Abstract
To comprehensively assess microbial diversity and abundance via molecular-analysis-based methods, procedures for sample collection, processing, and analysis were evaluated in depth. A model microbial community (MMC) of known composition, representative of a typical low-biomass surface sample, was used to examine the effects of variables in sampling matrices, target cell density/molecule concentration, and cryogenic storage on the overall efficacy of the sampling regimen. The MMC used in this study comprised 11 distinct species of bacterial, archaeal, and fungal lineages associated with either spacecraft or clean-room surfaces. A known cellular density of MMC was deposited onto stainless steel coupons, and after drying, a variety of sampling devices were used to recover cells and biomolecules. The biomolecules and cells/spores recovered from each collection device were assessed by cultivable and microscopic enumeration, and quantitative and species-specific PCR assays. rRNA gene-based quantitative PCR analysis showed that cotton swabs were superior to nylon-flocked swabs for sampling of small surface areas, and for larger surfaces, biological sampling kits significantly outperformed polyester wipes. Species-specific PCR revealed differential recovery of certain species dependent upon the sampling device employed. The results of this study empower current and future molecular-analysis-based microbial sampling and processing methodologies.
INTRODUCTION
The search for life and habitable conditions elsewhere in the solar system has led to a steady increase in the number of robotic probes and orbiters leaving Earth en route to other planetary systems. Planetary protection is a space science discipline that addresses concerns over the potential for forward contamination (i.e., the inadvertent contamination of extraterrestrial environments with terrestrial organic and biological matter) and backward contamination (i.e., the inadvertent exposure of the terrestrial biosphere to extraterrestrial material) during such endeavors (26). Current guidelines for the assessment of biological contamination of spacecraft surfaces throughout assembly, testing, and launch operations (ATLO) are overseen by the International Committee on Space Research (6) and are enacted by the National Aeronautics and Space Administration (NASA) to facilitate compliance with internationally agreed planetary protection policy, as developed by the Committee on Space Research (6). Most space-faring agencies (NASA included) also have their own policy that places specific implementation requirements on agency missions.
Planetary protection-geared approaches for monitoring of biological contamination of spacecraft and spacecraft assembly clean-room (SAC) surfaces at NASA are largely based on heritage data arising from studies supporting the Viking missions in the early 1970s. Scientists postulated that the probability of microorganisms surviving the journey to Mars and propagating upon arrival would be constrained by their hardiness with respect to the conditions they encountered. Therefore, NASA conservatively selected aerobic, heterotrophic mesophilic endospore-forming bacteria as the standard organisms by which to gauge the cleanliness of spacecraft and SAC surfaces. To be more specific, the enumeration of cultivable bacterial spores at the time of launch is compared to a maximum numerical limit. This mission-specific limit is derived from a combination of factors, including mission purpose, target planetary environment, and an assessed factor of risk that any organism transferred to said planetary environment would result in “harmful (forward) contamination” (24).
From the Viking era to the present, NASA has employed cotton swabs to collect biological materials from spacecraft and SAC surfaces. Recently, however, it has been shown that the sole use of a cotton swabbing device provides an inaccurate assessment of the total burden and spectrum of microbial diversity associated with SAC surfaces (M. Cooper et al., unpublished data). In addition, the National Research Council recently stated that NASA should “require the routine collection of (molecular) phylogenetic data to a statistically appropriate level to ensure that the microbial diversity of ATLO environments, and of all NASA spacecraft to be sent to Mars, is reliably assessed.” They also recommended that the space agency “require the systematic archiving of environmental samples for ATLO environments and for all spacecraft to be sent to Mars” (5).
While molecular analyses of microbial populations provide a more thorough phylogenetic assessment of spacecraft surfaces, such analysis regimens are not without pitfalls and limitations. Oftentimes, target biomolecules (e.g., rRNA genes, lipopolysaccharides) are not collected in great enough abundance to render downstream molecular analyses statistically significant and/or reproducible, thus requiring a more effective method for sample processing/concentration prior to analysis (17, 38). Furthermore, as sample collection protocols are modified to facilitate more effective downstream processing and analysis techniques, the efficiency and effectiveness of the entire procedure must be readdressed. Previous studies evaluating the effectiveness of sampling devices for the recovery of microorganisms from clean-room surfaces have used purified bacterial spores (8, 9, 11, 12, 29, 32, 34, 39) or pure cultures of vegetative cells (7) as positive controls. However, such controls do not accurately represent the highly diverse microbial populations which can be expected in nearly all environments, including clean rooms (10, 15, 30, 35). Additionally, whenever samples are collected for the purpose of molecular-analysis-based microbial monitoring, results will be affected not only by the presence of living cells but also by dead and decaying cells and spores, cellular debris, and indigenous biomolecules (e.g., nucleic acids, lipids, proteins, carbohydrates). Methodologies by which samples are collected, processed, and analyzed must be optimized to their respective targets to maximize the efficiency and accuracy of the final assessment.
To this end, suitable protocols and procedures involved with the collection, processing, and molecular analysis of contaminant biomolecules from clean-room surfaces were evaluated and standardized. Variables such as differences in sampling materials, concentration of target biomolecules postsampling, the effect of cryostorage on extracted molecules, etcetera, were individually tested, employing a model microbial community (MMC) that is relevant to low-biomass environments. To our knowledge, this is the first study to employ a complex MMC with relevant phylogenetic diversity for the testing and optimization of protocols for collecting, processing, and analyzing biomolecular samples from clean-room surfaces.
MATERIALS AND METHODS
MMC.
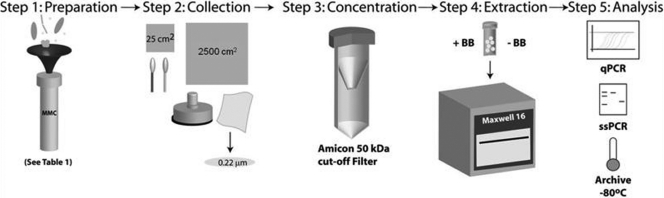
Microbes previously reported to be present and/or dominant on spacecraft and SAC surfaces (3, 16, 21–23, 27) were mixed together to achieve the MMC. These microbial components include bacteria, archaea, fungi, aerobes, anaerobes, spores, and non-spore-forming vegetative cells (Table 1). Whenever possible, strains isolated directly from spacecraft and SAC surfaces were included. Deinococcus radiodurans (ATCC 13939), Cupriavidus metallidurans CH34, Micrococcus luteus (ATCC 4698), Clostridium sporosphaeroides (DSM 1294), and Methanobacterium formicicum (DSM 1535), however, were procured from various culture collections (Table 1). All 11 strains used in this study were placed in the National Center for Agricultural Utilization Research (NRRL) culture collection, in Peoria, IL. The various steps involved in the standardization of procedures for MMC preparation, collection from surfaces, and subsequent processing and analysis are shown in Fig. 1.
Table 1.
