Abstract
A functional library of cytochrome P450 monooxygenases from Aspergillus oryzae (AoCYPs) was constructed in which 121 isoforms were coexpressed with yeast NADPH-cytochrome P450 oxidoreductase in Saccharomyces cerevisiae. Using this functional library, novel catalytic functions of AoCYPs, such as catalytic potentials of CYP57B3 against genistein, were elucidated for the first time. Comprehensive functional screening promises rapid characterization of catalytic potentials and utility of AoCYPs.
INTRODUCTION
Cytochrome P450 (CYP) enzymes constitute a large superfamily of heme-containing monooxygenases that are distributed among a wide variety of organisms. An evolutionary history of extensive divergence from a common ancestor is implied (14, 20, 28). The majority of CYPs are highly specialized and play crucial roles in meeting the metabolic requirements of particular organisms. They are involved in secondary metabolic pathways, such as detoxification of xenobiotics and synthesis of secondary metabolites (9, 30). Over the last few years, the sequence database of CYPs has greatly enlarged and continues to increase (27, 31). A compilation of CYP sequences could facilitate understanding of metabolic diversity and evolutionary history among living organisms. In addition to their biological importance, the catalytic functions of CYPs are of great interest to those involved in biotechnology (4, 5, 8, 13, 16, 42). Biocatalytic regiospecific and stereospecific oxidations by CYPs attract much attention in industry because (i) use of biocatalysts allows reduction in the number of tedious steps, such as blocking and deblocking, that are common in conventional chemical synthesis, (ii) a single product without by-product formation reduces the cost of downstream purification steps, and (iii) enantiomeric mixtures (rather than racemic mixtures) are strictly required in some fields, such as the pharmaceutical industry (12, 33, 39, 42). Therefore, a rational comprehensive approach toward elucidating catalytic potentials and utilities of CYPs is required to develop bioindustrial applications (8, 19, 34).
A wide variety of filamentous fungi are used to produce economically valuable consumer items. The filamentous fungus Aspergillus oryzae is one of the most widely utilized microorganisms. It has been used for more than 1,000 years in Japanese fermentation industries to produce indigenous products, such as sake (rice wine), miso (soybean paste), and shoyu (soy sauce). Besides traditional fermentation technologies, much recent research into A. oryzae has focused on production of recombinant enzymes and primary and secondary metabolites (1, 40). The whole genome of A. oryzae has been sequenced and is available to the public (21). Genomic data have the potential to provide a more thorough understanding of metabolic diversity and capability of A. oryzae. Recently, we identified 155 genes, including 13 putative pseudogenes of A. oryzae CYPs (AoCYPs), and cloned 121 full-length cDNAs encoding an open reading frame (25). Molecular and functional diversity of AoCYPs is presumably important to the metabolic capability of this fungus; however, biological functions and catalytic potentials remain obscure. A rational approach to find and exploit catalytic potentials of various CYP enzymes could facilitate a myriad of biocatalytic processes. In this study, we have aimed to develop a functional screening system for AoCYPs and to increase understanding of their functional information.
Heterologous expression of AoCYPs.
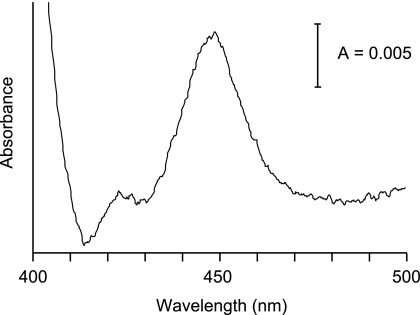
Using the isolated cDNAs, we attempted to express recombinant enzymes for a functional screening system of AoCYPs. For catalytic activity, CYPs must be associated with electron transfer systems providing reducing equivalents. Thus, systematic production of both CYPs and their redox partner(s) are important to facilitate comprehensive functional screening (19, 34). In this study, we employed S. cerevisiae as a host strain for heterologous expression because (i) a yeast expression system seemed appropriate to obtain recombinant eukaryotic CYP without genetic engineering of cDNA, (ii) S. cerevisiae has only three CYP genes, and their catalytic functions are well characterized (26), and (iii) S. cerevisiae is potentially useful for industrial applications as well as basic research. In addition, cytochrome P450 oxidoreductase (CPR) from S. cerevisiae is capable of donating reducing equivalents for various CYPs. It has been shown that a wide variety of eukaryotic CYPs, including Aspergillus spp., could be functional with CPR from S. cerevisiae (3, 23, 24, 32). Presumably, eukaryotic CYPs can exhibit catalytic activities with a heterologous CPR. Therefore, we constructed a yeast coexpression system of AoCYP and yeast CPR using pGYR vector (24, 35, 36) (see Material and Methods in the supplemental material). To elucidate heterologous expression of AoCYPs, we analyzed the CO difference spectrum of S. cerevisiae cells harboring the expression plasmid (29). Figure 1 depicts a CO difference spectrum of CYP628C1 expressed in S. cerevisiae. No peak was observed around 450 nm from a transformant harboring a plasmid without an AoCYP, indicating that the recombinant AoCYP enzyme was successfully expressed in S. cerevisiae in an active form. These results also implied that endogenous CYPs of S. cerevisiae did not interfere with spectroscopic analysis of recombinant AoCYPs due to their low levels of expression in this host strain (2). We have confirmed significant expression of 84 AoCYPs based upon CO difference spectra.
