Abstract
The fecal microbiome of cattle plays a critical role not only in animal health and productivity but also in food safety, pathogen shedding, and the performance of fecal pollution detection methods. Unfortunately, most published molecular surveys fail to provide adequate detail about variability in the community structures of fecal bacteria within and across cattle populations. Using massively parallel pyrosequencing of a hypervariable region of the rRNA coding region, we profiled the fecal microbial communities of cattle from six different feeding operations where cattle were subjected to consistent management practices for a minimum of 90 days. We obtained a total of 633,877 high-quality sequences from the fecal samples of 30 adult beef cattle (5 individuals per operation). Sequence-based clustering and taxonomic analyses indicate less variability within a population than between populations. Overall, bacterial community composition correlated significantly with fecal starch concentrations, largely reflected in changes in the Bacteroidetes, Proteobacteria, and Firmicutes populations. In addition, network analysis demonstrated that annotated sequences clustered by management practice and fecal starch concentration, suggesting that the structures of bovine fecal bacterial communities can be dramatically different in different animal feeding operations, even at the phylum and family taxonomic levels, and that the feeding operation is a more important determinant of the cattle microbiome than is the geographic location of the feedlot.
INTRODUCTION
The enteric microbiota of cattle affects animal health and food safety and is used as an indicator of fecal pollution, which can affect the types and concentrations of indicator organisms in recreational surface waters. The presence of pathogenic bacteria such as Escherichia coli O157:H7 in the bovine gastrointestinal tract has been linked to disease outbreaks due to the consumption of contaminated beef, milk, and drinking water (3). The average feedlot steer produces 1.62 kg of feces (dry matter) per day (2), resulting in more than 18 million metric tons of feces (dry matter) per year in the United States alone. When bovine fecal waste is moved from feedlot operations for land application as fertilizer or is accidentally discharged into the environment due to severe storms, hazardous events, or failure of onsite waste management practices, pathogenic members of this microbial community, such as E. coli O157:H7, Campylobacter jejuni, Salmonella spp., Leptospira interrogans, and Cryptosporidium parvum (5, 14, 22, 41, 44), can pose a serious public health risk.
Because of the enormous influence the fecal bacterial community of cattle has on the beef and dairy industry, the economy, and public health, a great deal of research has been conducted to characterize the effects of animal age, disease state, feeding practices, and antibiotic treatments on cattle fecal microorganisms. Many of the most comprehensive studies use DNA-based methodologies, such as sequencing of the full-length 16S rRNA gene (19, 37) and competitive hybridization (23, 48), to characterize bacterial communities. Next-generation pyrosequencing allows for the cost-efficient processing of hundreds of thousands of sequence reads in a single instrument run, enabling the characterization of both abundant and rare community members. The ability to produce detailed profiles based on next-generation sequencing of PCR amplicons from complex microbial communities of drinking water distribution systems (24), wastewater treatment systems (38, 45), sewage biosolids (6), and soils (34) has highlighted the benefits of this approach.
A recent microbial population study employed next-generation pyrosequencing technology to characterize fecal samples from 20 individuals from a single population of dairy cattle (18). A 600-bp 16S rRNA fragment was sequenced using eubacterial primers generating 1,732 to 3,224 pyrotags per fecal sample. That study successfully identified the dominant bacteria associated with commercial lactating dairy cattle, and 13 core genera were found among all animals tested in the study. Our work expands on that pyrosequencing-based study to address the following questions: (i) which taxa are most abundant in cattle feces, (ii) how much variability in bacterial community structure exists between individual animals from the same population, (iii) whether cattle fecal bacterial community structures are modulated by different animal management practices, and (iv) whether the concentration of starch in feces is a predictor of bacterial community structure.
MATERIALS AND METHODS
Fecal sample collection from cattle and DNA extraction.
Thirty rectal fecal samples were collected from six different cattle populations (5 individual samples per population) residing in four different geographic locations (Oconee County, GA; Larimer County, CO; Hamilton County, OH; and Clay County, NE) according to institutional animal care guidelines. All animals were adult beef cattle and had been subjected to their respective cattle feeding practices for a minimum of 90 days prior to sampling. Cows were visibly healthy, and no illnesses among these animals were reported subsequent to sample collection. Cattle populations were organized into three management groups: (i) the forage group, where >80% of feed consisted of plant material such as alfalfa, corn silage, and/or fescue, wheat, and rye grasses (the NE1 and USDA populations); (ii) the processed-grain group, where at least 75% of rations comprised steam-flaked, dry-rolled, and/or distiller's grain corn (CO1 and CO2); and (iii) the unprocessed-grain group, containing >75% whole-kernel corn (DK and NE2). Immediately following sample collection, fecal samples were sealed in sterile polypropylene containers, frozen to −20°C, and shipped overnight to Cincinnati, OH, for DNA extraction. Fecal samples were stored at −80°C for no longer than 12 weeks prior to DNA extraction. All DNA extractions were performed with the FastDNA kit for soils (Q-Biogene, Carlsbad, CA). The concentration and purity of each DNA extract were determined using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Determination of fecal starch concentrations.
Approximately 100 g (wet weight) of each fecal sample was shipped overnight to the Department of Animal Sciences of Colorado State University in Fort Collins for fecal starch measurements. The concentration of starch in each fecal sample was measured using an alpha-linked glucose polymer method (36). Triplicate measurements were performed for each sample.
DNA pyrosequencing.