Various characteristics of MMC constituents
| Sample no. | Microbe | Strain | Domain: phylum | Morphology | Culture conditions or referencea | Incubation time (h) | Other available source(s)b | Significance (reference[s]) |
|---|---|---|---|---|---|---|---|---|
| 1 | Aureobasidium pullulans | 28v1 | Eukarya: Ascomycota | Black yeast | TYG agar, 30°C | 86 | NRRL 58992 | Eukaryotic representative, facultative anaerobic spore former, original isolate (16) |
| 2 | Acinetobacter radioresistens | 50v1 | Bacteria: Gammaproteobacteria | Short rods | TYG agar, 32°C | 60 | NRRL B-59417 | Resistant to desiccation and radiation, aerobic non-spore former, original isolate (16) |
| 3 | Bacillus megaterium | KL-197 | Bacteria: Firmicutes | Rods | TYG agar, 32°C | 38 | NRRL B-59415 | Frequently isolated from SAC (15, 16), facultative anaerobic spore former, vegetative cells of original isolate |
| 4 | Bacillus pumilus | SAFR-032 | Bacteria: Firmicutes | Spores | See reference 13 | NAc | ATCC PTA-7603, NRRL B-30938 | Strains showing extraordinary UV resistance, frequently isolated from SAC (13), facultative anaerobic spore former, spores of original isolate |
| 5 | Deinococcus radiodurans | ATCC 13939 | Bacteria: Deinococcus-Thermus | Cocci, tetrad | TYG agar, 32°C | 60 | NRRL B-59418, DSM 20539 | Strains showing extraordinary gamma radiation resistance, sequences retrieved from spacecraft surfaces (3), aerobic non-spore former, isolate procured from culture collection |
| 6 | Microbacterium imperiale | 47v1 | Bacteria: Actinobacteria | Short rods | TYG agar, 32°C | 60 | NRRL B-59416 | Extremely hardy, difficult to extract DNA from cells (17), aerobic non-spore former, original isolate |
| 7 | Staphylococcus warneri | 82-4 | Bacteria: Firmicutes | Cocci | TYG agar, 32°C | 40 | NRRL B-59414 | Frequently isolated from SAC (16, 35), human associated, facultative anaerobic non-spore former, original isolate |
| 8 | Micrococcus luteus | ATCC 4698 | Bacteria: Actinobacteria | Rods | Nutrient agar, 32°C | 60 | NRRL B-59413, DSM 20030 | Common contaminant of indoor environments and SAC (16), air borne and human associated, aerobic non-spore former, original isolate |
| 9 | Cupriavidus metallidurans | CH34d | Bacteria: Betaproteobacteria | Rods | TYG agar, 32°C | 60 | ATCC 43123 | Ubiquitously isolated from SAC (22), heavy metal resistant, aerobic non-spore former |
| 10 | Clostridium sporosphaeroides | DSM 1294 | Bacteria: Firmicutes | Anaerobic rods | DSMZ medium 78, 37°C, anaerobic | 48 | ATCC 25781 | Representative of obligate anaerobic spore formers found in SAC (30, 35), isolate procured from culture collection, vegetative cells |
| 11 | Methanobacterium formicicum | DSM 1535 | Archaea: Euryarchaeota | Rods | DSMZ medium 119, 37°C, anaerobic | 72 | ATCC 33274 | Representative of broad (uncultivated) archaeal diversity of SAC (21, 23), anaerobic non-spore former, isolate procured from culture collection |
TYG, tryptone yeast extract glucose.
ATCC, American Type Culture Collection; NRRL, USDA culture collection; DSM, German collection of cell cultures; PTA, patented strain.
NA, not applicable since the spores were prepared prior to the investigation and the appropriate concentration from the original stock was used.
Received from Natalie Leys, Hoofd Onderzoeks Eenheid Microbiologie at SCK-CEN, Antwerp, Belgium.
Fig. 1.
Schematic representation of various procedures involved in the preparation of the MMC and its subsequent deposition onto metal surfaces, sample collection, processing, and analysis.
Preparation of the MMC.
All 11 strains in the MMC, with the exception of Bacillus pumilus SAFR-032, were streaked on appropriate culture media and incubated according to the growth conditions given in Table 1. B. pumilus spores were purified as previously described (13). Briefly, a single purified colony was inoculated into liquid nutrient broth sporulation medium, and after 2 to 3 days of incubation at 32°C, cultures were examined in wet mounts on an Olympus BX-60 phase-contrast microscope to determine the level of sporulation. Once the number of free spores in the culture was greater than the number of vegetative cells, spores were purified by treatment with lysozyme and washing with salt and detergents (13). Purified spores were resuspended in sterile deionized water, heat shocked (80°C for 15 min), and stored at 4°C in glass tubes.
For each strain, a single isolated colony was picked, inoculated into 50 ml of suitable broth, and incubated under appropriate growth conditions (Table 1). Petroff-Hausser microscopic counting chambers were used to verify the cell density of each strain. To circumvent problems associated with clumping as a result of washing and centrifuging, appropriate volumes (precalculated, necessary to achieve 105 cells/spores per MMC constituent) of cells or spores were removed from log-phase overnight broth culture and placed directly into 4 liters of phosphate-buffered saline (PBS, pH 7.2). After averaging 10 fields for each organism, the resulting MMC housed 1 × 106 cells and 1 × 105 (the standard error of the mean was used to assess variability) spores per ml (11 strains × 105 cells/spores per ml each). Following rigorous mixing for more than an hour, 1.2-ml aliquots of the MMC were aseptically dispensed into thousands of cryogenic vials and those not immediately analyzed were stored at −80°C.
Using the MMC as a positive control, prior to deposition onto stainless steel surfaces, a number of variables associated with sampling and processing were evaluated for their specific effects on the integrity and recovery of the MMC. The ramifications of the use of various concentrations (5 to 20%) of glycerol as a cryoprotectant, various buffered solutions (PBS or water) as suitable liquid media for deposition, ultrasonic energy to enhance the release of the MMC from sampling materials (31), and a method for concentrating large sample volumes were tested, and results were comparatively analyzed (see the supplemental material).
Deposition of the MMC onto a surrogate spacecraft hardware surface.
All stainless steel grade 304-020 (304 brushed stainless steel sheet, 20 gauge, 0.036 in. thick) coupons (25 cm2) and sheets (2,500 cm2) were precleaned with clean-room-grade polyester wipes (Coventry 6209 c-prime; Tech Spray, L.P., Amarillo, TX). Coupons and sheets were subjected to ultrasonic cleaning with alkaline detergent soap (815 GD; Brulin, Indianapolis, IN) at 160°F, followed by a deionized-water rinse, drying, and a final rinse with 97% ethyl alcohol (40). Subsequently, coupons and sheets were rinsed with deionized water and dried with clean, dry nitrogen. This precision cleaning procedure was effective in removing any cultivable microbes and genetic materials associated with the metal surfaces. All precision-cleaned coupons and sheets were packaged individually in sterile containers until the time of use. Aliquots of the MMC (40 × 1.2 ml) were thawed, pooled, and concentrated via centrifugation (15 min, 4,000 × g) using the Amicon 50-kDa filtration system (catalog no.UFC905024; Millipore, Billerica, MA). Finally, to circumvent crystal formation resulting from PBS precipitation on metal surfaces, the MMC was washed and resuspended in sterile water to a final volume of 48 ml prior to deposition.
Stainless steel coupons (25 cm2) and sheets (2,500 cm2) were aseptically removed from their sterile packaging and placed into individual sterile petri dishes or trays, respectively. One-milliliter aliquots of the MMC were deposited onto either coupons (20 × 50 μl drops) or sheets (50 × 20 μl drops) using a multichannel, multidispensing electronic pipette (E8-200; Rainin, Oakland, CA). All coupons and sheets, including those serving as sampling blanks, were covered and allowed to dry for 24 h. To minimize bias associated with sample collection, care was taken to ensure that the operator in charge of deposition was not also in charge of sampling.