Fig. 1.
CO difference spectrum of CYP628C1 expressed in S. cerevisiae.
Comprehensive functional screening of AoCYPs.
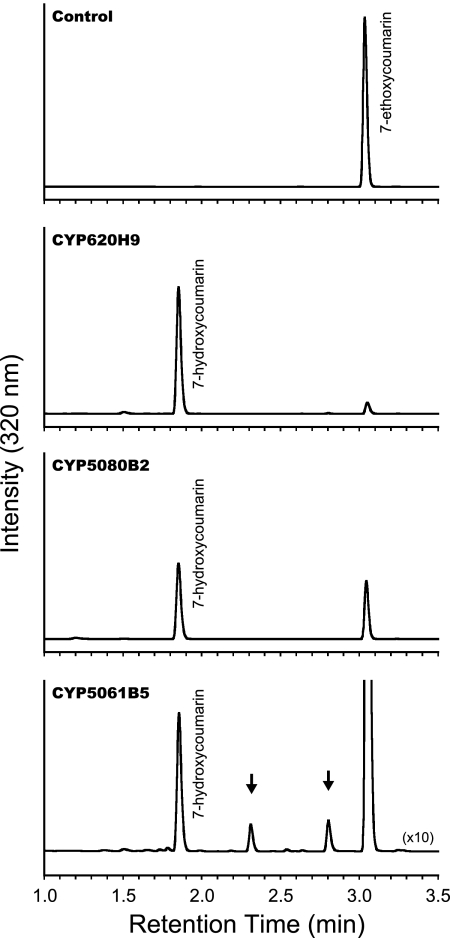
To develop a functional screening system, each transformant was separately inoculated into 0.5 ml culture medium and grown in 96-well DeepWell plates (see Materials and Methods in the supplemental material). Transformants accommodated in the 96-well plates were easily replicated and used for further experiments. To validate potential utility of the screening system, we initiated a functional survey using 7-ethoxycoumarin as a tentative model substrate. After 2 days of incubation, significant product formation was achieved by S. cerevisiae harboring expression plasmids of CYP57B3, CYP62C2, CYP68Q1, CYP531E1, CYP620G1, CYP620H1, CYP620H3, CYP620H9, CYP628C1, CYP675A2, CYP5061B5, CYP5078A5, and CYP5080B2. The product was identified as 7-hydroxycoumarin by comparison of the retention time on high-performance liquid chromatography (HPLC) with an authentic standard (Fig. 2). Moreover, several AoCYPs, such as 5061B5, yielded a minor unidentified product(s), in addition to the formation of 7-hydroxycoumarin (Fig. 2). Although further information should aim to elucidate their biochemical and biophysical properties, it can be hypothesized that the natural substrate(s) of these AoCYPs might be structurally related to coumarin, such as polyketide derivatives. It is thus a good example that non-target-driven screening could provide new insight into the fascinating biology and metabolic processes of A. oryzae. Interestingly, catalytic conversion of 7-ethoxycoumarin to 7-hydroxycoumarin was clearly demonstrated by the transformant harboring the CYP5080B2 expression plasmid; nevertheless, we could not confirm its expression by CO difference spectrum. Such observation was also found from several AoCYPs using different substrates. Combining the results of spectroscopic analysis and bioconversion, we could conclude that at least 92 AoCYPs were functionalized in S. cerevisiae. These results highlight the potential utility of our screening system.