Purified DNA from each fecal sample served as a template for the preparation of amplicon libraries from the hypervariable V6 region of the 16S rRNA coding region prior to pyrosequencing on a Roche GS FLX system. To help capture the full diversity of bacterial rRNA sequences currently described in molecular databases, a cocktail of five primers at the 5′ end and four primers at the 3′ end directed the amplification of V6 rRNA regions (26, 52). Fused primers that contained a unique five-nucleotide barcode between the 454 adapter-A sequence and the V6 rRNA primers were used to multiplex samples. Triplicate reactions for each PCR library minimized the impact of PCR errors generated during early cycles of amplification. The resulting amplicon libraries were purified with a Qiagen (Valencia, CA) MinElute PCR purification kit and were visualized on an Agilent Bioanalyzer, model 2100. Emulsion PCR was performed using the Roche protocols. The mixture of 16 different samples was then deposited in a picotiter plate for GS-FLX pyrosequencing. The selection of the V6 region for pyrotag analysis took advantage of (i) the availability of primers allowing low-bias coverage of most previously described bacteria (26, 52), (ii) the availability of a large database of V6 sequences from previous studies (1, 16, 26, 31, 52, 55, 58), and (iii) the small size of V6 amplification products (<120 nucleotides), which practically eliminates the likelihood of formation of chimeric molecules, minimizes the probability of accumulating sequencing errors in a single sequence read, and generates fewer high-order structures that may retard enzyme processivity (25, 54).
Quality trimming, taxonomic assignments, and operational taxonomic unit clustering.
Each pyrotag sequence read underwent a series of quality filters to trim and/or remove poor-quality data. Pyrotags were removed from the data set if the read contained any ambiguous base, if the read length was less than 50 nucleotides (the length of the shortest V6 sequence in the reference base), if the average quality score was less than 30, if there were any errors in the proximal primer sequence or barcode, or if the distal primer was not present (29). High-quality pyrotags were then binned into original sample groups using the five-nucleotide barcode keys and were assigned to taxa using the Global Assignment of Sequence Taxonomy (GAST) approach (28) and the SILVA database (43). Any sequences not identified as of bacterial origin, because they were either nonbacterial microbes, organelles, or of low quality, were excluded from all further analyses. The bacterial pyrotags were then clustered into operational taxonomic units (OTUs) at a 3% threshold using a method of single-linkage preclustering with average-linkage clustering (30).
Estimating richness.
The richness of each microbial community was estimated using the ACE (12), Chao1 (11), and CatchAll (8) methods. Rarefaction curves were generated using mothur (46). A Fitch tree was constructed using PHYLIP (21) to compare Yue-Clayton distances within and between individual fecal sample microbial populations.
Network analysis.
A network analysis approach was used to characterize the influences of geographic locale, management group, and fecal starch concentration on overall microbial community OTU structure and selected phyla, including Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. A total of 14,992 pyrotags (corresponding to the number of pyrotags in the smallest data set) were randomly selected from each DNA sample library and were used to assign OTUs at a 3% width to normalize data sets among all samples. Each bacterial network was visualized with Cytoscape, version 2.7 (50), using the edge-weighted spring embedded model (20). Edge weights were allowed to affect spring forcings based on the abundance of each OTU determined from normalized data, which was graphically illustrated by the thickness of the respective line in each plot. All network analyses were performed with reduced data sets to aid in pattern visualization. In the majority of analyses, all high-quality sequences were binned into OTUs as described above and were then sorted by OTU abundance (from most abundant to least abundant) for each fecal sample. A running sum of total sequences for each fecal sample was calculated, and only the OTUs present in the top 95% of the respective fecal sample data set were kept for further analyses. OTUs present in the top 95% of one fecal sample but not in another were added back into all samples. This data reduction method is referred to below as the 95% data set. In order to compare data sets containing rare OTUs to data sets containing only the most abundant OTUs, a second reduction method using the top 25% of sequences in each fecal sample, filtered as described above, was applied (referred to as the 25% data set).
The statistical significance of the network sample distribution patterns was assessed with t tests comparing within-group versus among-group plot distances for each defined grouping scheme (geographic locale, management group, and fecal starch concentration). Fecal starch concentrations from all 30 samples were categorized into four groups: ≤0.1 g/100 g feces (n = 9), 0.1 to 1.0 g/100 g feces (n = 9), 1.0 to 2.0 g/100 g feces (n = 5), and ≥2.0 g/100 g feces (n = 7).
Other statistical analyses.
Simple linear regression analysis was used to determine correlations between the relative abundances of key phyla and fecal starch concentrations with SigmaPlot, version 11 (Systat Software, Inc.).
RESULTS
Quality trimming of pyrosequencing tags.
Pyrosequencing of 30 cattle fecal samples yielded 732,096 pyrotags with read lengths ranging from 51 to 81 bp. Trimming filters identified 97,587 pyrotags as poor quality (see Table S1 in the supplemental material); these were removed from the data set, resulting in 634,509 high-quality pyrotags. An additional 622 pyrotags (0.08% of all reads) did not display significant matches to V6 rRNA regions in the SILVA database (43), presumably because they have no valid match to the reference database or they correspond to chimeras or to nonribosomal genomic regions. Of all high-quality bacterial V6 pyrotags (n = 633,877), GAST successfully annotated 99.7% to the phylum level, 99.4% to the class level, 96.1% to the order level, 94.8% to the family level, 49.2% to the genus level, and 0.7% to the species level.
Cluster analysis.
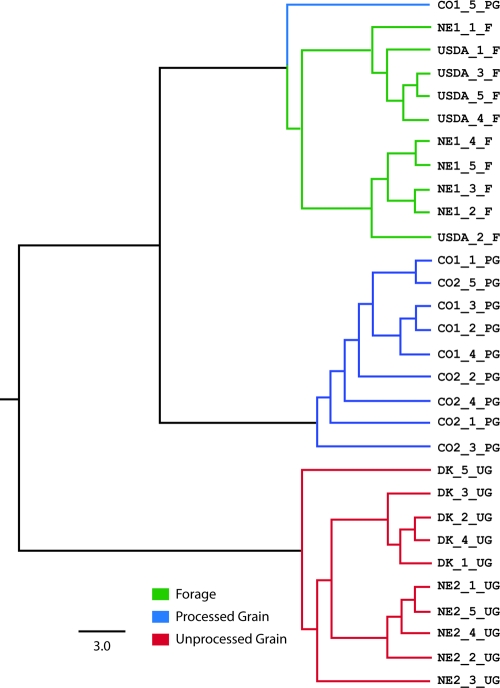
A Fitch tree based on Yue-Clayton distances between samples calculated at the phylum level indicates that individual fecal samples cluster by animal management grouping assignments (Table 1), except for sample CO1_5_PG (Fig. 1). This observation was used as justification for combining data sets into management groupings in subsequent analyses.