Sampling of MMC-spiked metal surfaces. (i) Small surface areas (25 cm2).
Coupons were sampled with either cotton (catalog no. 806-WCL; Puritan Medical Products, Guilford, ME) or nylon-flocked (catalog no. 552C.US; Copan Diagnostics Inc., Murrieta, CA) swabs, which were premoistened with sterile PBS immediately prior to sampling. Coupons were first sampled in a unidirectional horizontal manner while holding the swab at approximately a 30° angle to the surface. Swabs were rotated (ca. 120°) to present an area of the swab head that had not previously contacted the surface, and coupons were sampled in a unidirectional vertical manner. Finally, swabs were once again rotated (ca. 120°) and coupons were sampled in a unidirectional diagonal manner. Swab heads were aseptically severed from their shafts into individual 50-ml Falcon tubes containing 10 ml of sterile PBS. A negative control, whereby a swab was only premoistened with PBS, and a handling control, in which a swab was premoistened with PBS and exposed to the sampling environment, were also prepared.
(ii) Large surface areas (2,500 cm2).
Stainless steel sheets were sampled with either ITW Alpha polyester wipes (catalog no. TX1009; Texwipe, Kernersville, NC) or biological sampling kits (BiSKit, catalog no.BIS-40001A; Quicksilver Analytics Inc., Abingdon, MD). Wipes were rolled and placed into individual 50-ml glass test tubes containing 15 ml of PBS, which were autoclaved and allowed to cool prior to sampling (14). Wearing sterile gloves, we removed the wipe from the tube with sterile forceps and unrolled it, leaving it folded in quarters. Sheets were first sampled in a unidirectional horizontal manner, after which time wipes were turned over and used to sample the sheets in a unidirectional vertical manner. Finally, the wipe was inverted to expose a fresh surface and the sheet was sampled in a unidirectional diagonal manner. By this method, 3/4 of one side of the wipe was used in sampling the surface. Negative and handling controls were prepared as detailed above for coupons. All wipes were placed into individual sterile glass 500-ml bottles for processing.
BiSKits were moistened with 15 ml of sterile PBS, which was expelled from each unit and pooled to serve as the negative control. The collection bottle of each BiSKit was replenished with 15 ml of sterile PBS, and the entire module was inverted, allowing the macrofoam sponge to become saturated. The macrofoam sponge sampling portion of the BiSKit module was removed, and sheets were first sampled in a unidirectional horizontal manner. The BiSKit was then turned, and the sheets were sampled in a unidirectional vertical manner. Finally, the BiSKit was once again turned and sheets were sampled in a unidirectional diagonal manner. A handling control, in which a BiSKit was exposed to the environment but no surfaces, was also prepared.
Sample processing.
Each cotton or nylon-flocked swab was subjected to three rounds of biomolecule extraction processing. This consisted of vortex mixing (5 s, maximum speed), sonication in an ultrasonic bath (2 min, ∼25 kHz), decanting into a fresh sterile 50-ml tube, and replenishment of 10 ml of sterile PBS. In the end, each swab head yielded a total of 30 ml of biomolecule extraction eluent, which was then concentrated to approximately 400 μl via two rounds of Amicon centrifugal filtration (15 min, 4,000 × g).
Following the addition of 200 ml of sterile PBS, each polyester wipe-containing bottle was vortex mixed (5 s, maximum speed) and sonicated in an ultrasonic bath (2 min, ∼25 kHz). This was repeated three times, resulting in 600 ml of biomolecule extraction eluent, which was then subjected to membrane filtration (0.22-μm pore size). The 0.22-μm filter was aseptically transferred into a sterile 50-ml Falcon tube containing 10 ml of sterile PBS and vigorously agitated via vortex mixing (5 min, maximum speed). The resulting suspension was concentrated to approximately 400 μl via Amicon centrifugal filtration (15 min at 4,000 × g).
Each BiSKit was subjected to three rounds of biomolecule extraction processing. The macrofoam sponge sampling portion of each BiSKit was mated with its respective sample collection module, and with appropriate force, sample volumes were expelled into the collection bottle in accordance with the manufacturer's instructions. Sample volumes were transferred into a 50-ml Falcon tube, and an additional 15 ml of sterile PBS was added to the BiSKit collection bottle, which was then reattached for subsequent extractions. The resulting suspension (ca. 45 ml) was concentrated to approximately 400 μl via Amicon centrifugal filtration (50 kDa, 15 min at 4,000 × g).
The final concentrated biomolecule extract volume from each sampling device was resuspended in flowthrough from the final Amicon filtration to a total volume of 1.5 ml. Appropriate dilutions were then spread plated on suitable media, and CFU were enumerated after incubation under prescribed conditions (Table 1). Total cell/spore counts were also estimated via light microscopy as previously described (18). Five hundred microliters of this biomolecule extract volume was subjected to bead beating homogenization (using Lysing Matrix E and the FastPrep-24 Homogenizer for 1 min at a rate of 5.0 motions per s; MP Biomedicals, Solon, OH) and subsequently combined with sample portions (500 μl) that did not undergo bead beating (a total of 1 ml). This helped to ensure that the samples would contain DNA released from robust and labile cell types, as has been previously documented (17). This combined milliliter then gave rise to 100 μl of purified DNA following extraction via the Maxwell 16 Automated DNA/RNA Purification System (Promega, Madison, WI) in accordance with the manufacturer's instructions.
Sample analysis. (i) SYBR green quantitative PCR (qPCR).
rRNA gene copy numbers were quantified in triplicate using a CFX-96 instrument (Bio-Rad, Hercules, CA). Standards were created using known amounts of full-length Escherichia coli 16S rRNA genes incorporated into suitable vector plasmids (Invitrogen, Carlsbad, CA), and standard curves were repeated for each qPCR. rRNA gene standards, spanning 108 to 102 gene copies/μl, were generated by serially diluting rrn::pCR4-TOPO plasmids of known concentrations. Universal eubacterial primers (1369F and 1492R [36]) targeting the 16S rRNA gene and eukaryotic biased primers (NS91F and ITS51R [41]) targeting the internal transcribed spacer 1 region within the rRNA gene cluster, were employed for qPCR analysis. Each 25-μl reaction mixture consisted of 12.5 μl of Bio-Rad 2X iQ SYBR green Supermix, 10.5 μl of nuclease-free water (catalog no. W4502; Sigma, St. Louis, MO), 0.5 μl of forward primer 1369F (10 μM) or NS91F (10 μM), 0.5 μl of reverse primer 1492R (10 μM) or ITS51R (10 μM), and 1 μl of template DNA. Purified rRNA gene amplicon standards of known concentrations and Sigma nuclease-free water negative controls were included in all experiments. Reaction conditions for 16S rrn qPCR were as follows: denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s and combined annealing and extension at 55°C for 35 s. Reaction conditions for eukaryotic rrn qPCR were as follows: denaturation at 95°C for 5 min and 40 cycles of denaturation at 95°C for 15 s, annealing at 58°C for 20 s, and extension at 72°C for 15 s and 80°C for 5 s.
(ii) Species-specific PCR (ssPCR).
In order to assess the differential recovery of the various microbial phylotypes from both the stainless steel surfaces and sampling devices, ssPCR assays were engineered for each MMC strain. Species-specific primer sequences are provided in Table 2, alongside detailed thermal cycling conditions for each strain. None of the resulting primer combinations (Table 2) showed cross-reactivity with any of the other members of the MMC. Full-length (ca. 1,500-bp) rRNA gene amplicons were purified via gel excision and used as standards once concentrations were determined via microchannel-based electrophoresis (2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA) (28). All amplicons detected at a concentration of less than or equal to 0.25 ng/μl fell below the limits of statistically significant detection and were ignored.
