Abstract
The α subunit of E. coli RNAP plays an important role in the recognition of many promoters by binding to the A+T-rich UP element, a DNA sequence located upstream of the recognition elements for the ς subunit, the −35 and −10 hexamers. We examined DNA–RNAP interactions using high resolution interference and protection footprinting methods and using the minor groove-binding drug distamycin. Our results suggest that α interacts with bases in the DNA minor groove and with the DNA backbone along the minor groove, but that UP element major groove surfaces do not make a significant contribution to α binding. On the basis of these and previous results, we propose a model in which α contacts UP element DNA through amino acid residues located in a pair of helix–hairpin–helix motifs. Furthermore, our experiments extend existing information about recognition of the core promoter by ς70 by identifying functional groups in the major grooves of the −35 and −10 hexamers in which modifications interfere with RNAP binding. These studies greatly improve the resolution of our picture of the promoter–RNAP interaction.
Keywords: RNA polymerase, promoter, alpha subunit, UP element, minor groove, helix–hairpin–helix
Promoters in bacteria contain recognition sequences for RNA polymerase (RNAP, subunit composition α2ββ‘ςω) in up to three distinct regions: the −10 and −35 elements, both recognized by the ς70 subunit (for review, see Record et al. 1996; Gross et al. 1998), and the UP element, recognized by the α subunit (for review, see Gourse et al. 2000). The UP element, located just upstream of the −35 hexamer, was identified as a binding site for α in the rRNA promoter rrnB P1 of Escherichia coli, where it stimulates promoter activity ∼30-fold (Ross et al. 1993). UP elements have been identified in other promoters in E. coli, in other bacterial species, and in promoters transcribed by holoenzymes containing alternative ς factors (e.g., Moran et al. 1982; Newlands et al. 1992; Ross et al. 1993, 1998 Fredrick et al. 1995; Negre et al. 1997). The optimal (consensus) UP element sequence was identified by in vitro selection and contains alternating A and T tracts in two distinct subsites (proximal and distal; Estrem et al. 1998, 1999). Analysis of the E. coli genome sequence suggests that UP elements, consisting of either one or two subsites with no more than two mismatches to consensus, occur in more than one-third of stable RNA promoters, but are also found in about 4% of mRNA promoters (Estrem et al. 1999). Because sequences with much lesser degrees of match to consensus can still significantly affect promoter function (Ross et al. 1998), the number of promoters in which UP elements play some role in transcription is likely to be much larger. DNA–α interactions also occur adjacent to many activator protein-binding sites, although, in these cases, α interactions are often DNA sequence independent, the binding site(s) do not resemble the UP element consensus, and α binding does not occur in the absence of the bound activator (Hochschild and Dove 1998; Busby and Ebright 1999; Gourse et al. 2000).
The ∼37 kD α subunit contains two independently folded domains (Blatter et al. 1994). The amino-terminal domain (residues 8–233) is responsible for dimerization and interactions with β and β‘ (Hayward et al. 1991), and the carboxy-terminal domain (αCTD, residues 245–329) contains the DNA-binding determinants (Blatter et al. 1994). Genetic analyses have identified amino acid residues in the αCTD critical for DNA binding and UP element function (Gaal et al. 1996; Murakami et al. 1996). These residues are essential for cell viability (Gaal et al. 1996) and are highly conserved in prokaryotic α sequences. Amino acid sequence comparisons in conjunction with the solution structure of the αCTD, determined by nuclear magnetic resonance spectroscopy (Jeon et al. 1995; Gaal et al. 1996), suggest that α is a member of the helix–hairpin–helix (HhH) domain family of DNA-binding proteins (Shao and Grishin 2000).
Defining the interactions between components of the transcription initiation apparatus remains a central problem in molecular biology. Recently, there have been dramatic advances in our understanding of the structure of bacterial core RNAP at the atomic level (Zhang et al. 1999), but the αCTD was not resolved in this structure, and no information at the atomic level exists for complexes containing DNA bound to α or RNAP holoenzyme.
Footprinting and cross-linking studies have provided some information about the α–UP element interaction and the promoter–RNAP interaction. Footprints of RNAP on rrnB P1 and other promoters extend about 60 bp upstream of the transcription start site, and protection upstream of ∼−40 is attributable to interactions with α (Kolb et al. 1993; Ross et al. 1993). In the rrnB P1 UP element, DNA backbone regions centered at ∼−42 and ∼−52 are protected against hydroxyl radical cleavage by α or RNAP (Newlands et al. 1991; Estrem et al. 1998). However, a full understanding of the mode of α binding has been hindered by complicating features of its interactions with different promoter upstream regions. Not only can α bind to DNA in either a sequence-specific or a sequence-nonspecific manner, but α-dependent protection signals have also been observed upstream of −60, and sometimes multiple regions of protection have been observed in the same upstream region (Newlands et al. 1992; Kolb et al. 1993; Fredrick et al. 1995; Belyaeva et al. 1998). Comprehensive cross-linking studies further suggest that the two αCTDs may oscillate among different binding sites (like fingers on a keyboard; Naryshkin et al. 2000), or alternatively may bind to different sites in different molecules in the population. Variability in the position(s) of α binding sites is most likely attributable to the long, flexible tether linking the α amino- and carboxy-terminal domains (Blatter et al. 1994; Jeon et al. 1997), as well as to intrinsic DNA distortions and protein-induced DNA bending in initiation complexes containing activators (Gourse et al. 2000; Naryshkin et al. 2000).
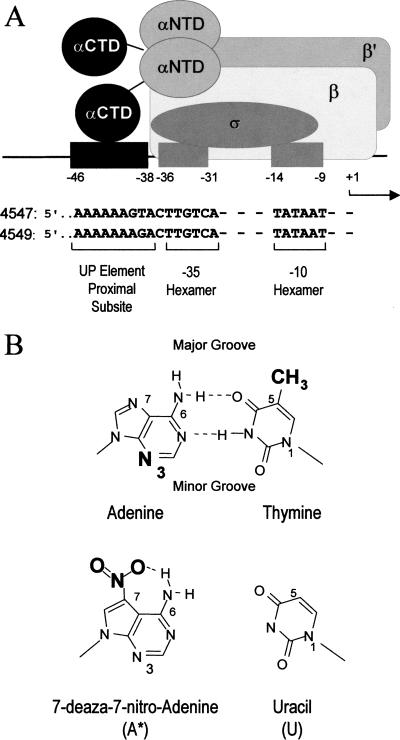
A binding site for a single α subunit (the consensus proximal subsite; see Fig. 1A) was defined by in vitro selection and by the use of RNAPs heterodimeric for α; [i.e., containing one wild-type α subunit and one α lacking its DNA-binding domain (ΔαCTD; Estrem et al. 1999)]. Only one αCTD was required for function of an UP element with only a proximal subsite and for protection of this subsite in hydroxyl radical footprints. The protected positions (about 4 bp on each DNA strand) are offset on the two strands by ∼3 bp, and are thus located in close proximity across the DNA minor groove (Estrem et al. 1999). Likewise, studies with a cross-linking agent tethered by a 10-Å linker indicated that αCTD interactions with a promoter upstream region lacking a functional UP element are centered on the DNA minor groove (Naryshkin et al. 2000). However, the footprinting and cross-linking studies do not provide sufficient information to build a structural model of the sequence-specific αCTD–UP element interaction; for example, proteins that bind to two successive major grooves are also centered on the minor groove. The previous studies do not distinguish whether the αCTD makes base contacts within a DNA groove, or whether these contacts, if they occur, are with the major groove or minor groove surfaces of the bases.
Figure 1.
Promoters and base analogs used in footprinting experiments. (A) RNAP binding to rrnB P1 with an UP element containing a single proximal subsite. The proximal subsite (black rectangle) is a binding site for one αCTD. The position of the other αCTD in this complex is not known. The ς subunit binds to the −10 and −35 hexamers (grey rectangles). The nontemplate strand sequences of the rrnB P1 −10 and −35 hexamers and the two UP elements used in this study (4547, 4549; Estrem et al. 1999) are shown. (B) Structures of an adenine:thymine (A:T) base pair and of the base analogs 7-deaza-7-nitro adenine (A*) and uracil (U). Surfaces facing the major and minor grooves are indicated for the A:T pair. Minor groove N3 position of A (bold) was probed in DMS footprints. Major groove positions N7 of A [modified in 7-deaza-7-nitro adenine to contain a nitro group on C7 (in bold); Min et al. 1996] and the C5 methyl group of T (in bold; not present in U) were probed in interference footprints using templates containing A* or U.
In this work, we address the basis for the specific interaction of α with UP element DNA. An understanding of bacterial promoter recognition and the architecture of transcription complexes containing activators requires detailed information about the αCTD–DNA interaction. Therefore, we carried out RNAP protection footprinting studies with dimethylsulfate (DMS) and interference footprinting studies with promoter fragments containing the thymine analog uracil or the adenine analog 7-deaza-7-nitro-adenine (A*; Fig. 1B). We also examined the effects of the minor groove-binding drug distamycin on α binding and on UP element-dependent transcription. These results and those from hydroxyl radical footprinting studies (Estrem et al. 1999) indicate that α binds to the DNA backbone across the minor groove and to bases within the minor groove. Together with genetic analyses of αCTD (Gaal et al. 1996; Murakami et al. 1996) and the recent proposal that α contains two HhH DNA binding motifs [an (HhH)2 domain; Shao and Grishin 2000)], our data suggest a model for the interaction of α with UP element DNA. In addition, our experiments provide detailed information about the proximity of the RNAP ς subunit to the major groove surface of thymine and adenine bases in the −35 and −10 hexamers.