Table 1.
Cattle fecal sample collection and management classificationa
| Population | Locale | Management group | Feeding regimenb |
|||
|---|---|---|---|---|---|---|
| % Grain | % Forage | Supplement | Ionophore | |||
| CO1 | CO | Processed grain | 80 | 15 | Yes | Yes |
| CO2 | 75 | 15 | Yes | Yes | ||
| DK | OH | Unprocessed grain | 94 | <1 | Yes | Yes |
| NE2 | NE | 76 | 20 | Yes | Yes | |
| NE1 | Forage | 0 | 100 | No | No | |
| USDA | GA | 15 | 80 | Yes | Yes | |
All samples were collected in 2008.
% Grain, the proportion of feed made up of steam-flaked, dry-rolled, whole-kernel, and/or distiller's grain corn; % forage, the proportion of feed made up of plant material commonly used for feed, such as alfalfa, corn silage and/or fescue, wheat, and rye grasses; supplement, a feed additive typically containing vitamins, minerals, urea, limestone, and salt; ionophore, an antimicrobial agent supplement including monensin or tylosin.
Fig. 1.
Fitch tree of distance estimates between individual fecal samples. Green branches mark samples from the forage management group. Blue and red branches mark samples from the processed- and unprocessed-grain groups, respectively.
Trends in pyrotag taxon annotation and clustering.
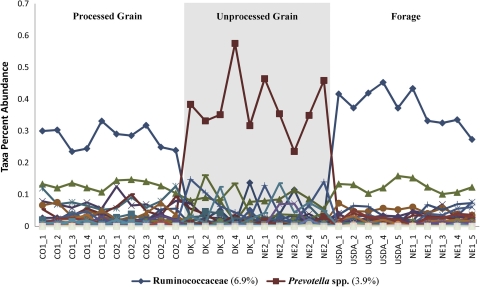
Of the 512 taxa defined by GAST, only 30 were shared across all individual data sets, making up 24.5% of all high-quality bacterial V6 pyrotags. Shared taxa spanned 5 different phyla: Actinobacteria (0.11% of all pyrotags; 0.67% of shared taxa), Bacteroidetes (5.7% of all; 13.3% of shared taxa), Cyanobacteria (0.08% of all; 3.33% of shared taxa), Firmicutes (17.5% of all; 73.3% of shared taxa), and Tenericutes (0.96% of all; 3.33% of shared taxa). The most abundant taxa included Ruminococcaceae (6.9% of all), Prevotella spp. (3.9%), Lachnospiraceae (3.2%), Peptostreptococcaceae (1.3%), and Turicibacter spp. (1.1%). A comparison of the percentages of abundance of all 512 taxa across each individual sample data set organized by management groups indicates that the majority of the taxa (99.6%; n = 510) do not fluctuate across different dietary practices. However, two taxa, Ruminococcaceae (6.9% of all high-quality bacterial V6 pyrotags) and Prevotella spp. (3.9%), demonstrate distinct patterns in response to feed rations (Fig. 2).
Fig. 2.
Multiple-line scatter plot showing percentages of GAST taxon abundance across all 30 individual samples organized by population and management groupings. Lines are coded by color and symbol for each GAST taxon assignment. A key for the two taxa that exhibit a management grouping response in the percentage of abundance is given below the plot. The overall percentage of abundance across the entire data set for each of these two taxa is given in parentheses.
Of the 9,201 OTUs (with a 3% difference level) identified with single-linkage preclustering, only 9 were shared among all individual sample data sets, making up 19.6% of all high-quality bacterial V6 pyrotags. All shared OTUs were from the Firmicutes phylum; they comprised 5 OTUs at the family level, including Peptostreptococcaceae (3 OTUs; 28,315 pyrotags) and Ruminococcaceae (2 OTUs; 66,392), and 4 OTUs at the genus level, consisting of Oscillibacter spp. (2 OTUs; 4,629 pyrotags), Clostridium spp. (1 OTU; 2,101 pyrotags), and Turicibacter spp. (1 OTU; 23,035 pyrotags).
Richness estimates.
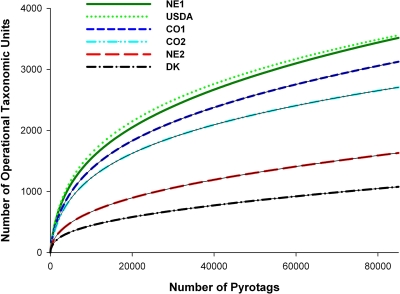
ACE, Chao1, and CatchAll values were calculated to estimate the minimum number of different OTUs in each population of cattle (Table 2) at a 3% width. The number of OTUs that were unique to a population ranged from 9,758 to 15,179. Figure 3 shows rarefaction curves for each cattle population generated from OTUs assigned at a 3% width.
Table 2.