Table 2.
ssPCR conditions
| Sample no., microbe, and primers | Primer sequences (5′–3′)a | Initial denaturation | Denaturation | Annealing | Extension | No. of cycles | Final extension | Amplicon size (bp) |
|---|---|---|---|---|---|---|---|---|
| 1, Aureobasidium pullulans 28v1,b ssAPf/ssAPr | AAT AAG GAT TGA CAG ATT GA/TAA GGT CTC GTT CGT TAT | 94°C, 4 min | 94°C, 50 s | 61°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 120 |
| 2, Acinetobacter radioresistens 50v1,c ssARf/ssARr | GCC TTA TGG TTG TAA AGC/CTT ATT CTG CGA GTA ACG | 94°C, 4 min | 94°C, 50 s | 53°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 94 |
| 3, Bacillus megaterium KL-197,c ssBMf/ssBMr | GTG CTA CAA TGG ATG GTA/AAT CCG AAC TGA GAA TGG | 94°C, 4 min | 94°C, 50 s | 52°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 85 |
| 4, Bacillus pumilus SAFR-032,c 600f/980r | TGA AGC ACT TGA GAA ATT/TGC TGC AAA GAA AAT | None | 95°C, 1 min | 58°C, 30 s | 72°C, 50 s | 35 | 72°C, 10 min | 380 |
| 5, Deinococcus radiodurans ATCC 13939,c ssDRf/ssDRr | CCA GAA GTC ACG AAT AAC/ATC CAG AAG CGA TAA ATC | 94°C, 4 min | 94°C, 50 s | 54°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 103 |
| 6, Microbacterium imperiale 47v1,c ssMIf/ssMIr | AAC GGC GTC TAA TAC TGG ATA TGA/AGC CCA TCC CAG ACC AAA | 94°C, 4 min | 94°C, 50 s | 54°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 83 |
| 7, Staphylococcus warneri 82-4,c ssSWf/ssSWr | ATA ACC TAC CTA TAA GAC T/ATC CAT CTA TAA GTG ACA | 94°C, 4 min | 94°C, 50 s | 58°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 124 |
| 8, Micrococcus luteus ATCC 4698,c ssMLf/ssMLr | TAA CCT GCC CTT AAC TCT/AAA CCG ATA AAT CTT TCC AA | 94°C, 4 min | 94°C, 50 s | 55°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 106 |
| 9, Cupriavidus metallidurans CH34m,d ssCMf/ssCMr | GGT GGA TGA TGT GGA TTA/ATC TCT GCT TCG TTA GTG | 94°C, 4 min | 94°C, 50 s | 64°C, 50 s | 72°C, 50 s | 36 | 72°C, 10 min | 86 |
| 10, Clostridium sporosphaeroides DSM 1294,c ssCSf/ssCSr | CAA GCA GTG GAG TAT GTG/CCT CGT TAG TTG GAT GTC | 94°C, 4 min | 94°C, 50 s | 53°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 84 |
| 11, Methanobacterium formicicum DSM 1535,c vssMFf/ssMFr | ATT GCT GGA GAT ACT ATT/GGG ATT ATA GGA TTT CAC | 94°C, 4 min | 94°C, 50 s | 52°C, 50 s | 72°C, 50 s | 35 | 72°C, 10 min | 89 |
All primers had an original concentration of 9 μM.
Reaction mixture (25 μl/sample) consisted of 12.5 μl 2× BioMix (catalog no. 25012; Bioline Inc., Taunton, MA), 10.3 μl nuclease-free water, 0.6 μl forward primer, 0.6 μl reverse primer, and 1 μl template DNA.
Reaction mixture (25 μl/sample) consisted of 12.5 μl 2× BioMix (catalog no. 25012; Bioline Inc., Taunton, MA), 9.5 μl nuclease-free water, 1 μl forward primer, 1 μl reverse primer, and 1 μl template DNA.
Reaction mixture (25 μl/sample) consisted of 12.5 μl 2× BioMix (catalog no. 25012; Bioline Inc., Taunton, MA), 8.5 μl nuclease-free water, 2 μl MgCl2 (50 mM), 1 μl forward primer, 1 μl reverse primer, and 1 μl template DNA.
Statistical analyses, controls, and lower detection limits of assays employed.
Care was taken to incorporate appropriate controls into each step of the sampling and analysis procedures discussed herein. Sterile, nuclease-free Sigma water and sterile PBS served as blanks to monitor the continued purity of reagents. The independent variables consisted of sampling device replicates (cotton and nylon-flocked swabs, BiSKits, and polyester wipes), their corresponding sampling blanks (active sampling of stainless steel sheets not seeded with the MMC), handling controls (sampling devices were exposed to the experimental environment), and negative controls (sampling materials with reagents). When needed, the MMC was used as a positive control with the medium blank serving as the corresponding negative control. Purified DNA arising from each MMC constituent served as a positive control for that strain's respective ssPCR assay, and samples that rendered no PCR amplification were spiked with positive-control DNA as a means of eliminating the possibility of false negatives resulting from inhibitory substances.
The cell density of microbes or concentration of 16S rRNA genes housed within the MMC prior to deposition on the metal surface was considered to be 100%. The recovery percentage of colony counts, microscopic counts, total gene copy numbers, and species-specific gene copy numbers yielded by various sampling materials was calculated by dividing resulting values by the initial values representative of 100% (MMC prior to deposition). To elucidate subtle differences in resulting efficiencies of recovery from the sampling materials tested in this study (cotton versus nylon-flocked swabs; BiSKits versus polyester wipes), statistical analyses were carried out at all stages of analysis with the SYSTAT VER. 13.00.05 statistical software suite (SYSTAT Software, Chicago, IL). Briefly, routine Pearson's chi-square tests were performed to determine whether the distributions of resulting data sets were statistically normal, and paired Student t tests were employed to test the statistically significant similarity between the means of the results being compared.
RESULTS
Effect of storage on the MMC.
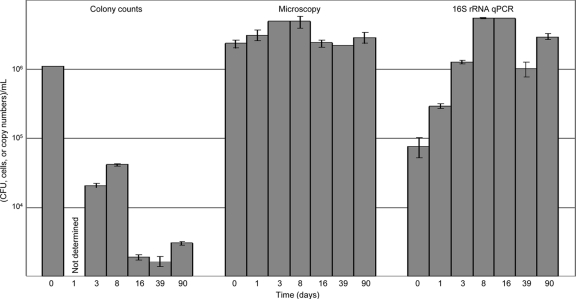
While storage for up to 90 days at −80°C had little or no effect on total cell/spore counts derived via Petroff-Hausser light microscopy counting (∼106 cells/spores per ml), the cultivability of MMC constituents decreased as much as 3 orders of magnitude (Fig. 2). When MMC aliquots were removed from −80°C storage at various time points (up to 90 days), subjected directly to DNA extraction protocols, and assessed via qPCR, the total DNA yield increased with increased storage time through 8 days, at which time it remained constant for up to 90 days at −80°C (Fig. 2).
Fig. 2.