Results
We carried out footprinting studies of RNAP on templates containing the rrnB P1 core promoter fused to UP elements consisting of only a single proximal subsite (UP elements 4547 or 4549; Estrem et al. 1999; see Fig. 1A). UP elements containing only a proximal subsite are the most frequent class of UP elements in the E. coli genome (Estrem et al. 1999). Most important, use of templates with only a proximal subsite simplifies interpretation to a single DNA–α interaction.
The UP elements used in these experiments are representative of the proximal subsite sequences identified by in vitro selection and can stimulate promoter activity >100-fold in vivo. Each contains an A tract extending from −46 to −41 but has a different sequence at positions −40 and −39 (Estrem et al. 1999). The region upstream of the proximal subsite in these promoters is not protected by RNAP and does not affect promoter function (Estrem et al. 1999).
Missing bases in the UP element interfere with RNAP binding
Limited information was available about the relative importance of individual positions in the UP elements, and no information was available about whether bases on the nontemplate strand, template strand, or both make contacts with α. As a first approach to understanding the role of specific bases in proximal subsite function, we carried out missing base interference footprinting (Brunelle and Schleif 1987). This method allows the importance of a single base at any promoter position to be determined by the selective depletion of templates lacking that base from the population of fragments able to form open complexes. Modified promoter fragment populations that contained on average one missing thymine per molecule were prepared by low frequency substitution of dUTP for TTP (Fig. 1B) followed by removal of the uracil base by use of uracil DNA glycosylase (UDG; Devchand et al. 1993). Only the base was removed, leaving the phosphodiester backbone intact.
Labeled promoter fragments containing missing bases were then incubated with RNAP. Because the α–UP element interaction increases the rate of association of the promoter with RNAP (Rao et al. 1994), the RNAP binding reaction was carried out at low RNAP concentration and in 20-fold molar excess of modified promoter fragment relative to RNAP, conditions in which complex formation was strongly dependent on the α–UP element interaction. The resulting promoter–RNAP complexes were separated from unbound DNA by electrophoresis on a nondenaturing gel, purified, subjected to strand cleavage at the abasic sites, and then fractionated on a high resolution denaturing gel. Missing bases that interfered with RNAP binding were under-represented in the strand cleavage profiles of bound DNA relative to their representation in the DNA population that was not incubated with RNAP.
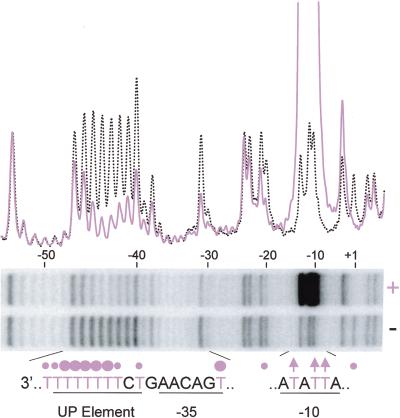
A typical missing T interference experiment carried out on a promoter with a consensus proximal subsite is shown in Figure 2. Each of the thymine positions in the proximal subsite (eight of nine positions on the template strand) was severely depleted in the RNAP-bound sample, indicating that a missing T at each of eight positions in the UP element interfered with RNAP binding. The strongest interference signals occurred in the center of the T tract (−41 to −45), whereas missing Ts upstream of the UP element sequence did not interfere with RNAP binding (Fig. 2 and data not shown). Identical results were obtained with an RNAP preparation containing only one αCTD (αI/αΔII heterodimeric RNAP; data not shown), indicating that, as expected from previous work (Estrem et al. 1999), missing Ts interfering with wild-type RNAP binding to the proximal subsite are attributable to interactions involving only one α subunit. No interference signals in the UP element were observed with RNAP lacking both αCTDs (data not shown).
Figure 2.
Missing thymine (T) base interference footprint of RNAP binding to rrnB P1 containing UP element 4549. The promoter fragment, with, on average, one missing T per molecule, was labeled in the template strand. RNAP–promoter complexes were isolated, and after strand cleavage at abasic sites, profiles were examined by gel electrophoresis. Gel lanes contain RNAP-bound populations (+) and control samples without RNAP (−). Superimposed profiles from scans of normalized lanes are aligned with the gel. (Purple solid line) RNAP-bound DNA fragments. (Black dotted line) control DNA fragments. UP element proximal subsite and −35 and −10 hexamer template strand sequences are shown; all Ts are in purple. (Circles) Missing Ts interfering with RNAP binding. (Arrows) missing Ts stimulating RNAP binding.
Missing Ts at some positions on each strand in the core promoter region also strongly affected RNAP binding (Fig. 2 and data not shown; summarized in Fig. 6, below). All missing Ts in the −35 hexamer and some in the spacer region between the −10 and −35 hexamers reduced RNAP binding. In contrast, most missing Ts in the −10 hexamer strongly stimulated RNAP binding (all but nontemplate strand position −9), most likely because unpaired nucleotides facilitate strand opening (see Fig. 2 and Discussion).
Figure 6.
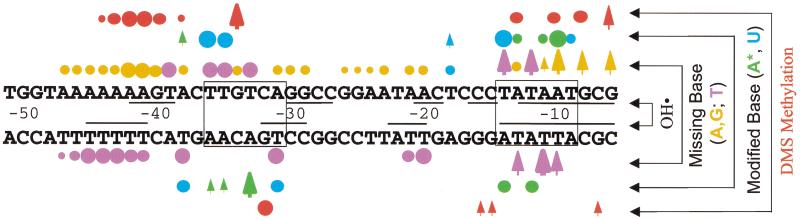
Summary of footprinting results (protection: DMS and hydroxyl radical; interference: missing base and modified base). Sequence is from rrnB P1 with UP element 4547 (−51 to −6). The −10 and −35 hexamers are boxed. Backbone positions protected by RNAP against hydroxyl radical cleavage (OH·): horizontal black lines above and below the sequences (data from Newlands et al. 1991; Estrem et al. 1999). Modified or missing base interference with RNAP binding (circles) or stimulation of RNAP binding (arrows). (Purple) Missing thymines; (orange) missing purines; (blue) uracil substitution; (green) A* substitution. (Red circles) Protection by RNAP against methylation by DMS; (red arrows) enhancement of methylation by bound RNAP. Sizes of symbols reflect the approximate degree of interference, stimulation, or protection.
Missing purine interference experiments (data not shown) with templates prepared by limited acid depurination (Maxam and Gilbert 1980), were used to examine the interaction of RNAP with the nontemplate strand. The conclusions from these experiments were similar to those from the missing T experiments: missing bases at the eight UP element positions that are purines, and at purine positions in the spacer, but not at control positions further upstream, resulted in reduced RNAP binding (for summary, see Fig. 6, below). Missing purines at positions −41 and −42 reduced RNAP binding the most. As reported previously (Li and McClure 1998), and as found in the missing T experiments, most missing purines in the −10 region (all but nontemplate strand −13) stimulated RNAP binding.
Major groove modifications affect RNAP binding to the core promoter but not to the UP element
The missing-base interference experiments indicated that each base in the UP element proximal subsite was important for α binding and suggested that bases in the center of the subsite (−41 to −45) are most critical. However, these results did not distinguish which base surfaces (major groove, minor groove, or both) are contacted by α, or alternatively, whether the interference signals might derive solely from indirect effects of missing bases on DNA backbone structure.
To determine whether α makes base contacts in the major groove, interference footprinting was carried out with promoter fragments containing adenine or thymine bases modified on their major groove surfaces. Modifications were introduced by limited incorporation of the nucleotide analogs dUTP or dA*TP (7-deaza-7-nitro dATP; Fig. 1B). U is identical to T except that it lacks the prominent major groove C5 methyl group. A* (Min et al. 1996) disrupts contacts to adenine at position 7 through steric exclusion and at position 6-NH through intramolecular hydrogen bonding. Both U and A* could potentially disrupt protein binding directly through loss of functional groups that contact amino acids, but, in addition, the adenine modification could potentially disrupt protein binding by steric hindrance. dUTP or dA*TP have not to our knowledge been used for footprints of RNAP, although they have been used in footprinting studies of other proteins (Pu and Struhl 1992; Devchand et al. 1993; Min et al. 1996).
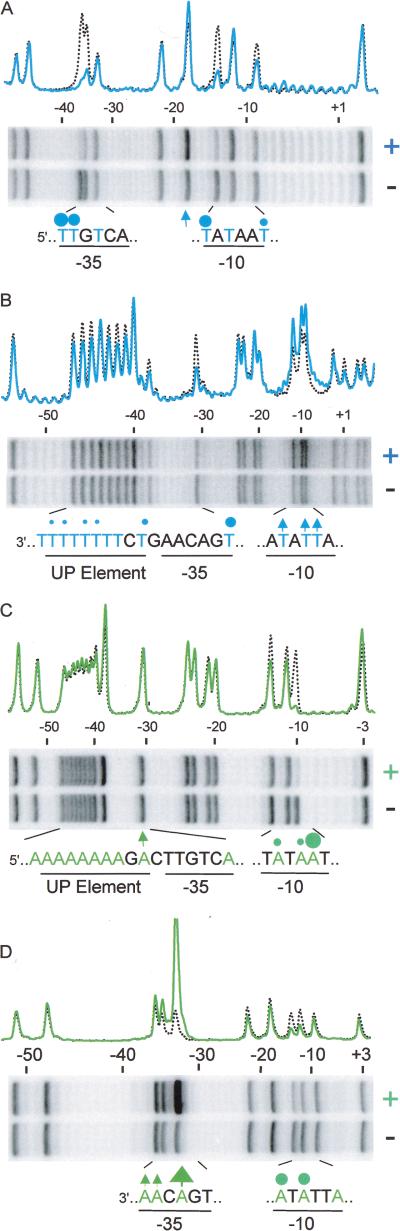
To carry out dUTP interference footprints, RNAP–promoter complexes were formed with templates containing, on average, one U substitution per molecule (Fig. 3A,B). Complexes were then gel isolated, and the DNA was purified, subjected to strand cleavage at modified bases, and fractionated on a denaturing gel. Uracil substitution at any of the eight thymine positions in the UP element had very little effect on RNAP binding (template strand, Fig. 3B), suggesting that α does not make critical contacts with major groove thymine methyl groups in the UP element. In contrast, U substitutions at any of three core promoter positions (−36, −35, and −14) strongly reduced or eliminated RNAP binding, and U substitutions at either of two other core promoter positions (−31 and −9) also reduced RNAP binding (Fig. 3A,B; for summary, see Fig. 6, below).