Summary of V6 pyrotag counts and richness estimates
| Management group and population | No. of V6 tags |
Richness estimateb (95% CI) by the following method: |
|||
|---|---|---|---|---|---|
| Totala | Unique | Chao1 | ACE | CatchAll | |
| Processed grain | |||||
| CO1 | 110,050 | 15,179 | 5,622 (5,307–5,990) | 6,434 (6,307–6,566) | 9,956 (8,345–12,095) |
| CO2 | 101,834 | 11,613 | 4,297 (4,069–4,567) | 4,737 (4,640–4,840) | 6,206 (5,538–7,042) |
| Unprocessed grain | |||||
| DK | 116,932 | 9,758 | 2,485 (2,223–2,818) | 3,718 (3,563–3,884) | 4,044 (3,386–4,906) |
| NE2 | 95,884 | 10,011 | 3,293 (2,992–3,665) | 3,976 (3,867–4,091) | 6,212 (4,894–8,078) |
| Forage | |||||
| NE1 | 94,793 | 14,448 | 5,643 (5,367–5,962) | 6,536 (6,413–6,664) | 7,967 (7,300–8,755) |
| USDA | 114,384 | 14,995 | 5,806 (5,543–6,112) | 6,386 (6,273–6,504) | 8,895 (7,869–10,188) |
Trim reads that passed quality controls.
Minimum number of different OTUs in each population of cattle, determined with a 3% width. CI, confidence interval.
Fig. 3.
Rarefaction curves for fecal bacterial communities. Each curve represents a single fecal sample. Curves are color coded based on animal management practice; red, green, and black lines indicate unprocessed grain, processed grain, and forage, respectively. OTUs are estimated at a 3% difference level.
Starch concentration gradient.
Starch concentrations in feces ranged from 0 to 21.9 g/100 g of manure. Cattle populations exhibited a gradient of starch concentrations, with mean starch concentrations (in grams per 100 grams of feces [dry weight]), from highest to lowest, as follows: DK, 11.13 ± 6.4; NE2, 2.75 ± 2.2; CO1, 0.89 ± 0.56; CO2, 0.86 ± 0.52; NE1, 0.09 ± 0.14; USDA, 0.02 ± 0.03.
Roles of geographic locale, management group, and fecal starch concentration.
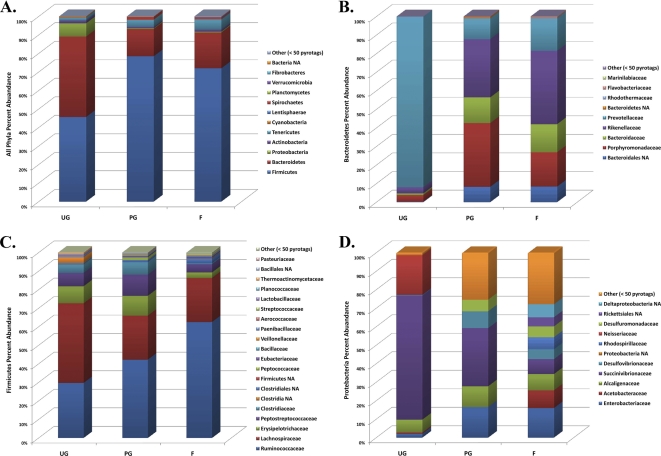
Several strategies were employed to explore the impact of geographic locale, management group, and fecal starch concentration on cattle fecal microbial community structure. The stacked histograms in Fig. 4A depict the frequencies of different phylum level OTUs defined at a 3% width, organized by management group. These results demonstrate that Firmicutes and Bacteroidetes are the most abundant phyla and that their percentages of abundance change dramatically from the forage and processed-grain groups to the unprocessed-grain group. Examination of OTUs constituting the Bacteroidetes, Firmicutes, and Proteobacteria at a family taxonomic level illustrates the shifts in population structure within each phylum (Fig. 4, panels B, C, and D, respectively).
Fig. 4.
Stacked histograms depicting the relative abundances of high-quality bacterial V6 pyrotags in different phyla for each animal management group: unprocessed grain (UG), processed grain (PG), and forage (F). Shown are the relative abundances of taxa within all phyla (A) and within the Bacteroidetes (B), Firmicutes (C), and Proteobacteria (D) based on family taxonomic ranks. OTUs are estimated at a 3% difference level. NA, no assignment.
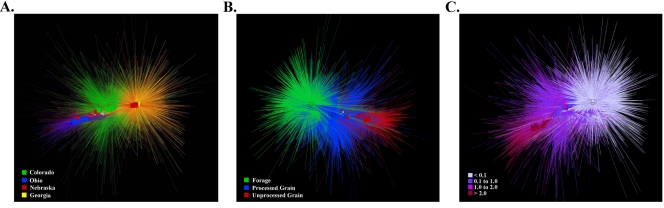
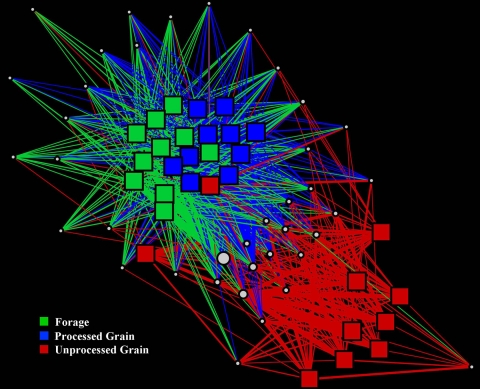
Network analyses of the 95% data set were performed to visualize the effects of the cattle populations' geographic locations (by state) (Fig. 5A), animal management groupings (processed grain, unprocessed grain, or forage) (Fig. 4B), and fecal starch concentrations (Fig. 5C). For a complete description of geographic locale and management grouping schemes, refer to Table 1. OTUs were organized into four groups for Fig. 4C based on fecal starch concentrations: <0.1, 0.1 to 1.0, 1.0 to 2.0, and >2.0 g/100 g dry feces. An additional network analysis was performed with the 25% data set to determine whether differences in management grouping are more closely associated with the more dominant OTUs than with the less-abundant and rare community members (Fig. 6). Network analysis of the 25% data set revealed no clear separation between the forage and processed-grain management group samples, although the unprocessed-grain samples showed a difference from the other two.
Fig. 5.