Fig. 2.
Screening of AoCYPs catalyzing 7-ethoxycoumarin conversions. Metabolic products observed from S. cerevisiae expressing no AoCYP (control), CYP620H9, CYP5080B2, and CYP5061B5, analyzed by HPLC at 320 nm. Arrows (in CYP5061B5) indicate unidentified products. The chromatogram for CYP5061B5 is plotted as a 10-fold-expanded scale.
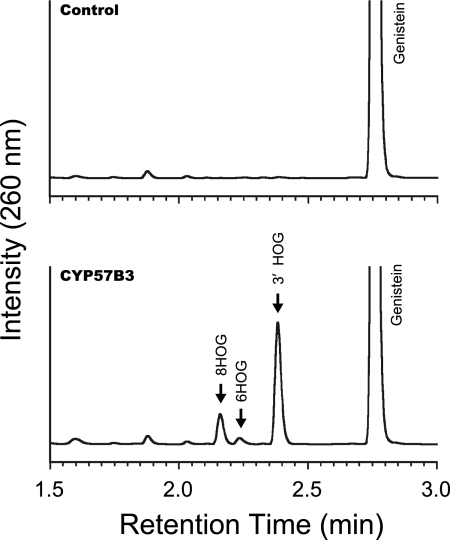
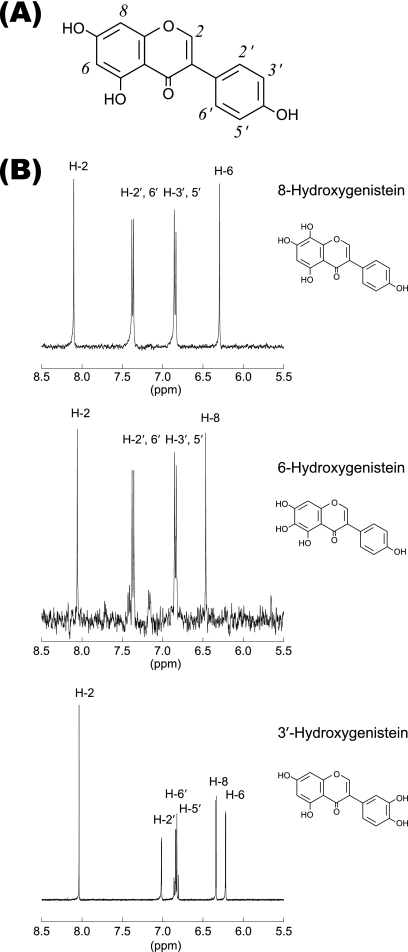
Using the functional screening system, we further carried out targeted functional screening of AoCYP to produce value-added isoflavonoids. Genistein is one of the major isoflavonoids found in soybean. Because of its biological and pharmacological activities, genistein derivatives are likely to have various useful applications (6, 10, 11, 15, 17, 41). When genistein was utilized as a substrate, significant product formations were achieved by S. cerevisiae expressing CYP57B3 (Fig. 3). Incorporation of a hydroxyl group was clearly demonstrated by LC-electrospray ionization (ESI)-mass spectrometry (MS) analysis (data not shown). Based upon 1H nuclear magnetic resonance (NMR) spectroscopic analysis, products were identified as 8-hydroxy-, 6-hydroxy-, and 3′-hydroxygenistein (Fig. 4). To the best of our knowledge, this is the first report describing catalytic activities of CYP57B3 against genistein and suggesting its possible roles in production of hydroxygenistein during soybean fermentation processes (6, 11). Based on sequence comparison, CYP57B3 seems phylogenetically close to a CYP from the plant pathogen Nectria haematococca; a fungus involved in biodegradation of phytoalexin pisatin (22). Although it is still unclear whether CYP57B3 exhibits catalytic activities against pisatin, it may play a biological role in the metabolism of phytoalexins with isoflavone skeletons. Biological and pharmacological potentials of the products have also been reported. 8-Hydroxygenistein shows much stronger antioxidative activity than genistein (11) and exhibits antiproliferative activity against cancer cells (15). 3′-Hydroxygenistein, known as orobol, exhibits unique pharmacological activities, such as enhancing sensitivity of human ovarian carcinoma cells against the anticancer drug paclitaxel (17) and inhibiting human immunodeficiency virus 1 integrase (41). Although hydroxylated genistein can be isolated from natural products, including fermented products, these natural compounds are limited in supply and problematic for practical utilization. Because the synthesis of isoflavones remains an important object (10, 37), it would be of great interest to utilize CYP57B3 for production of value-added rare isoflavonoids from genistein. However, further research efforts should aim to improve conversion efficiency for practical application. It has been reported that reaction efficiency can be influenced by redox partner(s) such that native combination of CYP-CPR from the same organism shows slightly better functionality than heterologous combination and several monooxygenase reactions are coupled with cytochrome b5 (7, 18, 30, 38). Further investigation and possible reengineering of CYP57B3 and its redox partner(s) should facilitate systematic understanding for the biotechnology sector.