Figure 3.
Interference footprints with the base analogs uracil (U; A,B) or 7-deaza-7-nitro adenine (A*; C,D). Promoter fragments (rrnB P1 with UP element 4549), containing, on average, one modified base per molecule, were labeled on the nontemplate strand (A,C) or the template strand (B,D). RNAP–promoter complexes were isolated, and samples were examined by gel electrophoresis after strand cleavage at modified bases. Superimposed profiles from scans of normalized lanes are aligned with gels; RNAP-bound samples (+) are indicated by solid blue lines (uracil; A,B) or green lines (A*; C,D), and control samples without RNAP (−) are black dotted lines. UP element, −35 and −10 hexamer sequences are shown, with all T positions in blue (A,B), and all A positions in green (C,D). Modified bases that affected RNAP binding are indicated: (blue or green circles) interference with binding; (blue or green arrows) stimulation of binding. Symbol sizes correlate with the approximate degree of interference or stimulation observed.
Similar experiments were carried out with promoter fragments containing A* substitutions (Fig. 3C,D). As in the U substitution experiments, A* at UP element positions had little or no effect on RNAP binding (nontemplate strand, Fig. 3C). However, A* substitutions had large effects on RNAP interactions with the core promoter. A* substitutions reduced binding at five of the six A residues in the two strands of the −10 hexamer (Fig. 3C,D; for summary, see Fig. 6, below); A* at nontemplate strand position −10 virtually eliminated binding of RNAP, and at template strand position −33 (in the −35 hexamer) strongly enhanced RNAP binding (see Discussion).
In summary, the U and A* interference experiments indicate that major groove base surfaces in the UP element play little if any direct role in RNAP binding, although they do play a major role in interactions between RNAP and the −10 and −35 elements (presumably by contacting the ς subunit; see Fig. 6, below and Discussion).
Distamycin inhibits α binding and UP element function
Hydroxyl radical footprints with RNAP and with purified α previously suggested that α contacts the DNA backbone along the minor groove in the UP element proximal subsite (Newlands et al. 1991; Estrem et al. 1999). To investigate the role of the minor groove in α binding, we used reagents that bind in the minor groove (distamycin) or that modify the N3 of adenine, which is located in the minor groove (dimethylsulfate, DMS).
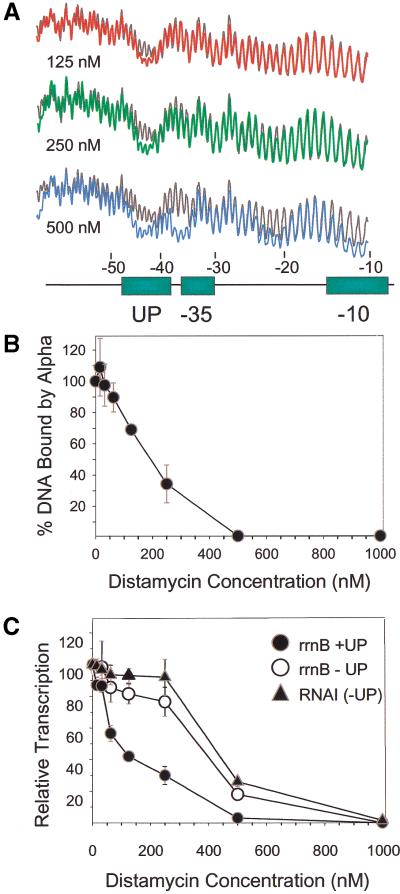
Distamycin, a small, well-characterized oligopeptide that binds within the minor groove of A+T-rich DNA (Coll et al. 1987; Churchill et al. 1990), was used to determine whether insertion of a small molecule in the minor groove of the UP element could compete with and prevent α binding to the promoter. Binding sites for distamycin in the promoter fragment were first characterized by hydroxyl radical footprinting to determine whether, as expected, the drug would bind preferentially to the UP element (Fig. 4A). Specific, but partial, protection in the UP element (positions −42 to −46) was observed at 125 nM distamycin. Protection of this site was more complete at 250 nM, but, at 500 nM, protection by distamycin also occurred at three other sites: in the −35 hexamer, in an A+T-rich region of the spacer (−20 to −24), and in the −10 hexamer (−10 to −14). The length of the protected regions, 4–5 bp, is consistent with the length reported previously in hydroxyl radical footprints with distamycin (Churchill et al. 1990).
Figure 4.
Distamycin experiments. (A) Scans of hydroxyl radical footprints of distamycin bound to a fragment containing rrnB P1 with UP element 4547. Profiles of distamycin-bound fragment (red, 125 nM distamycin; green, 250 nM: blue, 500 nM), are superimposed on profiles from the control fragment (no distamycin; grey lines). RNAP recognition elements (UP element, −35 and −10 hexamers), indicated by rectangles, are aligned with the scans. (B) Effect of pre-bound distamycin on the binding of purified α to a fragment containing rrnB P1 with UP element 4547. α binding was determined by a gel mobility shift assay; the percent of input DNA bound by α (averages with standard deviation from three separate experiments) is shown as a function of distamycin concentration. (C) Effect of pre-bound distamycin on transcription in vitro. Transcription from rrnB P1 with UP element 4547 (filled circles), rrnB P1 with no UP element (open circles), and from the RNA I promoter (filled triangles) was quantified from gel scans, and is expressed as a percentage of transcription from each promoter in the absence of distamycin. Results are the average (with range) from two independent experiments.
Because α and distamycin protect the same UP element positions against hydroxyl radical cleavage, the identity of the molecule binding to the UP element cannot be distinguished by footprinting when both the protein and the drug are present. However, purified α–UP element complexes migrate differently on gels from free or distamycin-bound DNA fragments. Therefore, we used electrophoretic mobility-shift assays to determine the effect of distamycin on binding of purified α to a promoter fragment.
The mobility shift assay was carried out with a concentration of α (2 μM dimer) where α bound specifically to the UP element (Ross et al. 1993). Formation of the α–DNA complex was inhibited by distamycin (Fig. 4B). Half-maximal inhibition occurred at ∼200 nM distamycin, a concentration where the drug bound specifically to the UP element and not to other regions of the promoter fragment (Fig. 4A). We conclude that distamycin competes with α for binding to the UP element, consistent with the model that α binds to the UP element minor groove. (At higher α concentrations, where α also binds to other DNA sites, higher concentrations of distamycin were required to inhibit α binding in mobility shift assays; data not shown).
Figure 4C shows the effects of a range of distamycin concentrations on UP element-dependent transcription in vitro. We examined expression from three different promoters, the rrnB P1 core promoter with a consensus proximal subsite, the rrnB P1 core promoter without an UP element, and the RNA I promoter (which also lacks an UP element) encoded by the plasmid vector. Transcription from the UP element-containing promoter was inhibited at concentrations of distamycin from 62.5 to 250 nM, whereas transcription from the promoters lacking UP elements was not affected, correlating well with the concentrations at which distamycin bound specifically to the UP element (Fig. 4A) and at which distamycin specifically inhibited α binding (Fig. 4B). We conclude that distamycin binding in the minor groove blocks UP element function in transcription. At higher distamycin concentrations (≥500 nM), transcription was inhibited from all three promoters (Fig. 4C), consistent with the observed binding of distamycin to core promoter sites. At these core promoter sites, high concentrations of distamycin may compete with RNAP for binding to DNA (e.g., at positions in the −10 region or in the spacer region that are protected by both distamycin and by RNAP in hydroxyl radical footprint; Fig. 4A and Newlands et al. 1991).
The UP element minor groove is protected by αCTD from modification by DMS
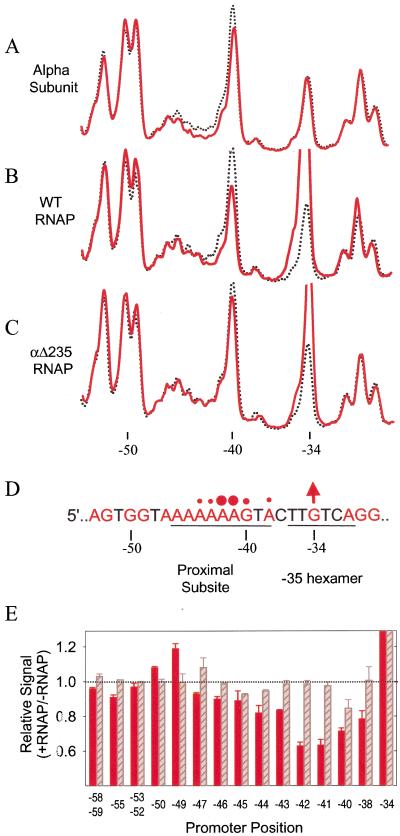
The distamycin experiments (Fig. 4) suggest that the DNA minor groove plays an important role in the binding of α to the UP element. To determine whether α contacts (or is in very close proximity to) specific bases within the minor groove, we asked whether prebound RNAP or purified α subunit protected the N3 position of adenines in the UP element minor groove (see Fig. 1B) against methylation by DMS. N3A methylation, as well as N7G methylation in the major groove, is detectable by elimination of the modified base at high temperature, followed by strand cleavage at the abasic site (Maxam and Gilbert 1980; see Materials and Methods).