Network analysis of the top 95% of all high-quality bacterial V6 pyrotags after binning into OTUs and sorting by OTU abundance (from most abundant to least abundant), categorized by the geographic origin of the sample (A), the animal management grouping (B), and the fecal starch gradient (C). Squares represent fecal samples, and circles represent individual OTUs. The size of each circle indicates the OTU abundance. The line color indicates the presence of an OTU in a sample category, and the line width indicates the abundance of an OTU in a sample.
Fig. 6.
Network analysis of the top 25% of all high-quality sequences after binning into OTUs and sorting by OTU abundance (from most abundant to least abundant), categorized by animal management practice. Squares represent fecal samples, and circles represent individual OTUs. The size of a circle indicates the OTU abundance. The line color indicates the presence of an OTU in a sample category, and the line width indicates the abundance of an OTU in a sample.
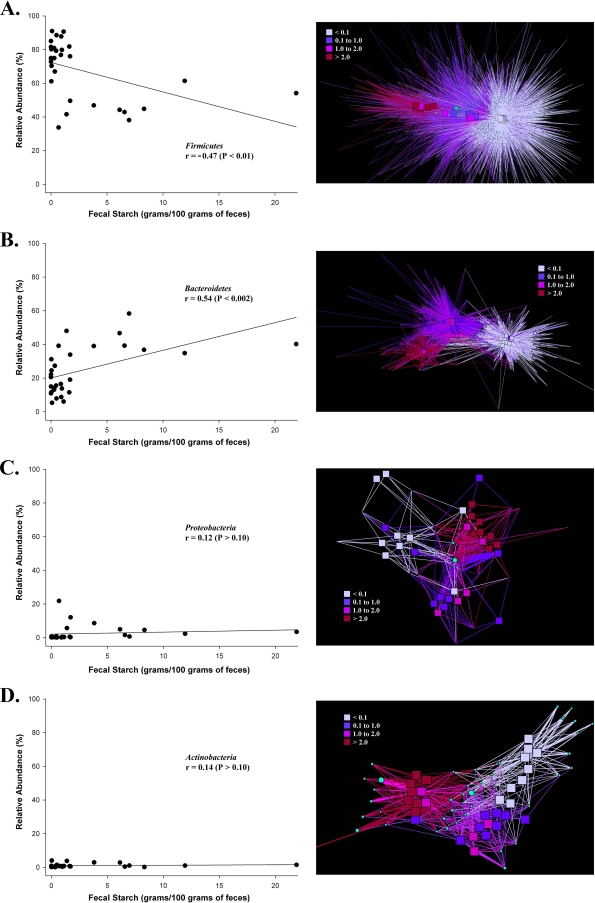
To characterize shifts in Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria populations among the different management groupings, two different analyses were performed. First, the fecal starch concentrations of samples were compared to the relative abundances of OTUs representative of each phylum by using simple linear regression analysis (Fig. 7). The regression plots indicate two different correlations: a positive response, where the relative abundance of Bacteroidetes increases with the fecal starch concentration (Fig. 7B), and a negative response, where the abundance of Firmicutes decreases with increasing fecal starch concentrations (Fig. 7B). No significant relationship was observed for the relative abundances of Proteobacteria and Actinobacteria (P > 0.10) across the fecal starch gradient (Fig. 7C and D). Second, a network analysis of the 95% data set within each phylum was conducted in order to visualize trends in population structure (Fig. 7).
Fig. 7.
Responses of Firmicutes (A), Bacteroidetes (B), Proteobacteria (C), and Actinobacteria (D) populations to fecal starch concentrations. Scatter plots depict the relative abundance of the phylum plotted against the fecal starch concentrations of the respective samples. The correlation coefficient (r) based on a simple linear regression model is given in each graph. Corresponding bacterial networks represent the top 95% of each phylum's high-quality sequences after binning and sorting by OTU abundance (from most abundant to least abundant), categorized by fecal starch concentration. Squares represent fecal samples, and circles represent individual OTUs. The size of a circle indicates the OTU abundance. The line color indicates the presence of an OTU in a sample category, and the line width indicates the abundance of an OTU in a sample.
DISCUSSION
General characteristics of bovine fecal microbial communities.
Deep sequencing of 30 individual fecal samples collected from six different cattle populations provides a detailed view of the cattle fecal microbiome for the samples tested. We detected members of 10 phyla of Bacteria (OTU count, >100) based on OTU assignments by single-linkage preclustering. The majority of the pyrotags belong to the Firmicutes (55.2% of 633,877 high-quality bacterial V6 pyrotags), Bacteroidetes (25.4%), Tenericutes (2.9%), and Proteobacteria (2.5%). These phyla were previously shown to constitute the majority of gut-associated phylotypes in a variety of different mammalian species (7, 18, 19, 35), suggesting that Firmicutes and Bacteroidetes (79.6% of all high-quality bacterial V6 pyrotags) in particular play a critical role in the microbial ecology of the mammalian gut, including the bovine gut. Other phyla represented were the Actinobacteria (0.73%), Spirochaetes (0.54%), Verrucomicrobia (0.19%), Cyanobacteria (0.15%), Fibrobacteres (0.02%), and Lentisphaerae (0.02%). Even though all of the bacterial phyla contain a diverse range of taxa, the metabolic potential of some phyla most likely allows some to dominate in bovine feces while others remain less abundant.
Useful information can also be gleaned from these data sets through the identification of shared (core) taxa based on GAST. The presence of “core” taxa across all samples implies that these microorganisms perform functions universal to the collective bovine fecal microbiome. A comparison of all taxa (3% width) from this study identified only 30 taxa that were present across the entire sample collection. The abundance of any given “core” taxon ranged from 0.035% to 6.9%, collectively representing 24.5% of all high-quality bacterial V6 pyrotags. The majority of the “core” taxa were classified as Firmicutes and Bacteroidetes (148,018 pyrotags; 23.4% of all high-quality bacterial V6 pyrotags). Members of these families are consistently identified in the bovine rumen and feces (4, 18, 19, 37), suggesting the importance and presence of a defined niche for these microorganisms.