Fig. 3.
HPLC analysis of genistein conversion catalyzed by CYP57B3. Metabolic products observed from S. cerevisiae expressing no AoCYP (control) and CYP57B3, analyzed by HPLC at 260 nm. 8HOG, 6HOG, and 3′ HOG indicate 8-hydroxy-, 6-hydroxy-, and 3′-hydroxygenistein, respectively.
Fig. 4.
Identification of hydroxylated genisteins. (A) Chemical structure of genistein. Protons in genisteins and derivatives are numbered. (B) 1H NMR spectra of hydroxylated genisteins produced by CYP57B3.
Through functional screening, the novel activities of AoCYPs were experimentally explored using a series of compounds, such as flavonoids, terpenoids, steroids, and pharmaceutical chemicals (Table 1). To the best of our knowledge, this is the first report experimentally demonstrating functional diversity of Aspergillus CYP. The functional screening system promises rapid and comprehensive characterization of the catalytic potential of AoCYPs, which could further provide novel insight into the biology and biotechnology. In this study, we have focused on the catalytic capability of CYP57B3 to produce value-added hydroxylated products from genistein. However, the screening system should be applicable for a myriad of compounds and facilitate investigation in industrial and biological fields. A thorough understanding of AoCYP functions will open the door for advanced fungal biology and biotechnology.
Table 1.
Catalytic potentials of AoCYPs against various compounds
| Substrate | AoCYP(s) |
|---|---|
| 7-Ethoxycoumarin | CYP57B3, CYP62C2, CYP68Q1, CYP531E1, CYP620G1, CYP620H1, CYP620H3, CYP620H9, CYP628C1, CYP675A2, CYP5061B5, CYP5078A5, CYP5080B2 |
| Genistein | CYP57B3 |
| Naringenina | CYP57B3, CYP62C2 |
| Testosteroneb | CYP65T4, CYP547C3, CYP595B1, CYP5061B5 |
| Dehydroabietic acidc | CYP5061B5 |
| Diclofenacd | CYP65AD1, CYP68Q1, CYP65AF1 |
HPLC analysis of naringenin conversion is shown in Fig. S2 in the supplemental material.
HPLC analysis of testosterone conversion is shown in Fig. S3 in the supplemental material.
HPLC analysis of dehydroabietic aicd conversion is shown in Fig. S4 in the supplemental material.
HPLC analysis of diclofenac conversion is shown in Fig. S5 in the supplemental material.
Supplementary Material
Acknowledgments
This research was supported in part by a Noda Institute for Scientific Research (NIRS) grant (to H.I.).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 4 March 2011.