Effect of low-temperature storage on MMC cellular cultivability and 16S rRNA gene integrity. Samples were stored at −80°C and processed at various intervals. CFU were counted after culture under appropriate conditions, total cells were enumerated microscopically with a Petroff-Hausser counting chamber, and 16S rRNA gene copy numbers were measured via qPCR. Error bars represent the standard error of the mean (n = 10).
Effects of sampling and processing variables on the MMC.
Using the MMC as a positive control, several processing techniques were thoroughly evaluated prior to the testing and evaluation of the different surface sampling regimens. Glycerol concentrations of >5% hindered the centrifugal filtration of sample volumes, presumably due to the clogging of filter pores (data not shown). In addition, results stemming from the use of two distinct types of suspension media (water and PBS) for deposition of the MMC onto stainless steel revealed a greater recovery (150-fold) of MMC-borne DNA when cells were suspended in water. Scanning electron microscopy showed a high degree of salt crystal formation on metal coupons laden with PBS-suspended MMC, whereas those seeded with water-suspended MMC had no visible crystals (see Fig. S1 in the supplemental material). In this study, ultrasonic energy, previously reported to facilitate the release of bacterial spores from sampling materials (31), did not dramatically hinder the cultivation of MMC strains (∼80% cultivable after sonication), nor did it compromise the integrity of sample DNA molecules (see the supplemental material). Finally, following concentration via Amicon 50-kDa centrifugal filters, ∼60% of the MMC DNA was recovered. However, additional washing of the filters did not result in a greater DNA yield.
Recovery of the MMC from stainless steel surfaces. (i) Small surface areas.
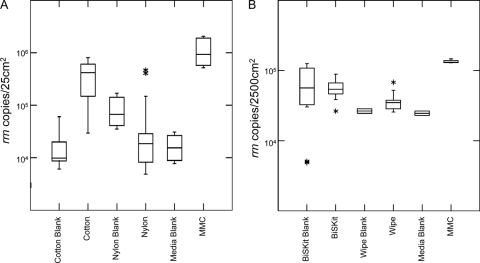
The efficiency of MMC constituent recovery from 25-cm2 stainless steel surface areas by cotton and nylon-flocked swabs was comparatively analyzed via qPCR (Fig. 3A). Using the mean MMC rRNA gene abundance as a point of comparison, cotton significantly outperformed nylon-flocked swabs (P < 0.00001, n = 30) with recovery percentages of 32.8% and 4.3%, respectively. While the signal-to-noise (noise is contaminant DNA) ratio associated with cotton swab sampling blanks was high (19.6), nylon-flocked sampling blanks had a much lower ratio (0.6). In addition, a wide range of values resulted among both cotton (104 to 105 rrn copies/25 cm2) and nylon-flocked (103 to 105 rrn copies/25 cm2) sampling replicates, possibly attributable to subtle variability in the composition of the swabbing matrices or the stainless steel surface.
Fig. 3.
Recovery of MMC constituents from metal surfaces by various sampling materials. Aliquots of the MMC were deposited onto metal surfaces, dried for 24 h, and collected with various sampling materials. Samples were analyzed via 16S rRNA qPCR, and results were visualized with box plots showing the mean and upper and lower quartiles for cotton and nylon-flocked swabs (A) and BiSKits and polyester wipes (B). Outliers (a greater-than-3-sigma deviation from the mean) are represented by asterisks. n = 30 measurements for each sampling material. Error bars depict the standard error of the mean in box plot format.
(ii) Large surface areas.
The efficiency of MMC constituent recovery from 2,500-cm2 stainless steel surface areas by BiSKit and polyester wipes was comparatively analyzed via qPCR (Fig. 3B). Using the mean MMC rRNA gene abundance as a point of comparison, BiSKit significantly outperformed wipes (P < 0.00001, n = 21) with recovery percentages of 41.9% and 26.7%, respectively. While the signal-to-noise (noise is contaminant DNA) ratio associated with polyester wipe sampling blanks was high (4.7), BiSKit sampling blanks had a much lower ratio (1.1). In addition, a wide range of values resulted among BiSKit sampling replicates (104 to 105 rrn copies/2,500 cm2), possibly attributable to subtle variability in the composition of the sampling matrices or the stainless steel surface.
ssPCR.
Species-specific oligonucleotide primers targeting the rRNA genes of each of the MMC constituents were developed alongside PCR thermal cycling regimens to assess the recovery of each MMC strain. Target PCR amplicons ranged in size from 83 bp (Microbacterium imperiale) to 380 bp (B. pumilus, Table 2), which enabled qPCR analysis of each strain's rRNA gene. The detection limit of ssPCR for each MMC constituent was established by serially diluting intact vegetative cells (from 107 to 101 cells or spores/ml) and subsequently subjecting each dilution to DNA purification and appropriate ssPCR conditions. These limits have, to date, varied extensively (101 to 106; Table 3). Notably, the ssPCR conditions used for B. pumilus SAFR-032 spores were highly sensitive, capable of amplifying 380-bp products from spore densities as low as 4.6 × 101 spores/ml. The other aerobic spore former, B. megaterium KL-197, yielded specific PCR products at 102 cells/ml. In contrast, detection limits for the anaerobic species C. sporosphaeroides, and M. formicicum (105 cells/ml) were determined from serially diluted DNA rather than intact cells since these anaerobic cultures did not yield uniform cell densities below 105 cells/ml.
Table 3.
Limits of detection of MMC constituents by ssPCR
| Sample no. | Microbe | Presence of amplicons at initial inoculum concn (cells/ml)a of: |
Detection limit (CFU/ml)b | |||||
|---|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 101 | |||
| 1 | Aureobasidium pullulans 28v1 | + | − | − | − | − | − | 9.0 × 106 |
| 2 | Acinetobacter radioresistens 50v1 | + | + | + | + | − | − | 3.4 × 103 |
| 3 | Bacillus megaterium KL-197 | + | + | + | + | + | − | 2.1 × 102 |
| 4 | Bacillus pumilus SAFR-032 | + | + | + | + | + | + | 4.6 × 101 |
| 5 | Deinococcus radiodurans ATCC 13939 | + | + | + | − | − | − | 1.6 × 104 |
| 6 | Microbacterium imperiale 47v1 | + | + | + | + | − | − | 5.6 × 103 |
| 7 | Staphylococcus warneri 82-4 | + | + | + | − | − | − | 4.3 × 104 |
| 8 | Micrococcus luteus ATCC 4698 | + | + | + | − | − | − | 4.5 × 104 |
| 9 | Cupriavidus metallidurans CH34 | + | + | + | + | − | − | 4.7 × 103 |
| 10 | Clostridium sporosphaeroides DSM 1294c | + | + | − | − | − | − | 7.7 × 105 |
| 11 | Methanobacterium formicicum DSM 1535c | + | + | − | − | − | − | 1.4 × 105 |
The number of cells was determined via microscopic counting. After serial dilution, DNA from the reaction mixtures was extracted using the Maxwell 16 system. See Materials and Methods for details.
The PCR set was scored positive (+) when the amplified products were ≥0.25 ng/μl, as quantified by Bioanalyzer.
DNA, instead of whole cells, was serially diluted for this strain.