DMS protection footprints were carried out with a promoter in which the UP element contained adenines at seven of nine positions in the nontemplate strand. Profiles from footprints with purified α, with wild-type RNAP, and with RNAP lacking the αCTD (αΔ235) are illustrated in Figure 5. In each case, the profile is compared with a control profile lacking protein. Methylation of several N3A positions was significantly reduced in the presence of purified α (Fig. 5A) or wild-type RNAP (Fig. 5B). Protection of the UP element from modification by DMS was not observed when RNAP lacked the αCTD (αΔ235 RNAP; Fig. 5C). Adenine residues are less reactive to DMS than guanine residues, and, therefore, the peak heights at A residues are lower than those for G even in the absence of bound protein, which tends to de-emphasize protection signals (Maxam and Gilbert 1980). However, the N3A protections by RNAP and α were observed reproducibly in many experiments with both native and reconstituted RNAPs (for summary, see Fig. 5D; Fig. 6). We determined the fraction of the N3A signals protected by RNAP (Fig. 5E). RNAP reduced the methylation of N3s at positions −41 and −42 by ∼40%, a value that is consistent with partial protection signals observed in other DMS footprints (e.g., Johnsrud et al. 1978; Craig and Nash 1984; Newlands et al. 1991). Protection of N3A by RNAP was also observed in UP elements from the native rrnB P1 and rrnD P1 promoters (data not shown).
Figure 5.
Methylation protection experiments with rrnB P1 containing UP element 4547. Footprints with purified α subunit (A), wild-type RNAP holoenyzme (B), or RNAP holoenyzme lacking the αCTD (αΔ235 RNAP) (C) bound to a fragment labeled on the nontemplate strand. Preformed complexes were methylated with DMS and samples analyzed by gel electrophoresis after strand cleavage at methylated purines. Superimposed gel scans are shown for protein-bound samples (red solid lines) and control DNAs (no protein; black dotted lines). (D) UP element and −35 hexamer region sequences of the nontemplate strand, with purines in red. Positions where bound protein affects the extent of methylation are indicated by circles (protection) or by an arrow (enhancement). (E) Extent of methylation at individual promoter positions in the presence of wild-type RNAP (red bars) or of RNAP lacking the αCTD (αΔ235 RNAP; hatched grey bars) relative to that of control DNA without RNAP. Average (with range) is shown from two independent experiments.
The major groove N7G at −40 was partially protected in RNAP complexes (Fig. 5). This protection was also observed (although to a lesser extent) with purified α and with RNAP lacking the αCTD (Fig. 5A,C). Thus, it is not clear whether the −40 N7G protection observed with wild-type RNAP is attributable to α or to ς interactions with the DNA or if it results from interactions between the two subunits in the vicinity of −40 (see Discussion).
Methylation of several N3A and N7G positions in the core promoter region was affected by the presence of bound RNAP (Figs. 5,6; data not shown). The observed protections or enhancements are consistent with those reported previously for DMS footprints of this and other promoters (Johnsrud 1978; Siebenlist et al. 1980; Duval-Valentin and Ehrlich 1986; Newlands et al. 1991). The results from the DMS protection footprints, from the U and A* interference footprints, and from previously reported hydroxyl radical footprints on this promoter are summarized in Figure 6.
Discussion
DNA features critical for α binding
Binding of one αCTD to the UP element proximal subsite produces a very small footprint whose features suggest interactions with the phosphodiester backbone and with bases in the minor groove (Fig. 7A). The DMS footprinting data indicate that α is in very close proximity to position N3 of adenine bases in the UP element minor groove. Protection of N3A positions upstream of the −35 hexamer was reported in early DMS footprints of RNAP on other promoters (e.g., Siebenlist et al. 1980; Duval-Valentin and Ehrlich 1986), although the role of α in promoter recognition was not known at the time of those experiments. X-ray structures of minor groove-binding proteins in complex with DNA have confirmed that protection of N3A positions in DMS footprints correlates with minor groove contacts [e.g., Hin recombinase (Glasgow et al. 1989; Feng et al. 1994) and IHF (Craig and Nash 1984; Rice et al. 1996)].
Figure 7.
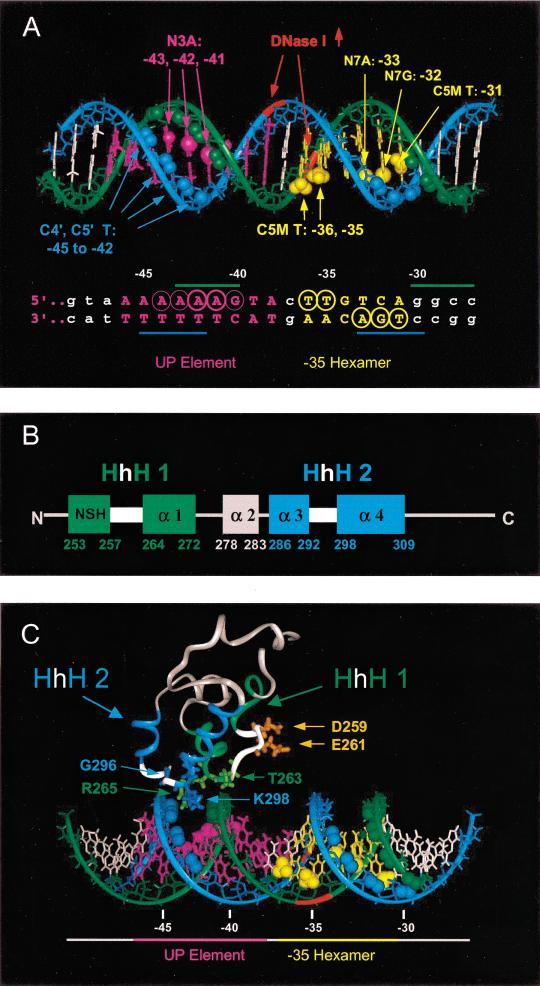
Positions of footprint signals and model for αCTD binding to UP element proximal subsite 4547. (A) Positions of footprint protection and interference signals in the proximal subsite and the −35 hexamer. Backbone phosphates are in ribbon form and deoxyribose and bases in stick form. Backbone positions are in green (nontemplate strand) or in blue (template strand), and bases are in magenta (UP element), yellow (−35 hexamer), or white. Positions of footprint signals (for summary, see Fig. 6) are represented as spheres, and include: (1) deoxyribose positions protected in hydroxyl radical footprints (C4‘, C5′ in UP element and downstream edge of −35 hexamer; green on nontemplate strand, blue on template strand; only a few signals are labeled); (2) purines with reduced or enhanced methylation in the presence of bound RNAP in DMS footprints (N3 of A at −41 to −43, magenta; N7 of G at −32, yellow); (3) −35 hexamer positions where uracil substitutions interfered with RNAP binding (shown as C5-methyl groups of thymines at −36, −35, and −31, yellow) or where A* substitution stimulated RNAP binding (shown as N7A at −33, yellow). Positions of enhanced cleavage in DNase I footprints of rrnB P1, in red along the backbone ribbon (−38, nontemplate strand; −35, template strand; Gourse 1988; Ross et al. 1993), are thought to reflect positions of DNA distortion in the RNAP–promoter complex. (B) Secondary structural features of αCTD involved in its (HhH)2 domain structure (Shao and Grishin 2000). (Green rectangles) α helices in HhH1; (blue rectangles) α helices in HhH2; (white rectangles) hairpin regions. Amino acid residues comprising α helices 1–4 (or nonstandard helix, NSH), are derived from the αCTD solution structure (Jeon et al. 1995; Gaal et al. 1996). (C) Model for αCTD binding to the UP element. DNA is rotated ∼90 degrees around the helix axis from the position in A. The αCTD solution structure (Jeon et al. 1995) is in ribbon form. α helices of the two HhH motifs (Shao and Grishin 2000) are green (HhH1) and blue (HhH2), and hairpin loops connecting the helices are white. Four residues required for binding to the UP element (R265, G296, K298, S299; Gaal et al. 1996), and two residues implicated in interactions with ς (D259, E261; see text) are in stick form. The αCTD is aligned with the DNA such that each HhH motif contacts the backbone of one of the DNA strands, as suggested by features of the RuvA–DNA complex (Ariyoshi et al. 2000). Side chains of R265 and K298 are shown fitting into the minor groove. Figures in A and C were prepared using Insight II software.
Our experiments using distamycin, which binds snugly in the minor groove of A+T-rich DNA (Coll et al. 1987), support the role of minor groove interactions in α binding. Distamycin protected essentially the same set of UP element residues as α in our hydroxyl radical footprints (Estrem et al. 1999; Figs. 4,6) and inhibited α binding to UP element DNA. Distamycin has been shown previously to interfere with minor groove binding of other proteins (e.g., HMG Y/I; Reeves and Nissen 1990).