In addition, the characterization of the percentages of abundance of all taxa in the different bovine management groupings (Fig. 2) suggests that the population structures of microorganisms from the family Ruminococcaceae and the genus Prevotella can shift dramatically from one management group to another. Indeed, many members of the Ruminococcaceae and Prevotella spp. are common inhabitants of the gastrointestinal tracts and feces of many mammals, including the rumina of cattle (15, 42, 56). The shifts in the abundances of these microorganisms in bovines fed forage, processed-grain, or unprocessed-grain rations in this study suggest that members of these taxa are much more diverse than initially reported and harbor the metabolic potential to thrive across a broad range of feed sources.
A more complete characterization of bovine fecal bacterial community composition afforded by pyrosequencing can also help address current research gaps concerning odor emissions and the shedding of fecal indicators used for recreational water quality testing. For example, a recent study assessing the performance of Bacteroidales genetic markers associated with bovine fecal pollution reported a dramatic decrease in the abundance of target DNA in animals fed predominately processed-grain rations (49). Pyrosequencing of fecal samples from animals fed unprocessed grains indicates that the abundance of Bacteroidetes OTUs is 2.4 times higher than that in animals fed forage rations (Fig. 4A) and, at first glance, suggests that Bacteroidales bovine-associated genetic markers should be easier to detect, not more difficult. However, a closer examination of the Bacteroidetes OTUs at a family taxonomic level reveals important information (Fig. 4B); the relative abundance of Prevotellaceae in animals fed unprocessed grain rations is more than 10 times higher than that in animals fed exclusively forage rations. In addition, fecal bacterial communities from animals fed unprocessed grains exhibit a severe decrease in community richness (Fig. 3 and Table 2), which is attributed largely to the emergence of the Prevotellaceae OTUs (Fig. 4B). In other words, the reported decrease in bovine-associated genetic marker abundance in animals fed increasing amounts of grain rations (49) is most likely due to a shift in population structure where Prevotellaceae, not Bacteroides spp., dominate the Bacteroidetes population.
Influences of geographic location and management practices.
In order to explore the notion that geographic location and animal management practices potentially influence fecal microbial community structure, bovine samples were organized into groups based on the geographic locale (by state) from which samples were collected or the animal management practice as determined by the percentages of forage and grain rations given (Table 1). It should be noted that groupings are not representative of all geographic locations or cattle feeding practices; instead, these approximate classifications were intended to help identify general trends. Multiple lines of evidence suggest that bovine management practices play an integral role in fecal microbial community structure.
The data suggest that animals subjected to similar management practices are more closely associated with one another than with animals from different groups. In contrast, a network analysis where OTUs from each bovine sample are organized by geographic location (Fig. 5A) clearly indicates that populations from the same geographic location (i.e., NE1 and NE2) subjected to different management practices are not similar (P ≥ 0.10). Instead, similarity between individuals from the same population is most likely due to shared management practices (Fig. 5B) (P ≤ 0.01) rather than to site-specific attributes, such as water source, elevation, humidity, or other factors closely associated with a particular geographic location. The degree of similarity among individuals is somewhat surprising in view of findings from other studies reporting large differences in community structure between individual bovine fecal samples (19). Fecal samples from the same management groupings also exhibit remarkably similar patterns of richness (Fig. 1 and 3; Table 2), suggesting that animal management practices strongly influence fecal microbial ecology. The uniformity in OTU composition from bovine fecal samples collected from the same population and from populations subjected to the same management practices could be extended to other agriculturally important animals, such as swine and poultry, where large numbers of animals are confined to the same place and subjected to similar feed rations, antibiotic treatments, and supplements. This is encouraging for researchers interested in microbially mediated generation of greenhouse gases, odor emission, animal productivity, and food safety, because the respective key metabolic processes are most likely linked to particular management practices rather than to factors associated with geography. On the other hand, this finding also serves as a cautionary note that different management practices can dramatically alter the bacterial community structure and, most likely, metabolic potential.
Starch concentration as a predictor of bacterial community structure.
The digestive tract of a bovine is ideally suited for the fermentation of starch and sugars from fibrous plant materials (27). Bovines themselves do not produce the required fiber-degrading enzymes; instead, they harbor fungi, protozoa, and bacteria that can. Thus, bovine digestive physiology is dictated largely by the presence of fibrous materials in the rumen. If bovines are fed fiber-deficient diets, such as finishing beef grain rations (processed and unprocessed grains), normal digestive processes can be disrupted, leading to the accumulation of fermentation acids, lowering the ruminal pH (32, 47, 51). In turn, these changes in sugar and starch sources, combined with shifts in pH, alter the digestive habitat, resulting in a fecal bacterial community composition that can impact methane production (2), pathogen shedding (17), and the abundance of microorganisms harboring bovine-associated genes used for recreational water quality testing (49).
An important component of many bovine feeding practices is the addition of antimicrobial compounds called ionophores. Ionophores such as monensin and tylosin are commonly fed to feedlot and grazing cattle to improve feed efficiency (10). These antimicrobial compounds have been shown to alter the bacterial community composition of the digestive microbiota, including members of the Bacteroidales group, such as Prevotella ruminicola and Prevotella bryantii (9, 13, 39, 40). One of the most dramatic shifts in population structure documented in this study occurred in the Bacteroidetes phylum, where the abundance of Prevotellaceae OTUs increased by a factor of 10 (Fig. 4B) in animals fed unprocessed grain, suggesting that these bacteria may be sensitive to ionophore treatments. However, both unprocessed-grain populations (DK and NE2) and the USDA population from the forage group had ionophores included in their rations (Table 1). Therefore, it seems unlikely that the presence or absence of an ionophore is the sole explanation for shifts in the population compositions of Firmicutes, Bacteroidetes, and Proteobacteria (Fig. 4).