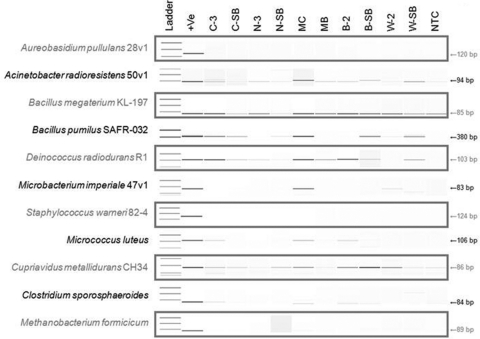
Trends in the differential recovery of MMC constituents achieved by various sampling devices were evaluated via standard and quantitative ssPCRs (Table 4). Traditional ssPCRs were performed with DNA extracts arising from each sampling replicate, and resulting amplicons were visualized with an Agilent 2100 Bioanalyzer. As was observed with general rRNA gene qPCR, cotton swabs proved more effective than nylon-flocked swabs in collecting MMC-derived DNA from stainless steel surfaces (66.2% versus 48.1% occurrence, respectively; Table 4) following upon ssPCR analysis. Similarly, BiSKits outperformed polyester wipes in the recovery of MMC constituents from large surfaces (55.8% versus 40.3% incidence, respectively; Table 4). B. megaterium KL-197 was the only MMC strain whose rRNA gene was recovered from every sampling device replicate (Fig. 4 and Table 4). The rRNA genes of other desiccation-tolerant strains, such as D. radiodurans R1 and Acinetobacter radioresistens 50v1, were retrieved in the majority of swab and BiSKit sampling replicates (Fig. 4 and Table 4). Aureobasidium pullulans 28v1 and Staphylococcus warneri 82-4 were the only MMC constituents whose DNA was not recovered, regardless of the sampling matrix employed (Fig. 4 and Table 4). Coincidentally, the detection limits for these strains were among the highest within the MMC (9.0 × 106 and 4.3 × 104, respectively). Finally, effective recovery of the C. sporosphaeroides rRNA gene was limited to both swabbing materials from small surface samples, as BiSKits and polyester wipes failed to yield amplifiable DNA from large surfaces (Fig. 4 and Table 4).
Table 4.
Presence/absence of specific MMC species from several sampling material replicates
| Microbea | PCR amplicon size (bp) | Presence/absence of microbesb after sampling with: |
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cotton swab replicates |
Nylon-flocked swab replicates |
BiSKit replicates |
Polyester wipe replicates |
||||||||||||||||||||||||||
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | N1 | N2 | N3 | N4 | N5 | N6 | N7 | B1 | B2 | B3 | B4 | B5 | B6 | B7 | W1 | W2 | W3 | W4 | W5 | W6 | W7 | ||
| Aureobasidium pullulans 28v1 | 120 | ||||||||||||||||||||||||||||
| Acinetobacter radioresistens 50v1 | 94 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| Bacillus megaterium KL-197 | 85 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Bacillus pumilus SAFR-032 | 380 | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||||||
| Deinococcus radiodurans ATCC 13939 | 103 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||
| Microbacterium imperiale 47v1 | 83 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||
| Staphylococcus warneri 82-4 | 124 | ||||||||||||||||||||||||||||
| Micrococcus luteus ATCC 4698 | 106 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| Cupriavidus metallidurans CH34 | 86 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||
| Clostridium sporosphaeroides DSM 1294 | 84 | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||||
| Methanobacterium formicicum DSM 1535 | 89 | + | + | ||||||||||||||||||||||||||
A. pullulans and S. warneri were not detected in any of the sampling materials.
The PCR set was scored positive (+) when the amplified products were ≥0.25 ng/μl as quantified by the Agilent Bioanalyzer. The percent occurrence was calculated based on positive amplification from the total number of assays. For example, of the total of 77 samples (11 species × 7 replicates per sampling device), percent incidence is the percentage of 77 samples where DNA was detected.
Fig. 4.
ssPCR gel electrophoresis results for each MMC constituent. Selected sample replicates and sampling blanks of each sampling device, MMC aliquots, and media blanks were subjected to ssPCR for each MMC strain. ssPCR products were visualized via microchannel-based electrophoresis with an Agilent 2100 Bioanalyzer. Abbreviations: +Ve, positive control (DNA from cultured cells); C-3, cotton swab replicate 3; C-SB, cotton swab sampling blank; N-3, nylon-flocked swab replicate 3; N-SB, nylon-flocked swab sampling blank; MC, MMC (unseeded); MB, medium blank used for MMC; B-2, BiSKit replicate 2; B-SB, BiSKit sampling blank; W-2, polyester wipe replicate 2; W-SB, polyester wipe sampling blank; NTC, no-template control.
DISCUSSION
The goal of this study was to evaluate and standardize protocols and procedures involved with the collection, processing, and molecular analysis of contaminant biomolecules from SAC surfaces. While several protocols are in place for the sampling of low-biomass surfaces (1, 2, 4, 11, 19, 25, 29), few employ molecular-analysis-based methods, and most experiment only with one or two test organisms. In this study, a consortium of bacterial spores and cells representing all 3 domains of microbial life was engineered as a proxy for the diverse, low-abundance microbial populations typical of clean-room environments. This MMC was used to test and optimize the numerous stages involved in molecular microbial diversity analysis. To the best of our knowledge, this is the first study to employ a complex MMC of diverse phylogenetic relevance to the testing and optimization of protocols for collecting, processing, and analyzing biomolecular samples from clean-room surfaces.
Using the MMC as a positive control, a complex suite of sample handling and processing techniques was thoroughly evaluated prior to the testing and evaluation of different sampling regimens. The use of cryoprotectant chemicals (e.g., dimethyl sulfoxide, glycerol) is well known to protect microbial cells and allow increased viability and cultivability following prolonged storage at low temperatures (20). However, as the MMC was intended to mimic the complex array of live, dead, and degraded material found on clean-room surfaces, no attempt to preserve the integrity of MMC organisms was made once the MMC was mixed. Instead, care was taken to protect the molecular analyses from the seemingly unavoidable presence of contaminant nucleic acid residues associated with cryoprotectant reagents. Furthermore, glycerol concentrations suitable for maintaining cellular cultivability (20) impeded the centrifugal filtration of large sample volumes. Ergo, the use of such chemicals was intentionally avoided for this study and the MMC was stored at −80°C in PBS only. Similarly, while PBS was used for storing the MMC, it was found to hinder the recovery of MMC biomolecules. PBS and other buffered solutions may help prevent cellular death and degradation resulting from osmotic stress, but such protection comes with a cost: the precipitation and deposition of salt crystals onto the metal surfaces induce changes in surface chemistry and hinder the recovery of microbes (150-fold lower than with water-suspended MMC) (see Fig. S1 in the supplemental material). Furthermore, such protection from drying and other osmotic stresses is not representative of the conditions within clean-room environments. Therefore, the MMC was harvested via centrifugal filtration from PBS and resuspended in sterile water (with accepted consequences for osmotically sensitive MMC strains) immediately prior to its deposition onto metal surfaces.