Several backbone positions on each strand in the UP element proximal subsite are protected by RNAP from hydroxyl radical cleavage (Estrem et al. 1999; for summary, see Figs. 6 and 7A). Hydroxyl radicals react primarily with the hydrogens on positions C4 and C5 on the minor groove side of deoxyribose (Balasubramanian et al. 1998). Therefore, the UP element base and backbone positions protected by α are located in very close proximity (Fig. 7A). Although not tested in our experiments, the O2 positions of thymine bases in the minor groove would also be candidates for direct hydrogen bond contacts with α.
Our results suggest that α does not make significant major groove contacts. Substitution of U or A* for T or A, respectively, in the UP element had little if any effect on RNAP binding to the promoter (Fig. 3). Although the partial protection of major groove position N7G at −40 against DMS methylation (Fig. 5) may indicate close proximity of RNAP to this position, other considerations suggest that the identity of this residue is not critical to UP element function. G is not strictly conserved at −40 in UP element proximal subsites (Estrem et al. 1999), and no steric hindrance to RNAP binding was observed when A* was substituted for A at this same position in the UP element (Fig. 3). In addition, although some degree of protection of N7G at −40 was observed with purified α, protection was also observed with RNAP lacking the αCTD. Thus, it is not clear whether it is α or ς that is in close proximity to the N7G at −40 in the context of RNAP holoenzyme. Furthermore, the reduced methylation of the N7G at −40 could be attributable to altered DNA structure resulting from RNAP binding rather than from direct protein contact. The latter interpretation is supported by the strong enhancement of DNase I cleavage at position −38 observed in footprints of RNAP on rrnB P1 and other promoters (Fig. 7A; Gourse 1988; Ross et al. 1993; Craig et al. 1995).
Although our footprints indicated that α contacts only a small subset of base surfaces, a missing base at any position in the UP element interfered very strongly with RNAP binding (Fig. 2). The strongest of the missing base signals on the nontemplate strand (−41 to −43) coincide with positions protected by RNAP in DMS footprints, suggesting that the effects of missing bases reflect, in part, the loss of minor groove base contacts. However, distortion of the normal structure of the UP element proximal subsite could also account for the effect of the missing bases, since structural changes can occur up to 4 bp from an abasic site (Wang et al. 1997). The proximal subsite contains a long A tract (7–8 residues), and unusual stuctural features of the A tract (e.g., narrow minor groove width and/or propeller twisting of bases; Shatzky-Schwartz et al. 1997) could play an important role in recognition by α. Thus, at least some of the interference to RNAP binding caused by missing bases could be attributable to effects on a structure important to recognition by α, rather than to loss of direct base contacts.
In contrast with our results, it was reported that missing bases in the UP elements of the ace or the rrnB P1 promoters stimulated (rather than inhibited) RNAP binding (Negre et al. 1997). We suggest that the A+T-rich UP element region containing an abasic site may have been recognized as a −10 hexamer rather than as an α-binding site, accounting for the observed stimulation of RNAP binding in these experiments. We and others (Li and McClure 1998) have observed that missing bases in the −10 hexamer strongly stimulate RNAP binding (Figs. 2,6).
The results of our footprinting studies are consistent with the effects on transcription of mutations in the UP element proximal subsite (Estrem et al. 1999). Substitution of C:G for A:T pairs at several UP element positions (−41 to −45) reduced transcription in vivo and in vitro, with the strongest effects (>10-fold) occurring at positions −41, −42, and −43. The 2-amino group of guanine at these positions could sterically hinder α binding in the minor groove; alternatively, effects on α binding of a C:G substitution could result from disruption of A-tract structure (Koo et al. 1986). Interestingly, a T:A rather than an A:T pair at position −42 (in UP element 4542; Estrem et al. 1999) did not reduce UP element function. This substitution would preserve hydrogen bonding capabilities in the minor groove (with the O2 of thymine).
A model for αCTD interaction with an UP element subsite
Our results indicating that α binds to the UP element minor groove are consistent with the recent observation that the αCTD contains an (HhH)2 domain. (Shao and Grishin 2000). On the basis of this observation, on our biochemical data, and on our previous genetic identification of the αCTD DNA-binding surface (Gaal et al. 1996; Murakami et al. 1996), we suggest a preliminary model for how α might interact with its binding site (Fig. 7C).
The HhH motif was identified initially in the structure of the DNA repair enzyme Endonuclease III (Thayer et al. 1995), and has since been identified in DNA-binding proteins involved in replication, recombination, and repair (e.g., DNA polymerases, ligases, and glycosylases; Doherty et al. 1996; Shao and Grishin 2000). The HhH motif contains a pair of anti-parallel α helices connected by a hairpin loop that are present in either one or two copies [the (HhH)2 domain; Shao and Grishin 2000]. DNA binding, in examples considered thus far, is not sequence specific (Doherty et al. 1996). In the rat DNA polymerase β–DNA complex (Pelletier et al. 1996) and the E. coli RuvA–DNA complex (Ariyoshi et al. 2000), amino acid residues in the loop and in the amino-terminus of the second α helix make nonspecific hydrogen bond contacts to the DNA backbone. RuvA binds across the minor groove of duplex DNA, with each of the repeated HhH motifs in a RuvA monomer contacting the backbone of one of the two DNA strands.
The RNAP αCTD contains a short nonstandard helix (NSH) followed by four α helices (Jeon et al. 1995; Gaal et al. 1996). In the (HhH)2 domain interpretation of αCTD (shown in schematic form in Fig. 7B), the first HhH motif contains the NSH (residues ∼253–257), a loop (∼258–263), and a second α helix (264–272) referred to as α helix 1. The second HhH contains α helix 3 (286–292), a loop (∼293–297), and α helix 4 (298–309). The two HhH motifs are separated by α helix 2. The positions of substitutions in αCTD that result in strong defects in DNA binding (Gaal et al. 1996; Murakami et al. 1996) affect residues that, by analogy to (HhH)2 domains in other proteins (Rafferty et al. 1998; Ariyoshi et al. 2000), would be predicted to make DNA contacts (e.g., R265 in the amino terminus of the second helix in HhH1, G296 in the loop, and K298, S299 in the amino terminus of the second helix in HhH2).
Based on these considerations, and by analogy to the RuvA–DNA structure (Ariyoshi et al. 2000), we suggest that each of the HhH motifs in the αCTD makes backbone contacts with one of the DNA strands, at the positions that are protected by the αCTD in hydroxyl radical footprints (Fig. 7C). The length of the α-binding site is similar to the length of DNA covered by one RuvA monomer (Ariyoshi et al. 2000).
Unlike other HhH-motif proteins, the αCTD demonstrates sequence specificity in DNA binding. We suggest that this sequence specificity could derive from (at least) two characteristics of the UP element DNA sequence. First, the narrow width of the minor groove in an A tract (the preferred α-binding site) could be important for simultaneous binding of α to both DNA strands. It has been proposed previously that effects of sequence on DNA structure could influence binding of HhH motif proteins (Rafferty et al. 1998).
Second, our data suggest that the specificity of α binding to an A-tract sequence derives in part from base contacts in the minor groove (Fig. 5). These contacts (to N3A and potentially to O2T) could occur through hydrogen bonds with the basic side chains of residues R265 and K298. Base-specific contacts in the minor groove of A+T-rich DNA sequences have been demonstrated for a number of other DNA-binding proteins. Frequently these interactions are made by short peptide motifs containing arginine residues that contact bases in the minor groove and the DNA backbone along the minor groove [e.g., AT-hooks in the chromatin-associated protein HMG-I(Y) (Huth et al. 1997), and extended arm sequences in the Hin recombinase (Feng et al. 1994) and in homeodomains (Kissinger et al. 1990)]. Because several substitutions in the αCTD cause defects in both specific and nonspecific DNA binding (Savery et al. 1998), we suggest that the same DNA-binding motif is responsible for interactions with UP element and non-UP element DNA sequences.
In the model for the α–UP element interaction presented in Figure 7C, we have chosen a specific orientation for αCTD with respect to the UP element sequence, although in other contexts the orientation could be reversed. The orientation shown is suggested by the results of genetic experiments that implicate α residues 259 and 261 in interactions with the ς subunit at promoters containing the UP element proximal subsite (Gourse et al. 2000). This orientation was also suggested by the identification of residues potentially involved in ς–α interactions in the lac promoter–CRP–RNAP complex (Busby and Ebright 1999; Gourse et al. 2000).
α binding to UP elements with two subsites
In the present study, we focused on interactions of α with a single consensus binding site, the proximal subsite. However, we have also performed footprinting experiments on full (i.e., two subsite) UP elements. The distal subsite in the full UP element responded similarly in protection and interference experiments to the proximal site by itself (W. Ross and R.L. Gourse, data not shown). However, the proximal subsite in the full UP element responded differently from the proximal subsite in the absence of the distal subsite. In the context of the full UP element, missing bases in the proximal subsite did not interfere with RNAP binding (W. Ross and R.L. Gourse, data not shown), and the proximal subsite was not fully protected in hydroxyl radical footprints (Newlands et al. 1991; Estrem et al. 1998).
These results suggest that the proximal subsite plays a different role when alone than it does in a full UP element. These results are also consistent with three lines of genetic evidence: (1) Proximal subsite sequences identified in binding-site selections were not identical in the presence versus absence of the consensus distal subsite (Estrem et al. 1998, 1999). (2) Proximal subsite mutations had larger effects on transcription when the consensus distal subsite was absent (Estrem et al. 1998; S. Estrem and R.L. Gourse, unpubl.). (3) Certain αCTD and ςCTD substitutions reduced transcription from promoters containing only a proximal subsite, but not from promoters with full UP elements (W. Ross, A. Mertens, B. Paul, D. Schneider, J. Toplin, M. Burgess, and R.L. Gourse, unpubl.).