A more likely explanation for shifts in microbial community structure in grain-fed animals may be the abundance and digestibility of starch in grains compared to that in fibrous plant materials associated with a forage diet. The concentration of starch in feces from unprocessed-grain-fed animals was 67 times higher than that in forage-fed cattle. Fecal starch concentrations from animals fed processed grain were reduced ∼8.5-fold. In addition, network analysis suggests that the ability of cattle to absorb starch during digestion has a direct and predictable impact on the fecal bacterial community structure (Fig. 5C) and that phylum subpopulations respond in different ways across a fecal starch gradient (Fig. 7). For example, network analysis combined with phylum OTU abundance data and starch concentrations indicate that Firmicutes change in composition across a starch gradient (P ≤ 0.01), as well as decreasing in abundance (Fig. 7A). In contrast, Bacteroidetes change in composition (P ≤ 0.01) but increase in abundance (Fig. 7B). Bacteroidetes also exhibit a clear separation between management practices. This is not surprising, considering that members of this phylum are known to benefit their hosts by aiding in the digestion of complex carbohydrates such as starch (33, 53, 57). Actinobacteria exhibit an entirely different behavior pattern: OTU abundance does not change significantly (P ≥ 0.10), but their composition does (P ≤ 0.01), suggesting a different response potential to the starch level in feces (Fig. 7D). Proteobacteria do not change significantly in OTU abundance (P ≥ 0.10) or in composition (P ≥ 0.05) across the starch gradient (Fig. 7C), indicating that other factors are important in structuring this population.
Starch is the major energy component of grains and, as such, the primary source of energy in grain diets. The amount of starch in bovine feces has been linked to odor emissions (2) and to changes in ruminal microbial species composition (7, 32, 47). It should be noted that the grain test populations in this study do not represent all grain types or all processing methods. Further work is required to determine whether particular grain sources combined with specific grain-processing practices, such as moistening, heating, or mechanical pressure, influence the ecology of bovine fecal bacterial communities. Nonetheless, this study indicates that there are fundamental differences between the fecal microbial communities of animals fed different sources of starch and that bacterial phyla respond to a starch gradient in different ways. In addition, the pyrosequencing data from this study should provide important baseline information for further studies to examine other factors, which may also affect bovine fecal microbial community structures.
Utility of deep-sequencing data sets.
The pyrosequencing technique used in this study can provide a robust description of bacterial community structure across a sample set and offers several advantages over traditional full-length sequencing strategies. The most obvious benefit is the ability to process hundreds of thousands of sequence reads in parallel. The depth of sequencing afforded by a pyrosequencing approach allows not only for the characterization of the dominant bacterial community but also for that of the less-abundant and rare community members (52). This study represents the largest bovine fecal barcoding pyrosequencing effort to date in terms of number of individual samples and the depth of sequencing (≥14,992 reads per sample). The added value of characterizing low-abundance community members is clearly illustrated by a comparison of the 95% (Fig. 5B) and 25% (Fig. 6) network data sets. With the 95% data set, which includes 1,716 OTUs in the network, there is a clear disassociation between the forage, processed-grain, and unprocessed-grain management groups (P ≤ 0.05). However, when only the more-abundant OTUs are included (25% data set; 38 OTUs), there is no clear distinction between the forage and processed-grain management groups (P ≥ 0.10), suggesting that it is the less-abundant and possibly rare community members that are responsible for the differences between these two management practices. Based on these findings, it is evident that the continued application of deep-sequencing approaches will promote the discovery of less-abundant and rare community members and will help provide a better understanding of the importance of these microorganisms.
Supplementary Material
ACKNOWLEDGMENTS
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described here. It has been subjected to the Agency's peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Andersson A. F., et al. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Archibeque S. L., Miller D. N., Freetly H. C., Ferrell C. L. 2006. Feeding high-moisture corn instead of dry-rolled corn reduces odorous compound production in manure of finishing beef cattle with decreasing performance. J. Anim. Sci. 84:1767–1777 [DOI] [PubMed] [Google Scholar]
- 3. Armstrong G. L., Hollingsworth J., Morris J. G. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29–51 [DOI] [PubMed] [Google Scholar]
- 4. Balarajan R., et al. 1991. Health risks associated with bathing in sea water. BMJ 303:1444–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benenson A. S. 1995. Control of communicable diseases manual, 16th ed. American Public Health Association, Washington, DC [Google Scholar]
- 6. Bibby K., Viau E., Peccia J. 2010. Pyrosequencing of the 16S rRNA gene to reveal bacterial pathogen diversity in biosolids. Water Res. 44:4252–4260 [DOI] [PubMed] [Google Scholar]
- 7. Brulc J. M., et al. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U. S. A. 106:1948–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bunge J. 2011. Estimating the number of species with CatchAll. Pac. Symp. Biocomput. 2011:121–130 [DOI] [PubMed] [Google Scholar]
- 9. Callaway T. R., Adams K. A., Russell J. B. 1999. The ability of “low G+C gram-positive” ruminal bacteria to resist monensin and counteract potassium depletion. Curr. Microbiol. 39:226–230 [DOI] [PubMed] [Google Scholar]
- 10. Callaway T. R., et al. 2003. Ionophores: their use as ruminant growth promotants and impact on food safety. Curr. Issues Intest. Microbiol. 4:43–51 [PubMed] [Google Scholar]
- 11. Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Statist. 11:265–270 [Google Scholar]
- 12. Chao A., Lee S.-M. 1992. Estimating the number of classes via sample coverage. J. Am. Statist. Assoc. 87:210–217 [Google Scholar]