For the purposes of this evaluative study and in accordance with previously published findings, it was important to consider the amount of time the MMC spent in a completely desiccated state while dried onto the metal surface. By applying tape-lifting and/or polyvinyl alcohol coating techniques, effective removal of bacterial spores from metal surfaces was demonstrated, with recoveries as high as 90% for up to 18 months of desiccation (37). With regard to vegetative cells, however, previous unpublished studies have shown that the recovery of vegetative cells from metal surfaces was negatively influenced by increasing drying times on metal surfaces or swab heads (from a few to 24 h). In order to accurately mimic the conditions present in a humidity-controlled clean room (relative humidity of ∼40 to 55%, facility dependent), the MMC was deposited and allowed to dry for 24 h on the metal surface prior to sample collection. There were several variables to examine with regard to the processing of collected MMC samples. Subjecting sampling devices to a sonic bath upon the addition of resuspension solution (sterile water or PBS) has been shown to promote the release of cells and spores from cotton, foam, and polyester matrices (31). In this study, sonication did not dramatically hinder the cultivation of MMC strains (∼80% cultivable after sonication), nor did it compromise the integrity of sample DNA molecules. As different sampling devices required different volumes of resuspension fluid (∼45 to 600 ml) for optimum recovery, following sonication, the sample eluates underwent a centrifugal concentration step with Amicon filters. This concentration of cells, spores, and all other materials of biotic origin proved very effective and facilitated molecular biological downstream analysis, which is not possible at lower concentrations. The ∼ 60% recovery of DNA achieved using Amicon centrifugal filters was superior to all other available protocols. Previously, DNA loss as high as 90% was reported when ∼10 ml of sample was directly subjected to standard phenol-chloroform-based DNA extraction procedures, likely resulting from the direct processing of large sample volumes (>10 ml) without prior concentration (17, 33). Other studies have also confirmed the necessity of centrifugation or filtration of large sample volumes prior to downstream DNA processing and analysis (4). The Amicon centrifugal filtration procedure advocated in this study proved effective and yielded reproducible results for sample volumes of up to 45 ml. When processing the ∼600-ml samples collected with polyester wipes, the entire volume was first subjected to 0.22-μm membrane filtration and then further concentrated via Amicon centrifugal filtration.
To achieve thorough and effective lysis of the hardiest and most diverse microbial cell walls and outer membranes, both automated and manual alkaline lysis-based DNA purification procedures can be augmented with some form of mechanical abrasion (33). A simple bead-beating step prior to nucleic acid extraction protocols was necessary to lyse open the extremely resilient actinobacterial cells and even bacterial spores from environmental samples (17, 42). However, bead beating or any other form of mechanical abrasion could inadvertently shear and/or significantly degrade large nucleic acid polymers from less resilient organisms (17). To avoid biases in this regard, the samples collected in this study were split into two equivalent portions. One portion was subjected to bead beating prior to undergoing nucleic acid extraction, and the other portion was not subjected to mechanical abrasion prior to the purification of nucleic acids. Both portions were pooled together and subjected to automated DNA extraction as one sample volume (17). As was previously recommended, automated methods are preferred to traditional manual phenol-chloroform-based DNA extraction techniques and were employed during this study so as to maximize reproducibility and avoid technician-based variability (17).
Recently, a comparative evaluation of cotton swabs with other swab varieties concluded that nylon-flocked swabs were superior to cotton swabs for the collection of Bacillus sp. spores from stainless steel and other surfaces (29). In a similar vein, the use of nylon-flocked swabs resulted in more-efficient recovery of cultivable contaminants from pharmaceutically relevant surfaces (7). To date, the literature is devoid of reports similar to that of the present investigation, which focused on evaluating procedures for the effective and efficient collection and processing of not only intact microorganisms but a wide spectrum of biomaterials (from cells to nucleic acid remnants) from clean-room surfaces. This study found nylon-flocked swabbing material to be comparable to cotton for removing intact cultivable cells and spores from small (∼25-cm2) metal surfaces (≤1% recovery). However, cotton swabs consistently yielded a higher recovery of rRNA gene copies than nylon-flocked swabs (10-fold more). BiSKit sampling modules were superior (2-fold more) to NASA-certified polyester wipes for retrieving DNA molecules from larger surface areas, perhaps a consequence of the highly porous nature of the BiSKit macrofoam versus the tightly woven fibers within the polyester wipe.
Even though all of the sampling devices of a given type originated from the same lot, both cotton and nylon-flocked swabs suffered from remarkably high variability in resulting qPCR values for sampling blanks and across sample replicates (Fig. 3). Such inconsistency (up to 2 logs in the case of nylon-flocked swab replicates) could be indicative of a notable degree of variation in sampling material composition within the same lot and merits further investigation. In addition, the signal-to-noise ratios obtained with both nylon-flocked swabs and BiSKits were surprisingly low, suggestive of indigenous DNA associated with each sampling device.
Though laborious, ssPCR proved invaluable in assessing the differential recovery of MMC constituents by the various sampling matrices tested. In addition, this molecular technique was not at all affected by the considerable levels of indigenous DNA associated with sampling materials. As was observed with general rRNA gene qPCR, cotton swabs and BiSKits were superior to their respective counterparts in collecting MMC-borne DNA from metal surfaces. As could be expected, several of the MMC constituents most frequently detected via ssPCR were among those still cultivable after 90 days of storage at −80°C. These bacterial species were apparently the most resistant to desiccation and cryogenic storage. Additionally, it is not surprising that all sampling devices failed to recover ssPCR-amplifiable levels of DNA from A. pullulans 28v1 since the limit of detection of this eukaryotic strain was 9.0 × 106 gene copies. Since ∼1,300-bp regions of the 18S-internal transcribed spacer gene cluster were able to be amplified with as little as ∼103 CFU/reaction mixture (pure culture), it is apparent that ssPCR conditions must be further optimized to assess eukaryotic cells in this manner.
These results stress the importance of selecting and tailoring sample collection and processing approaches based on the desired target within the sample and the level of detection/resolution required. If the relevance of a given investigation is limited to the presence of intact, cultivable cells, then, based on other studies, nylon-flocked swabs would likely be the better sampling device. If, on the other hand, a survey is more interested in the entire spectrum of microbial diversity on a given surface, then the preferred sampling instrument would be either cotton swabs or BiSKits, depending on the surface area. The findings of this study tend to support those of previous work and hint at the need to readdress the use of standard methodologies over state-of-the-art, biomolecule-friendly sample collection and processing regimens.
Supplementary Material
ACKNOWLEDGMENTS
Part of the research described in this publication was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with NASA. This research was funded by the Mars Program Office.
We are grateful to F. Karouia, NASA Ames, for participating in the distribution of the MMC, J. Barengoltz for statistical analysis, and N. Leys for donating C. metallidurans CH34. We also appreciate the valuable advice and encouragement received from J. A. Spry, K. Buxbaum, C. Conley, and G. Kminek.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 11 March 2011.