These results suggest that the two subsites do not contribute equally to the binding of RNAP in promoters containing full UP elements. Possible explanations (see also Estrem et al. 1999) could include: (1) Binding of αCTD to the proximal subsite could be weaker, less specific, or nonspecific in the context of both subsites. (2) The proximal subsite in the context of a full UP element could serve primarily a structural role, rather than a role in α binding. (3) The longer, symmetric distal subsite sequence could be a binding site for both α subunits in the context of a full UP element, thereby reducing or excluding occupancy of the proximal subsite by α.
RNAP contacts with the −35 hexamer
Our interference footprints yielded considerable information about the role of specific major groove base surfaces in binding RNAP to the core promoter. These data, together with the DMS protection data, confirm and extend previous information about the interaction of the ς subunit with the −35 and −10 elements.
Of the 12 bases present in the −35 hexamer of rrnB P1 (six on each DNA strand), information about 10 bases was obtained with the base-specific reagents used, and of these 10, five appear to be in close proximity to or in contact with RNAP (Figs. 6,7). Particularly strong interference signals were observed when U was substituted for T at positions −35 and −36, indicating probable interactions of ς with the C5 methyl groups of these thymines that are critical for RNAP binding. Because A* substitutions at the same positions on the template strand did not interfere with RNAP binding, we suggest that the strong conservation of T:A pairs at these positions in promoter −35 hexamers (Hawley and McClure 1983) reflects interactions with the thymine bases.
These results are consistent with previous observations. Mutations of the thymines in the nontemplate strand at the upstream end of the −35 hexamer have strong phenotypes (Youderian et al. 1982), and 5-bromouracil substitutions at these positions in lacUV5 are protected by RNAP against UV-induced strand cleavage (Siebenlist et al. 1980). The region of ς70 that interacts with the −35 hexamer (region 4.2) is thought to contain a helix–turn–helix motif (HTH; Record et al. 1996; Gross et al. 1998). Although we do not know which amino acid residues in region 4.2 interact with the thymine methyl groups at −35 and −36, they are likely to be located in the HTH recognition helix, carboxy-terminal to the arginine residues at positions 584 and 588 that are implicated in contacting the central region of the −35 hexamer (for review, see Gross et al. 1998). R588 was proposed to contact position −34. However, because the major groove surface of −34 G does not appear to be in contact with RNAP (methylation of this G is increased, not reduced, by the presence of prebound RNAP; Fig. 5; Siebenlist et al. 1980), we suggest that the template strand C, rather than the nontemplate strand G, may contact RNAP and be responsible for the conservation of the G:C pair at −34.
Our results suggest that ς also contacts three template strand bases at the downstream end of the −35 hexamer. A* at position −33 strongly stimulated RNAP binding. This is a nonconsensus position in rrnB P1 (T:A rather than A:T), and the presence of the 7-nitro group on the modified base might provide a contact to RNAP that serves as an alternative to that provided by thymine at the same position in a consensus −35 hexamer, even though the nitro and methyl groups are chemically dissimilar. Additional template strand contacts in the −35 hexamer are suggested by partial protection of N7G at −32 against methylation by DMS (Newlands et al. 1991; see also Johnsrud 1978; Duval-Valentin and Ehrlich 1986), and by partial interference resulting from U substitution at −31. Hydroxyl radical footprints indicate that RNAP also interacts with the DNA backbone along the adjacent minor groove in the spacer at the downstream edge of the −35 hexamer (Newlands et al. 1991; Figs. 6,7). Hydroxyl radical protections (Newlands et al. 1991) and the effects of missing bases at several positions in the spacer region, particularly around −20 (Fig. 6), suggest that the conformation of this region may be important for RNAP binding.
RNAP contacts with the −10 hexamer
In the −10 hexamer, major groove modifications at several positions interfered with the formation of a stable open complex at rrnB P1. Because open complex formation involves recognition of the −10 region as duplex DNA, followed by strand separation and sequence-specific interaction with the nontemplate strand (Roberts and Roberts 1996; Marr and Roberts 1997; Qiu and Helmann 1999; Fenton et al. 2000), the interference signals observed in our footprints could result from disruption of contacts with duplex DNA, single-stranded DNA, or both. Our footprinting results should be useful for future interpretation and modeling of the interaction of ς region 2.4 with the −10 hexamer.
Specifically, we found that U substitution at two of the three nontemplate strand thymines reduced RNAP binding, indicating that ς may directly contact the C5 methyl group of these Ts (rrnB P1 positions −14 and −9, corresponding to positions −12 and −7 in lacUV5 and λPR‘; Marr and Roberts 1997; Fenton et al. 2000). A* substitution for five of the six adenines in the −10 hexamer interfered with RNAP binding. Because A* contains a C7-nitro group in place of N7, interference could result from loss of direct contacts or from steric hindrance (Min et al. 1996). In either case, our data indicate that RNAP is in close proximity to these bases.
The inhibitory effect of A* was particularly strong at nontemplate strand position −10 (−8 in other promoters), although this is not one of the most highly conserved −10 hexamer positions (Hawley and McClure 1983). A* at −10 could conceivably sterically hinder the approach of RNAP to the adjacent highly conserved thymine or might interfere with critical base stacking interactions of the separated strand (Qiu and Helmann 1999). Strong effects of A* substitutions were also observed at rrnB P1 template strand positions −14 and −12 (−12 and −10 in other promoters).
Although an A at nontemplate strand position −13 (−11 in other promoters) is very highly conserved and plays a critical role in promoter function, the substitution of A* at −13 had a relatively small effect on RNAP binding (Fig. 3). This A is important for single-stranded oligonucleotide binding and fork-junction binding (Marr and Roberts 1997; Guo and Gralla 1998; Qiu and Helmann 1999) and is thought to nucleate strand opening, perhaps through base stacking interactions with aromatic residues in ς region 2.4 (Fenton et al. 2000; Panaghie et al. 2000). Recognition of −13A by ς apparently does not involve critical contacts to the functional groups altered by the A* modification.
Missing bases at all but two of the positions tested in the −10 hexamer (all except −13A, −9T) stimulated RNAP binding, presumably by facilitating the strand separation process. These results are consistent with those reported previously for other promoters (Li and McClure 1998). In addition, a missing base at template strand +1 strongly interfered with RNAP binding to the rrnB P1 promoter (data not shown). This negative effect reflects the requirement for incorporation of the initiating two nucleotides, ATP and CTP, to stabilize the rrnB P1 open complex, and is not a phenomenon general to promoters with longer-lived open complexes; that is, the effect of a missing base at +1 of rrnB P1 does not indicate a requirement for a specific base contact to RNAP at this position.
Conclusion
The work reported here provides information about the functional consequences of specific DNA–protein contacts in the initiation complex. Such data will be important for interpretation of atomic level structures of the promoter–RNAP complex that may become available in the future. We have shown that the α subunit of RNAP binds to UP element DNA using minor groove as well as backbone contacts, and we have proposed a model for αCTD binding to DNA. These data should also facilitate modeling of αCTD interactions with DNA in promoter complexes containing transcription activators. Our results suggest further that the contacts made by α to the UP element and by ς70 to the −35 hexamer are in very close proximity. Thus, αCTD–ς70 interactions could play a role in the function of the UP element proximal subsite. Finally, our results identify functional groups in the −10 and −35 hexamers that are involved in the interaction with the ς subunit and thus increase the resolution of our picture of the RNAP–core promoter interaction.
Materials and methods
Plasmid DNAs
DNA fragments containing rrnB P1 promoters with different sequences in the UP element region were inserted as EcoRI (−66 with respect to the transcription start site) to HindIII (+50) fragments into pRLG770 (Estrem et al. 1999). Plasmids contained consensus proximal subsite 4547 [(−46) 5′-AAAAAAGTA-3′ (−38)] in pRLG4213, consensus proximal subsite 4549 [(−46) 5′-AAAAAAAGA-3′ (−38)] in pRLG3289, full consensus UP element 4192 [(−59) 5′-GGAAAATTTTTTTTCAAAAGTA-3′ (−38)] in pRLG3278, or an upstream sequence lacking UP element function (SUB) in pRLG4210. DNA sequences flanking the promoters were described previously (Estrem et al. 1998, 1999).
RNAP and α purification
Wild-type native RNAP was a gift from Dr. Robert Landick (University of Wisconsin). Wild-type α subunit was purified as described (Gaal et al. 1996). RNAPs containing wild-type or mutant α subunits lacking the carboxy-terminal domain (αΔ235) and containing an amino-terminal hexahistidine tag were reconstituted from isolated subunits (Gaal et al. 1996), purified by nickel chromatography (Qiagen) and by centrifugal filtration (Centricon YM-100; 100,000 MW cutoff). RNAPs heterodimeric for α (containing one wild-type α and one αΔ235) were purified as described (Estrem et al. 1999).
Missing base interference footprinting
Promoter fragments were prepared by 15 cycles of PCR amplification using primers that flanked the promoter in the vector sequence (upstream primer 5′-CCGCGGATCCGTATCAC GAGGCCCTTTCG-3′; downstream primer 5′-GCGCTACG GCGTTTCACTTC-3′). PCR reactions (50 μL) contained 0.4 μg of plasmid DNA, 20 pmoles of each primer, 200 μM amounts of each dNTP (Pharmacia), 1.5 mM MgCl2, and 2.5 units of Taq DNA polymerase (Promega) in a buffer supplied by the manufacturer. For preparation of fragments containing missing thymine bases, the PCR reactions also contained 2 μM dUTP (Pharmacia; Devchand et al. 1993).