- 13. Chen M., Wolin M. J. 1979. Effect of monensin and lasalocid-sodium on the growth of methanogenic and rumen saccharolytic bacteria. Appl. Environ. Microbiol. 38:72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Covert T. C. 1999. Salmonella, p. 107–110 In Waterborne pathogens: manual of water supply practices, M48. American Water Works Association, Denver, CO [Google Scholar]
- 15. Dehority B. A., Grubb J. A. 1977. Characterization of the predominant bacteria occurring in the rumen of goats (Capra hircus). Appl. Environ. Microbiol. 33:1030–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dethlefsen L., Huse S. M., Sogin M. L., Relman D. A. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diez-Gonzalez F., Callaway T. R., Kizoulis M. G., Russell J. B. 1998. Grain feeding and the dissemination of acid-resistant Escherichia coli from cattle. Science 281:1666–1668 [DOI] [PubMed] [Google Scholar]
- 18. Dowd S. E., et al. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Durso L. M., et al. 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 76:4858–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eades P. 1984. A heuristic for graph drawing. Congressus Numerantium 42:149–160 [Google Scholar]
- 21. Felsenstein J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle, WA [Google Scholar]
- 22. Fricker C. 1999. Campylobacter, p. 87 In Waterborne pathogens: manual of water supply practices, M48. American Water Works Association, Denver, CO [Google Scholar]
- 23. Galbraith E. A., Antonopoulus D. A., White B. A. 2004. Suppressive subtractive hybridization as a tool for identifying genetic diversity in an environmental metagenome: the rumen as a model. Environ. Microbiol. 6:928–937 [DOI] [PubMed] [Google Scholar]
- 24. Hong P., et al. 2010. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Appl. Environ. Microbiol. 76:5631–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huber J. A., et al. 2009. Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Environ. Microbiol. 11:1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huber J. A., et al. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100 [DOI] [PubMed] [Google Scholar]
- 27. Hungate R. E. 1966. The rumen and its microbes. Academic Press, New York, NY [Google Scholar]
- 28. Huse S. M., et al. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huse S. M., Huber J. A., Morrison H. G., Sogin M. L., Welch D. M. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huse S. M., Welch D. M., Morrison H. G., Sogin M. L. 2010. Ironing out the wrinkles in the rare biosphere. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keijser B. J. F., et al. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 87:1016–1020 [DOI] [PubMed] [Google Scholar]
- 32. Klieve A. V., et al. 2003. Establishing populations of Megasphaera elsdenii YE 34 and Butyrivibrio fibrisolvens YE 44 in the rumen of cattle fed high grain diets. J. Appl. Microbiol. 95:621–630 [DOI] [PubMed] [Google Scholar]
- 33. Kuwahara T., et al. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. U. S. A. 101:14919–14924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ley R. E., et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacRae J. C., Armstrong D. G. 1968. Enzyme methods for determination of α-linked glucose polymers in biological materials. J. Sci. Food Agric. 19:578–581 [Google Scholar]
- 37. McGarvey J. A., Hamilton S. W., DePeters D. J., Mitlehner F. M. 2010. Effect of dietary monensin on the bacterial population structure of dairy cattle colonic contents. Appl. Microbiol. Biotechnol. 85:1947–1952 [DOI] [PubMed] [Google Scholar]
- 38. McLellan S. L., Huse S. M., Mueller-Spitz S. R., Andreishcheva E. N., Sogin M. L. 2010. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol. 12:378–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newbold C. J., Wallace R. J. 1989. Changes in the rumen bacterium, Bacteroides ruminicola, grown in the presence of the ionophore, tetronasin. Asian-Australasian J. Anim. Sci. 2:452–453 [Google Scholar]
- 40. Newbold C. J., Wallace R. J., Walker N. D. 1993. The effect of tetronasin and monensin on fermentation, microbial numbers and the development of ionophore-resistant bacteria in the rumen. J. Appl. Bacteriol. 75:129–134 [DOI] [PubMed] [Google Scholar]
- 41. Okhuysen P. C., Chappell J. H., Crabb J. H., Sterling C. R., DuPont H. L. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275–1278 [DOI] [PubMed] [Google Scholar]
- 42. Orpin C. G., Mathiesen S. D., Greenwood Y., Blix A. S. 1985. Seasonal changes in the ruminal microflora of the high-arctic Svalbard reindeer (Rangifer tarandus platyrhynchus). Appl. Environ. Microbiol. 50:144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pruimboom-Brees I. M., et al. 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc. Natl. Acad. Sci. U. S. A. 97:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sanapareddy N., et al. 2009. Molecular diversity of a North Carolina wastewater treatment plant as revealed by pyrosequencing. Appl. Environ. Microbiol. 75:1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schloss P. D., Handlesman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwartzkopf-Genswein K. S., et al. 2004. Effect of feed delivery fluctuations and feeding time on ruminal acidosis, growth performance, and feeding behavior of feedlot cattle. J. Anim. Sci. 82:3357–3365 [DOI] [PubMed] [Google Scholar]
- 48. Shanks O. C., Santo Domingo J. W., Lamendella R., Kelty C. A., Graham J. E. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shanks O. C., et al. 2010. Performance assessment of cattle-associated PCR and quantitative real-time PCR assays targeting Bacteroidales genes. Appl. Environ. Microbiol. 76:1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shannon P., et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Slyter L. L. 1976. Influence of acidosis on rumen function. J. Anim. Sci. 43:910–929 [DOI] [PubMed] [Google Scholar]
- 52. Sogin M. L., et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Spence C., Wells W. G., Smith C. J. 2006. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: regulation by carbon source and oxygen. J. Bacteriol. 188:4663–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Suzuki M. T., Giovannoni S. J. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Turnbaugh P. J., et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Varel V. H., Dehority B. A. 1989. Ruminal cellulolytic bacteria and protozoa from bison, cattle-bison hybrids, and cattle fed three alfalfa-corn diets. Appl. Environ. Microbiol. 55:148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu J., et al. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076 [DOI] [PubMed] [Google Scholar]
- 58. Zhang H., et al. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.