REFERENCES
- 1. Abe K., Gomi K., Hasegawa F., Machida M. 2006. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia 162:143–153 [DOI] [PubMed] [Google Scholar]
- 2. Akiyoshi-Shibata M., et al. 1991. Expression of rat liver vitamin D3 25-hydroxylase cDNA in Saccharomyces cerevisiae. FEBS Lett. 280:367–370 [DOI] [PubMed] [Google Scholar]
- 3. Artigot M. P., et al. 2009. Molecular cloning and functional characterization of two CYP619 cytochrome P450s involved in biosynthesis of patulin in Aspergillus clavatus. Microbiology 155:1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernhardt R. 2006. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 124:128–145 [DOI] [PubMed] [Google Scholar]
- 5. Chang M. C. Y., Eachus R. A., Trieu W., Ro D.-K., Keasling J. D. 2007. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 3:274–277 [DOI] [PubMed] [Google Scholar]
- 6. Chang T.-S., Ding H.-Y., Tai S. S.-K., Wu C.-Y. 2007. Metabolism of the soy isoflavones daidzein and genistein by fungi used in the preparation of various fermented soybean foods. Biosci. Biotechnol. Biochem. 71:1330–1333 [DOI] [PubMed] [Google Scholar]
- 7. Chemler J. A., Lim C. G., Daiss J. L., Koffas M. A. G. 2010. A versatile microbial system for biosynthesis of novel polyphenols with altered estrogen receptor binding activity. Chem. Biol. 17:392–401 [DOI] [PubMed] [Google Scholar]
- 8. Chigu N. L., et al. 2010. Cytochrome P450 monooxygenases involved in anthracene metabolism by the white-rot basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 87:1907–1916 [DOI] [PubMed] [Google Scholar]
- 9. Demain A. L., Fang A. 2000. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 69:1–39 [DOI] [PubMed] [Google Scholar]
- 10. Dixon R. A., Ferreira D. 2002. Genistein. Phytochemistry 60:205–211 [DOI] [PubMed] [Google Scholar]
- 11. Esaki H., Onozaki H., Morinitsu Y., Kawakishi S., Osawa T. 1998. Potent antioxidant isoflavones isolated from soybeans fermented with Aspergillus saitoi. Biosci. Biotechnol. Biochem. 62:740–746 [DOI] [PubMed] [Google Scholar]
- 12. Gavrilescu M., Chisti Y. 2005. Biotechnology—a sustainable alternative for chemical industry. Biotechnol. Adv. 23:471–479 [DOI] [PubMed] [Google Scholar]
- 13. Gillam E. M. J. 2008. Engineering cytochrome P450 enzymes. Chem. Res. Toxicol. 21:220–231 [DOI] [PubMed] [Google Scholar]
- 14. Gotoh O. 1993. Evolution and differentiation of P450 genes, p. 255–272 In Omura T., Ishimura Y., Fujii-Kuriyama Y. (ed.), Cytochrome P-450, 2nd ed. Kodansha, Tokyo, Japan [Google Scholar]
- 15. Hirota A., Taki S., Kawaii S., Yano M., Abe N. 2000. 1,1-Diphenyl-2-picrylhydrazyl radical-scavenging compounds from soybean miso and antiproliferative activity of isoflavones from soybean miso toward the cancer cell lines. Biosci. Biotech. Biochem. 64:1038–1040 [DOI] [PubMed] [Google Scholar]
- 16. Ichinose H., Wariishi H., Tanaka H. 1999. Biotransformation of recalcitrant 4-methyldibenzothiophene to water-extractable products using lignin-degrading basidiomycete Coriolus versicolor. Biotechnol. Prog. 15:706–714 [DOI] [PubMed] [Google Scholar]
- 17. Isonishi S., Saitou M., Saitou M., Yasuda M., Tanaka T. 2007. Differential regulation of the cytotoxicity activity of paclitaxel by orobol and platelet derived growth factor in human ovarian carcinoma cells. Oncol. Rep. 18:195–201 [PubMed] [Google Scholar]
- 18. Lamb D. C., Kelly D. E., Manning N. J., Kaderbhai M. A., Kelly S. L. 1999. Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 462:283–288 [DOI] [PubMed] [Google Scholar]
- 19. Lamb D. C., et al. 2002. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2). J. Biol. Chem. 277:24000–24005 [DOI] [PubMed] [Google Scholar]
- 20. Lewis D. F. L., Watson E., Lake B. G. 1998. Evolution of the cytochrome P450 superfamily: sequence alignments and pharmacogenetics. Mutat. Res. 410:245–270 [DOI] [PubMed] [Google Scholar]
- 21. Machida M., et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161 [DOI] [PubMed] [Google Scholar]