PCR products were purified by use of a Qiaquick PCR Purification Kit (Qiagen), digested with the appropriate restriction enzyme [HindIII (+50) for nontemplate strand labeling or BamHI (primer-encoded) for template strand labeling], and 3′-end-labeled with Sequenase (USB) and [α-32P]dATP for HindIII sites, or [α-32P]dGTP for BamHI sites. Labeled fragments were gel purified, eluted by diffusion into 0.2 M NaCl, 20 mM Tris-Cl (pH 7.4), 1 mM EDTA, and concentrated by use of Elutip D columns (Schleicher and Schuell). To prepare fragments with, on average, one missing thymine base per molecule, end-labeled fragments containing dUTP were then treated with uracil DNA glycosylase (UDG; NEB; 0.75 units for 10 min at 37°C in 100 μL with buffer supplied by the manufacturer) to remove the uracil base, leaving the DNA backbone intact. To prepare fragments with, on average, one missing purine base per molecule, end-labeled fragments (prepared without modified nucleotides) were treated with formic acid as described previously (Maxam and Gilbert 1980).
To carry out the missing base interference footprint, RNAP (0.5 nM) was incubated with a 20-fold excess of end-labeled DNA fragment (10 nM) containing either a missing thymine base or a missing purine, for 5 min at 22°C in 100 μL of 10 mM Tris-Cl (pH 7.9), 30 mM KCl, 10 mM MgCl2, 1 mM DTT, 500 μM ATP, 50 μM CTP. Heparin (final 10 μg/mL) was added, and RNAP–promoter complexes (∼5% of input DNA) were separated from unbound DNA by electrophoresis on a 5% acrylamide gel in 0.5× Tris-Borate-EDTA (TBE). Because DNA was kept in excess to promote competition for RNAP, the unbound promoter fraction did not provide useful information and was not analyzed. Control DNA used for comparison to the RNAP-bound population was the input DNA population without incubation with RNAP.
RNAP-bound DNA was eluted by diffusion, and purified by use of Elutip D columns as described above. Then, strand cleavage was carried out at the abasic sites by incubation in 100 μL of 1 M piperidine at 90°C for 30 min. Fragments were ethanol precipitated, then separated by electrophoresis on 9.5% acrylamide–7 M urea gels. Dried gels were scanned by PhosphorImaging using a Storm system (Molecular Dynamics). Data points in each lane were normalized to correct for differences in loading. Normalization factors were derived from comparisons of portions of the profiles that were distant from the promoter and assumed not to be in contact with RNAP. Normalized profiles were graphed using Sigma Plot (Jandel Scientific).
Modified base interference footprinting
Promoter fragments for modified base interference footprinting were prepared by PCR as described above, but reactions also contained either 2 μM dUTP (Pharmacia) for modified thymine containing fragments, or 30 μM 7-deaza-7-nitro dATP (dA*TP; see below for synthesis) for modified adenine containing fragments. Fragments were 3′-end-labeled and gel purified as described above.
RNAP–promoter fragment complexes were formed under conditions described above for missing base interference footprinting, gel isolated and the DNA eluted and purified as described. Strand cleavage at modified adenine (A*) bases was then carried out by incubation with 1 M piperidine, as described above. DNA fragments containing dUTP were treated with UDG (above) to remove the uracil base prior to the strand cleavage at the resulting abasic sites (see above). Then, fragments were analyzed by denaturing gel electrophoresis as described.
Synthesis of 7-deaza-7-nitro-2‘-deoxyadenosine triethylammonium 5′-triphosphate (dA*TP)
7-Deaza-7-nitro dATP (dA*TP) was synthesized using modifications of the procedure described previously (Min et al. 1996). A suspension of 7-deaza-7-nitro-2‘-deoxyadenosine (Min et al. 1996; Seela et al. 1997; 23.2 mg, 78.4 μmoles, coevaporated twice from pyridine and dried under high vacuum overnight) in trimethylphosphate (0.32 mL) was treated at 0°C with POCl3 (8.1 μL, 86.3 μmoles), and stirred for 20 min at 0°C to produce a yellow solution. This solution was treated dropwise with 0.5 M (Bu3NH)2H2P2O7 [solution in dimethylformamide (0.78 mL, 0.39 mmoles); prepared according to Moffatt 1964] and Bu3N (0.08 mL, 0.32 mmoles), stirred vigorously for 1.5 min, poured into 10 mL of ice-cold 1.5 M triethylammonium bicarbonate (TEAB at pH 7.5), and lyophilized. The residue was dissolved in 10 mL of water and purified by ion-exchange chromatography [DEAE Sephadex A25, 9 × 2.5 cm, linear gradient of TEAB (0.05 to 1.5 M at pH 7.5, 1500 mL) at a flow rate of 5 mL/min]. The triphosphate was eluted at 0.6–0.7 M TEAB. Lyophilization (three times from water) gave dA*TP (40.6 mg, 55%) as a yellow solid. Analytical HPLC: tR 18.27 min. UV (H2O): 364, 276. 31P-NMR (162 MHz, D2O): −7.19 [d, J=20.1, P(γ)]; −10.62 [d, J=20.9, P(α)]; −22.07 [t, J≈19.5, P(β)]. MS-ES (negative mode): C11H16N5O14P3 (535): 534 (100, [M − H]−).
Footprinting with DMS
Promoter fragments used in DMS protection footprints were obtained by HindIII digestion of pRLG4213 or pRLG3289 and labeling of the 3′ end of the nontemplate strand using Sequenase (USB) and [α-32P]dATP (Newlands et al. 1991). Then, labeled DNA was digested with AatII (vector-encoded site is 75 bp upstream of EcoRI), and the fragment was gel isolated, eluted, and purified by use of Elutip D columns as described above.
Promoter fragments (<1 nM) were pre-incubated with wild-type or α mutant RNAP (∼10 nM) or with purified α subunit (∼2 μM) for 10 min at 22°C in 50 μL of the buffer used above for interference footprinting. Then, the DNA was methylated by addition of 1 μL of DMS for 30 sec at 22°C, followed by addition of 25 μL of cold 1.5 M β-mercaptoethanol and 0.3 M NaOAc, extraction with 75 μL of cold phenol, precipitation with ethanol, and reprecipitation from 0.3 M NaOAc. The DNA pellet was resuspended in 30 μL of 10 mM sodium phosphate (pH 6.8), 1 mM EDTA, and methylated purines (A,G) were released by incubation at 92°C for 15 min (Maxam and Gilbert 1980). Strand cleavage at abasic sites was carried out by addition of 3 μL of 1 N NaOH and further incubation at 92°C for 30 min. Samples were precipitated with ethanol, and examined by electrophoresis, gel scanning, and quantitation as described above.
Hydroxyl radical footprinting to detect distamycin-binding sites
An end-labeled promoter fragment containing rrnB P1 with UP element 4547 (prepared as described for DMS footprinting above) was incubated at 22°C for 10 min with 125, 250, or 500 nM distamycin A (Sigma) in 150 mM NaCl, 20 mM Tris-Cl (pH 7.4), 1 mM EDTA, and 1 μg/mL sonicated calf thymus DNA. OH· cleavage was carried out as described previously (Newlands et al. 1991), and gel electrophoresis and scanning of gels was performed as described above.
Gel mobility shift assay with the α subunit
An end-labeled promoter fragment (∼0.1 nM; rrnB P1 with proximal subsite 4547) was preincubated for 10 min at 22°C with 15.6–1000 nM distamycin A in 10 μL of buffer containing 150 mM NaCl, 20 mM Tris-Cl (pH 7.4), 1 mM EDTA, 10% glycerol, and 1 μg/mL sonicated calf thymus DNA. Purified α subunit (2 μM) was then added for 10 min, and electrophoresis was carried out at 4°C on prechilled 6% acrylamide gels containing 0.5× TBE and 10% glycerol. Bands containing the α–DNA complex were quantified by PhosphorImaging, and values were expressed as a percentage of the fragment population bound in the absence of distamycin.
In vitro transcription
Plasmid DNAs (60 ng of pRLG4213 or pRLG4210) were incubated with distamycin A (15.6–1000 nM) for 10 min at 22°C in 25 μL of transcription buffer containing 150 mM NaCl, 10 mM Tris-Cl (pH 7.9), 10 mM MgCl2, 1 mM DTT, 100 μg/mL bovine serum albumin, 500 μM ATP, 50 μM CTP, 50 μM GTP, 10 μM UTP, and 4 μCi [α-32P]UTP (NEN). Transcription was initiated by addition of RNAP (2 nM) for 15 min, and the reaction was terminated by addition of 25 μL of stop solution (Ross et al. 1998). Samples were analyzed by electrophoresis on 5% acrylamide–7 M urea gels and quantified after gel scanning with a PhosphorImager.
Acknowledgments
We thank Melanie Barker, Richard Ebright, Tamas Gaal, and Ruth Saecker for comments on the manuscript and Richard Ebright for calling dA*TP to our attention. We also thank Tamas Gaal for preparation of purified α subunit and Bob Landick for RNAP holoenzyme. This work was supported by GM37048 to R.L.G. from the National Institutes of Health. A.E. gratefully acknowledges support from the Swiss National Science Foundation (postdoctoral fellowship, 1998–1999) and from Professor Gregory L. Verdine (Harvard University).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgourse@bact.wisc.edu; FAX (608) 262 9865.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.870001.
