubMed] [Google Scholar]
- 3. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappinscott H. M. 1995. Microbial biolofilms. Annu. Rev. Microbiol. 49:711–745 [DOI] [PubMed] [Google Scholar]
- 4. Courpon-Claudinon A., et al. 23 July 2010. Bacteremia caused by third-generation cephalosporin-resistant Escherichia coli in France: prevalence, molecular epidemiology and clinical features. Clin. Microbiol. Infect. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 5. Davis S. A., Gordon D. M. 2002. The influence of host dynamics on the clonal composition of Escherichia coli populations. Environ. Microbiol. 4:306–313 [DOI] [PubMed] [Google Scholar]
- 6. Diard M., et al. 2007. Caenorhabditis elegans as a simple model to study phenotypic and genetic virulence determinants of extraintestinal pathogenic Escherichia coli. Microbes Infect. 9:214–223 [DOI] [PubMed] [Google Scholar]
- 7. Escobar-Páramo P., et al. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085–1094 [DOI] [PubMed] [Google Scholar]
- 8. Farmer J. J., et al. 1985. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon D. M., Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586 [DOI] [PubMed] [Google Scholar]
- 10. Gordon D. M., FitzGibbon F. 1999. The distribution of enteric bacteria among Australian mammals. Microbiology 145:2663–2671 [DOI] [PubMed] [Google Scholar]
- 11. Gordon D. M., Stern S. E., Collignon P. J. 2005. The influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151:15–23 [DOI] [PubMed] [Google Scholar]
- 12. Huys G., Cnockaert M., Janda J. M., Swings J. 2003. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 53:807–810 [DOI] [PubMed] [Google Scholar]
- 13. Johnson J. R., et al. 2006. Experimental mouse lethality of Escherichia coli isolates in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150 [DOI] [PubMed] [Google Scholar]
- 14. Leclerc H., Mossel D. A. A., Edberg S. C., Struijk C. B. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201–234 [DOI] [PubMed] [Google Scholar]
- 15. Liao J. J. Z., Duan F. 2006. Calibrating the concentration from a serial dilution process. J. Chemometrics 20:294–301 [Google Scholar]
- 16. Mahapatra A., Mahapatra S., Mahapatra A. A. 2005. Escherichia fergusonii: an emerging pathogen in South Orissa. Indian J. Med. Microbiol. 23:204. [DOI] [PubMed] [Google Scholar]
- 17. Meng J., Schroeder C. M. 2007. Escherichia coli, p. 1–26 In Simjee S. (ed.), Foodborne diseases, 2007. Humana Press, Totowa, NJ [Google Scholar]
- 18. Murphy H. M., Payne S. J., Gagnon G. A. 2008. Sequential UV- and chlorine-based disinfection to mitigate Escherichia coli in drinking water biofilms. Water Res. 42:2083–2092 [DOI] [PubMed] [Google Scholar]
- 19. Oaks J. L., et al. 2009. Infection and mortality in wild and domestic birds due to Escherichia albertii: description of an emerging avian pathogen. Emerg. Infect. Dis. 16:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okada S., Gordon D. M. 2001. Host and geographical factors influence the thermal niche of enteric bacteria isolated from Australian mammals. Mol. Ecol. 10:2499–2513 [DOI] [PubMed] [Google Scholar]
- 21. Picard B., et al. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pörtner H. O., et al. 2006. Trade-offs in thermal adaptation: the need for a molecular to ecological integration. Physiol. Biochem. Zool. 79:295–313 [DOI] [PubMed] [Google Scholar]
- 23. Power M. L., Littlefield-Wyer J., Gordon D. M., Veal D. A., Slade M. B. 2005. Phenotypic and genotypic characterisation of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631–640 [DOI] [PubMed] [Google Scholar]
- 24. Regha K., Satapathy A. K., Ray M. K. 2005. RecD plays an essential function during growth at low temperature in the Antarctic bacterium Pseudomonas syringae Lz4W. Genetics 170:1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reisner A., Krogfelt K. A., Klein B. M., Zechner E. L., Molin S. 2006. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J. Bacteriol. 188:3572–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Savageau M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732–744 [Google Scholar]
- 27. Sherley M., Gordon D. M., Collignon P. J. 2000. Variations in antibiotic resistance profile in Enterobacteriaceae isolated from wild Australian mammals. Environ. Microbiol. 2:620–631 [DOI] [PubMed] [Google Scholar]
- 28. Skurnik D., et al. 2005. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents Chemother. 49:3062–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skurnik D., et al. 2006. Effect of human vicinity on antimicrobial resistance and integrons in animal fecal Escherichia coli. J. Antimicrob. Chemother. 57:1215–1219 [DOI] [PubMed] [Google Scholar]
- 30. Tenaillon O., Skurnik D., Picard B., Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 31. Walk S. T., Alm E. W., Calhoun L. M., Mladonicky J. M., Whittam T. S. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274–2288 [DOI] [PubMed] [Google Scholar]
- 32. Walk S. T., et al. 2009. Cryptic lineages of the Escherichia genus. Appl. Environ. Microbiol. 75:6534–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]