- 22. Maloney A. P., VanEtten H. D. 1994. A gene from the fungal plant pathogen Nectria haematococca that encodes the phytoalexin-detoxifying enzyme pisatin demethylase defines a new cytochrome P450 family. Mol. Gen. Genet. 243:506–514 [DOI] [PubMed] [Google Scholar]
- 23. Martel C. M., et al. 2010. Complementation of a Saccharomyces cerevisiae ERG11/CYP51 (sterol 14a-demethylase) doxycycline-regulated mutant and screening of the azole sensitivity of Aspergillus fumigatus isoenzymes CYP51A and CYP51B. Antimicrob. Agents Chemother. 54:4920–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murakami H., Yabusaki Y., Sakaki T., Shibata M., Ohkawa H. 1990. Expression of cloned yeast NADPH-cytochrome P450 reductase gene in Saccharomyces cerevisiae. J. Biochem. 108:859–865 [DOI] [PubMed] [Google Scholar]
- 25. Nazir K. H. M. N. H., Ichinose H., Wariishi H. 2010. Molecular characterization and isolation of cytochrome P450 genes from the filamentous fungus Aspergillus oryzae. Arch. Microbiol. 192:395–408 [DOI] [PubMed] [Google Scholar]
- 26. Nelson D. R. 1999. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 369:1–10 [DOI] [PubMed] [Google Scholar]
- 27. Nelson D. R. 2009. The cytochrome p450 homepage. Hum. Genomics 4:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nelson D. R., et al. 1993. The P450 superfamily—update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 12:1–51 [DOI] [PubMed] [Google Scholar]
- 29. Omura T., Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239:2379–2385 [PubMed] [Google Scholar]
- 30. Ortiz de Montellano P. R. 2005. Cytochrome P450: structure, mechanism, and biochemistry, 3rd ed. Kluwer Academic/Plenum, New York, NY [Google Scholar]
- 31. Park J., et al. 2008. CFGP: a Web-based, comparative fungal genomics platform. Nucleic Acids Res. 36:D562–D571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Renaud J. P., et al. 1993. Recombinant yeast in drug metabolism. Toxicology 82:39–52 [DOI] [PubMed] [Google Scholar]
- 33. Ro D. K., et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943 [DOI] [PubMed] [Google Scholar]
- 34. Sabbadin F., et al. 2010. LICRED: a versatile drop-in vector for rapid generation of redox-self-sufficient cytochrome P450s. ChemBioChem 11:987–994 [DOI] [PubMed] [Google Scholar]
- 35. Sakaki T., Akiyoshi-Shibata M., Yabusaki Y., Ohkawa H. 1992. Organella-targeted expression of rat liver cytochrome P450c27 in yeast: genetically engineered alteration of mitochondrial P450 into a microsomal form creates a novel functional electron transport chain. J. Biol. Chem. 267:16497–16502 [PubMed] [Google Scholar]
- 36. Sakaki T., Shinkyo R., Takita T., Ohta M., Inouye K. 2002. Biodegradation of polychlorinated dibenzo-p-dioxins by recombinant yeast expressing rat CYP1A subfamily. Arch. Biochem. Biophys. 410:91–98 [DOI] [PubMed] [Google Scholar]
- 37. Sato S., Hiroe K., Kumazawa T., Onodera J.-I. 2006. Total synthesis of two isoflavone C-glycosides: genistein and orobol 8-C-beta-d-glucopyranosides. Carbohydr. Res. 341:1091–1095 [DOI] [PubMed] [Google Scholar]
- 38. Schenkman J. B., Jansson I. 2003. The many roles of cytochrome b5. Pharmacol. Ther. 97:139–152 [DOI] [PubMed] [Google Scholar]
- 39. Schmid A., et al. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258–268 [DOI] [PubMed] [Google Scholar]
- 40. Taylor M. J., Richardson T. 1979. Applications of microbial enzymes in food systems and in biotechnology. Adv. Appl. Microbiol. 25:7–35 [DOI] [PubMed] [Google Scholar]
- 41. Tewtrakul S., Subhadhirasakul S., Cheenpracha S., Karalai C. 2007. HIV-1 protease and HIV-1 integrase inhibitory substance from Eclipta prostrata. Phytother. Res. 21:1092–1095 [DOI] [PubMed] [Google Scholar]
- 42. Urlacher V. B., Eiben S. 2006. Cytochrome P450 monooxygenases: perspectives for synthetic application. Trends Biotechnol. 24:324–330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.