References
- Ariyoshi M, Nishino T, Iwasaki H, Shinagawa H, Morikawa K. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc Natl Acad Sci. 2000;97:8257–8262. doi: 10.1073/pnas.140212997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva TA, Rhodius VA, Webster CL, Busby SJ. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: Organisation of the RNA polymerase alpha subunits. J Mol Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- Blatter EE, Ross W, Tang H, Gourse RL, Ebright RH. Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994;78:889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- Brunelle A, Schleif RF. Missing contact probing of DNA–protein interactions. Proc Natl AcadSci. 1987;84:6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby SJ, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Churchill ME, Hayes JJ, Tullius TD. Detection of drug binding to DNA by hydroxyl radical footprinting. Relationship of distamycin binding sites to DNA structure and positioned nucleosomes on 5S RNA genes of Xenopus. Biochem. 1990;29:6043–6050. doi: 10.1021/bi00477a023. [DOI] [PubMed] [Google Scholar]
- Coll M, Frederick CA, Wang AH, Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci. 1987;84:8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig ML, Suh W-C, Record MT., Jr HO· and DNase I probing of Eς70 RNA polymerase-λPR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochem. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- Craig NL, Nash HA. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Devchand PR, McGhee JD, van de Sande JH. Uracil-DNA glycosylase as a probe for protein–DNA interactions. Nucleic Acids Res. 1993;21:3437–3443. doi: 10.1093/nar/21.15.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP. The helix–hairpin–helix DNA-binding motif: A structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval-Valentin G, Ehrlich R. Interaction between E. coli RNA polymerase and the tetR promoter from pSC101: Homologies and differences with other E. coli promoter systems from close contact point studies. Nucleic Acids Res. 1986;14:1967–1983. doi: 10.1093/nar/14.5.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Gaal T, Ross W, Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci. 1998;95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Ross W, Gaal T, Chen ZWS, Niu W, Ebright RH, Gourse RL. Bacterial promoter architecture: Subsite structure of UP elements and interactions with the C-terminal domain of the RNA polymerase α subunit. Genes & Dev. 1999;13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JA, Johnson RC, Dickerson RE. Hin recombinase bound to DNA: The origin of specificity in major and minor groove interactions. Science. 1994;263:348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- Fenton MS, Lee SJ, Gralla JD. Escherichia coli promoter opening and −10 recognition: Mutational analysis of ς70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Caramori T, Chen YF, Galizzi A, Helmann JD. Promoter architecture in the flagellar regulon of Bacillus subtilis: High-level expression of flagellin by the sigma D RNA polymerase requires an upstream promoter element. Proc Natl Acad Sci. 1995;92:2582–2586. doi: 10.1073/pnas.92.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Ross W, Blatter EE, Tang H, Jia X, Krishnan VV, Assa-Munt N, Ebright RH, Gourse RL. DNA-binding determinants of the α subunit of RNA polymerase: Novel DNA-binding domain architecture. Genes & Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- Glasgow AC, Bruist MF, Simon MI. DNA-binding properties of the Hin recombinase. J Biol Chem. 1989;264:10072–10082. [PubMed] [Google Scholar]
- Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: The role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- Gross C A, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of ς factors in transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward RS, Igarashi K, Ishihama A. Functional specialization within the α-subunit of Escherichia coli RNA polymerase. J Mol Biol. 1991;221:23–29. doi: 10.1016/0022-2836(91)80197-3. [DOI] [PubMed] [Google Scholar]
- Hochschild A, Dove SL. Protein–protein contacts that activate and repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nature Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- Jeon Y H, Negishi T, Shirakawa M, Yamazaki T, Fujita N, Ishihama A, Kyogoku Y. Solution structure of the activator contact domain of the RNA polymerase alpha subunit. Science. 1995;270:1495–1497. doi: 10.1126/science.270.5241.1495. [DOI] [PubMed] [Google Scholar]
- Jeon YH, Yamazaki T, Otomo T, Ishihama A, Kyogoku Y. Flexible linker in the RNA polymerase alpha subunit facilitates the independent motion of the C-terminal activator contact domain. J Mol Biol. 1997;267:953–962. doi: 10.1006/jmbi.1997.0902. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci. 1978;75:5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: A framework for understanding homeodomain–DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Kolb A, Igarashi K, Ishihama A, Lavigne M, Buckle M, Buc H. E. coli RNA polymerase, deleted in the C-terminal part of its α-subunit, interacts differently with the cAMP–CRP complex at the lacP1 and at the galP1 promoter. Nucleic Acids Res. 1993;21:319–326. doi: 10.1093/nar/21.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo HS, Wu HM, Crothers DM. DNA bending at adenine-thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Li XY, McClure WR. Stimulation of open complex formation by nicks and apurinic sites suggests a role for nucleation of DNA melting in Escherichia coli promoter function. J Biol Chem. 1998;273:23558–23566. doi: 10.1074/jbc.273.36.23558. [DOI] [PubMed] [Google Scholar]
- Marr MT, Roberts JW. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Min C, Cushing TD, Verdine GL. Template-directed interference footprinting of protein-adenine contacts. J Am Chem Soc. 1996;118:6116–6120. [Google Scholar]
- Moffatt JG. A general synthesis of nucleoside-5′ triphosphates. Can J Chem. 1964;42:599–604. [Google Scholar]
- Moran CP, Jr, Lang N, LeGrice SFJ, Lee G, Stephens M, Sonenshein AI, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Murakami K, Fujita N, Ishihama A. Transcription factor recognition surface on the RNA polymerase alpha subunit is involved in contact with the DNA enhancer element. EMBO J. 1996;15:4358–4367. [PMC free article] [PubMed] [Google Scholar]
- Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Negre D, Bonod-Bidaud C, Oudot C, Prost JF, Kolb A, Ishihama A, Cozzone AJ, Cortay JC. DNA flexibility of the UP element is a major determinant for transcriptional activation at the Escherichia coli acetate promoter. Nucleic Acids Res. 1997;25:713–738. doi: 10.1093/nar/25.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlands JT, Ross W, Gosink KK, Gourse RL. Factor-independent activation of Escherichia coli rRNA transcription. II. characterization of complexes of rrnB P1 promoters containing or lacking the upstream activator region with Escherichia coli RNA polymerase. J Mol Biol. 1991;220:569–583. doi: 10.1016/0022-2836(91)90101-b. [DOI] [PubMed] [Google Scholar]
- Newlands JT, Josaitis CA, Ross W, Gourse RL. Both fis-dependent and factor-independent upstream activation of the rrnB P1 promoter are face of the helix dependent. Nucleic Acids Res. 1992;20:719–726. doi: 10.1093/nar/20.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaghie G, Aiyar SE, Bobb KL, Hayward RS, deHaseth PL. Aromatic amino acids in region 2.3 of Escherichia coli ς70 participate collectively in the formation of an RNA polymerase–promoter open complex. J Mol Biol. 2000;299:1217–1230. doi: 10.1006/jmbi.2000.3808. [DOI] [PubMed] [Google Scholar]
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase β complexed with DNA: Implications for catalytic mechanism, processivity, and fidelity. Biochem. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- Pu WT, Struhl K. Uracil interference, a rapid and general method for defining protein-DNA interactions involving the 5-methyl group of thymines: The GCN4–DNA complex. Nucleic Acids Res. 1992;20:771–775. doi: 10.1093/nar/20.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Helmann JD. Adenines at −11, −9 and −8 play a key role in the binding of Bacillus subtilis EςA RNA polymerase to −10 region single-stranded DNA. Nucleic Acids Res. 1999;27:4541–4546. doi: 10.1093/nar/27.23.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafferty JB, Ingleston SM, Hargreaves D, Artymiuk PJ, Sharples GJ, Lloyd RG, Rice DW. Structural similarities between Escherichia coli RuvA protein and other DNA-binding proteins and a mutational analysis of its binding to the holliday junction. J Mol Biol. 1998;278:105–116. doi: 10.1006/jmbi.1998.1697. [DOI] [PubMed] [Google Scholar]
- Rao L, Ross W, Appleman JA, Gaal T, Leirmo S, Schlax PJ, Record MT, Jr, Gourse RL. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- Record MT, Jr, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ. Escherichia coli RNA polymerase (E670), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt FC, editor. Escherichia coli and Salmonella, cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 792–820. [Google Scholar]
- Reeves R, Nissen MS. The AT-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF–DNA complex: A protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Roberts JW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Ross W, Aiyar SE, Salomon J, Gourse RL. Escherichia coli promoters with UP elements of different strength: Modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, Gourse RL, Busby SJW. Transcription activation at class II CRP-dependent promoters: Identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela, F., Rosemeyer, H., Zulauf, M., Chen, Y., Kastner, G., and Reuter, H. 1997. 7-Nitro-7-deaza-2‘-deoxyadenosine and 8-methyl-7-deaza-2‘-deoxyguanosine: Pyrrolo[2,3-d]pyrimidine nucleosides with different sugar conformations. Liebigs Ann. Annalen-Recueil 2225–2230.
- Shao X, Grishin NV. Common fold in helix–hairpin–helix proteins. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatzky-Schwartz M, Arbuckle ND, Eisenstein M, Rabinovich D, Bareket-Samish A, Haran T E, Luisi BF, Shakked Z. X-ray and solution studies of DNA oligomers and implications for the structural basis of A-tract-dependent curvature. J Mol Biol. 1997;267:595–623. doi: 10.1006/jmbi.1996.0878. [DOI] [PubMed] [Google Scholar]
- Siebenlist U, Simpson RB, Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KY, Parker SA, Goljer I, Bolton PH. Solution structure of a duplex DNA with an abasic site in a dA tract. Biochem. 1997;36:11629–11639. doi: 10.1021/bi971464l. [DOI] [PubMed] [Google Scholar]
- Youderian P, Bouvier S, Susskind MM. Sequence determinants of promoter activity. Cell. 1982;30:843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 angstrom resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]