REFERENCES
- 1. Brown G. S., et al. 2007. Evaluation of a wipe surface sample method for collection of Bacillus spores from nonporous surfaces. Appl. Environ. Microbiol. 73:706–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown G. S., et al. 2007. Evaluation of vacuum filter sock surface sample collection method for Bacillus spores from porous and non-porous surfaces. J. Environ. Monit. 9:666–671 [DOI] [PubMed] [Google Scholar]
- 3. Bruckner J. C., Osman S., Conley C., Venkateswaran K. 2008. Space microbiology: planetary protection, burden, diversity and significance of spacecraft associated microbes, p. 52–65 In Schaechter M. (ed.), Encyclopedia of microbiology. Elsevier, Oxford, United Kingdom [Google Scholar]
- 4. Buttner M. P., Cruz-Perez P., Stetzenbach L. D. 2001. Enhanced detection of surface-associated bacteria in indoor environments by quantitative PCR. Appl. Environ. Microbiol. 67:2564–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Committee on Preventing the Forward Contamination of Mars, National Research Council 2006. Preventing the forward contamination of Mars. The National Academies Press, Washington, DC [Google Scholar]
- 6. COSPAR 2002. Planetary protection policy, October 2002, as amended, March 2008. COSPAR, Houston, TX [Google Scholar]
- 7. Dalmaso G., Bini M., Paroni R., Ferrari M. 2008. Qualification of high-recovery, flocked swabs as compared to traditional rayon swabs for microbiological environmental monitoring of surfaces. PDA J. Pharm. Sci. Technol. 62:191–199 [PubMed] [Google Scholar]
- 8. Edmonds J. M., et al. 2009. Surface sampling of spores in dry-deposition aerosols. Appl. Environ. Microbiol. 75:39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Estill C. F., et al. 2009. Recovery efficiency and limit of detection of aerosolized Bacillus anthracis Sterne from environmental surface samples. Appl. Environ. Microbiol. 75:4297–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghosh S., Osman S., Vaishampayan P., Venkateswaran K. 2010. Recurrent isolation of extremo-tolerant bacteria from the clean room where Phoenix spacecraft components are assembled. Astrobiology 10:325–335 [DOI] [PubMed] [Google Scholar]
- 11. Hodges L. R., Rose L. J., O'Connell H., Arduino M. J. 2010. National validation study of a swab protocol for the recovery of Bacillus anthracis spores from surfaces. J. Microbiol. Methods 81:141–146 [DOI] [PubMed] [Google Scholar]
- 12. Hodges L. R., Rose L. J., Peterson A., Noble-Wang J., Arduino M. J. 2006. Evaluation of a macrofoam swab protocol for the recovery of Bacillus anthracis spores from a steel surface. Appl. Environ. Microbiol. 72:4429–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kempf M. J., Chen F., Kern R., Venkateswaran K. 2005. Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology 5:391–405 [DOI] [PubMed] [Google Scholar]
- 14. Kirschner L. E., Puleo J. R. 1979. Wipe-rinse technique for quantitating microbial contamination on large surfaces. Appl. Environ. Microbiol. 38:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. La Duc M. T., et al. 2007. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean-room environments. Appl. Environ. Microbiol. 73:2600–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. La Duc M. T., Nicholson W., Kern R., Venkateswaran K. 2003. Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility. Environ. Microbiol. 5:977–985 [DOI] [PubMed] [Google Scholar]
- 17. La Duc M. T., Osman S., Venkateswaran K. 2009. Comparative analysis of methods for the purification of DNA from low-biomass samples based on total yield and conserved microbial diversity. J. Rapid Methods Autom. Microbiol. 17:350–368 [Google Scholar]
- 18. Lawrence J. R., Korber D. R., Hoyle B. D., Costerton J. W., Caldwell D. E. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewandowski R., Kozlowska K., Szpakowska M., Stepinska M., Trafny E. A. 2010. Use of a foam spatula for sampling surfaces after bioaerosol deposition. Appl. Environ. Microbiol. 76:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X., Ricke S. C. 2004. Comparison of cryoprotectants for Escherichia coli lysine bioavailability assay culture. J. Food Process. Preserv. 28:39–50 [Google Scholar]
- 21. Moissl C., Bruckner J., Venkateswaran K. 2008. Archaeal diversity analysis of spacecraft assembly facilities. ISME J. 2:115–119 [DOI] [PubMed] [Google Scholar]
- 22. Moissl C., La Duc M. T., Osman S., Dekas A. E., Venkateswaran K. 2007. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol. Ecol. 61:509–521 [DOI] [PubMed] [Google Scholar]
- 23. Moissl-Eichinger C. 2011. Archaea in artificial environments: their presence in global spacecraft clean rooms and impact on planetary protection. ISME J. 5:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. National Aeronautics and Space Administration 1967. Harmful contamination: moon treaty United Nations treaty on principles governing the activities of states in the exploration and use of outer space, including the moon and other celestial bodies. Article IX, UN doc. A/RES/222/(XXI), TIAS no. 6347). United Nations Office for Outer Space Affairs, Vienna, Austria [Google Scholar]
- 25. National Aeronautics and Space Administration 1980. NASA standard procedures for the microbiological examination of space hardware. Office of Space Science, National Aeronautics and Space Administration, Washington, DC [Google Scholar]
- 26. National Aeronautics and Space Administration 2005. Planetary protection provisions for robotic extraterrestrial missions. NPR 8020.12C, April 2005 National Aeronautics and Space Administration, Washington, DC [Google Scholar]
- 27. Newcombe D. A., LaDuc M. T., Vaishampayan P., Venkateswaran K. 2008. Impact of assembly, testing, and launch operations on the airborne bacterial diversity within a spacecraft assembly facility clean-room. Int. J. Astrobiol. 7:223–236 [Google Scholar]
- 28. Panaro N. J., et al. 2000. Evaluation of DNA fragment sizing and quantification by the Agilent 2100 bioanalyzer. Clin. Chem. 46:1851–1853 [PubMed] [Google Scholar]
- 29. Probst A., Facius R., Wirth R., Moissl-Eichinger C. 2010. Validation of a nylon-flocked-swab protocol for efficient recovery of bacterial spores from smooth and rough surfaces. Appl. Environ. Microbiol. 76:5148–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Probst A., et al. 2010. Diversity of anaerobic microbes in spacecraft assembly clean rooms. Appl. Environ. Microbiol. 76:2837–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puleo J. R., Favero M. S., Petersen N. J. 1967. Use of ultrasonic energy in assessing microbial contamination on surfaces. Appl. Microbiol. 15:1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rose L., Jensen B., Peterson A., Banerjee S. N., Srduino M. J. 2004. Swab materials and Bacillus anthracis spore recovery from nonporous surfaces. Emerg. Infect. Dis. 10:1023–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Sanderson W. T., et al. 2002. Surface sampling methods for Bacillus anthracis spore contamination. Emerg. Infect. Dis. 8:1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stieglmeier M., Wirth R., Kminek G., Moissl-Eichinger C. 2009. Cultivation of anaerobic and facultatively anaerobic bacteria from spacecraft-associated clean rooms. Appl. Environ. Microbiol. 75:3484–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suzuki M. T., Taylor L. T., DeLong E. F. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. App. Environ. Microbiol. 66:4605–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tauscher C., Schuerger A. C., Nicholson W. L. 2006. Survival and germinability of Bacillus subtilis spores exposed to simulated Mars solar radiation: implications for life detection and planetary protection. Astrobiology 6:592–605 [DOI] [PubMed] [Google Scholar]
- 38. Vaishampayan P., Osman S., Andersen G., Venkateswaran K. 2010. Molecular methods to monitor microbial community composition of the phoenix spacecraft assembly clean room. Astrobiology 10:499–508 [DOI] [PubMed] [Google Scholar]
- 39. Valentine N. B., et al. 2008. Evaluation of sampling tools for environmental sampling of bacterial endospores from porous and nonporous surfaces. J. Appl. Microbiol. 105:1107–1113 [DOI] [PubMed] [Google Scholar]
- 40. Venkateswaran K., Chung S., Allton J., Kern R. 2004. Evaluation of various cleaning methods to remove Bacillus spores from spacecraft hardware materials. Astrobiology 4:377–390 [DOI] [PubMed] [Google Scholar]
- 41. White T. J., Bruns T. D., Lee S. B., Taylor J. W. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322 In Innis N., Gelfand D., Sninsky J., White T. (ed.), PCR protocols and applications—a laboratory manual. Academic Press, New York, NY [Google Scholar]
- 42. Wilson K. H., et al. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.