Abstract
The current standard of care for hepatitis C virus (HCV)-infected patients consists of lengthy treatment with interferon and ribavirin. To increase the effectiveness of HCV therapy, future regimens will incorporate multiple direct-acting antiviral (DAA) drugs. Recently, the HCV-encoded NS5A protein has emerged as a promising DAA target. Compounds targeting NS5A exhibit remarkable potency in vitro and demonstrate early clinical promise, suggesting that NS5A inhibitors could feature in future DAA combination therapies. Since the mechanisms through which these molecules operate are unknown, we have used NS5A inhibitors as tools to investigate their modes of action. Analysis of replicon-containing cells revealed dramatic phenotypic alterations in NS5A localization following treatment with NS5A inhibitors; NS5A was redistributed from the endoplasmic reticulum to lipid droplets. The NS5A relocalization did not occur in cells treated with other classes of HCV inhibitors, and NS5A-targeting molecules did not cause similar alterations in the localization of other HCV-encoded proteins. Time course analysis of the redistribution of NS5A revealed that the transfer of protein to lipid droplets was concomitant with the onset of inhibition, as judged by the kinetic profiles for these compounds. Furthermore, analysis of the kinetic profile of inhibition for a panel of test molecules permitted the separation of compounds into different kinetic classes based on their modes of action. Results from this approach suggested that NS5A inhibitors perturbed the function of new replication complexes, rather than acting on preformed complexes. Taken together, our data reveal novel biological consequences of NS5A inhibition, which may help enable the development of future assay platforms for the identification of new and/or different NS5A inhibitors.
INTRODUCTION
Hepatitis C virus (HCV) is a global health concern; recent estimates suggest that 2.2 to 3% of the world's population, equivalent to 130 to 170 million individuals, are chronically infected with the virus (13, 31). These patients are at risk of developing debilitating liver diseases such as cirrhosis and hepatocellular carcinoma (1). Furthermore, current models suggest that the burden of HCV-associated disease is set to rise for the next 20 years (6). There is no HCV vaccine; the current standard of care (SOC) involves lengthy treatments with ribavirin and injected pegylated interferon, which exhibit variable efficacies and are associated with severe, and sometimes life-threatening, side effects. Encouragingly, many direct-acting antiviral (DAA) molecules are in clinical development, and the most advanced (telaprevir and boceprevir) will probably be used to treat HCV-infected patients in 2011 (19, 29, 42, 43, 61). However, caution should be employed against overoptimism; attrition rates are high during drug development, and the first drugs will be given in combination with, not instead of, the current SOC. Therefore, the continued development of additional treatments is needed, especially since it is widely acknowledged that to limit the emergence of drug-resistant viral variants, effective therapeutic strategies for HCV will consist of multiple DAAs (50).
A multitude of screening campaigns has revealed many diverse and interesting chemical compounds capable of specifically inhibiting HCV RNA replication. Many of these compounds target the HCV-encoded nonstructural (NS) proteins (NS3, NS4A, NS4B, NS5A, and NS5B), which are required for HCV genome synthesis (3, 37). To instigate HCV genome replication, the NS proteins interact with viral genomes and certain host-encoded factors to form multiprotein assemblies termed “replication complexes” (RCs), which are sites of viral RNA synthesis derived from the endoplasmic reticulum (ER) (8, 14, 45, 53). In HCV-infected cells, RCs are juxtaposed to intracellular lipid storage organelles termed lipid droplets (LDs), which are coated with the HCV capsid protein (core) and probably serve as platforms to accept replicated genomes from RCs to initiate virion assembly (26, 44, 53). Of considerable interest are inhibitors that target the HCV-encoded NS5A protein. These inhibitors were originally discovered from the screening of cells containing HCV subgenomic replicons against libraries of small molecules and were identified as NS5A inhibitors by utilizing a strategy termed “chemical genetics” (12, 32). NS5A-targeting inhibitors are notable for their unprecedented potency in cell-based HCV replication assays: 50% inhibitory concentrations (IC50s) in the low-picomolar range have been reported for this class of inhibitors (32). Moreover, they demonstrate pangenotype activity and exhibit early clinical promise (12, 32). Thus, NS5A inhibitors are attractive candidates for inclusion in future HCV DAA combination therapies since, in theory, they could partner with any other DAA mechanistic class. However, assigning NS5A as the target of a defined series of small molecules is not straightforward, since no direct screening assays for a definitive NS5A function exist, and no specific binding of the compound to purified NS5A in vitro has been reported. Furthermore, although NS5A is essential for HCV RNA replication (3, 36, 37, 58) and is also required for virion morphogenesis (2, 23, 44, 57), its precise roles in the HCV life cycle are unknown. NS5A does exhibit a number of biological properties that probably enable it to perform a multitude of roles within the virus replicative cycle. These include the capacity to exist as differentially phosphorylated species (9, 22, 34, 57), the ability to bind HCV RNA (10, 21, 24), the facility to interact with other HCV-encoded proteins (7, 40, 52), and the propensity to bind certain host factors (16, 46, 63, 64). Structurally, NS5A is divided into three domains. Domains II and III appear unfolded when examined in isolation (17, 18, 33), whereas domain I of NS5A has been crystallized in alternative dimeric structures (38, 60). These findings suggest that NS5A may exhibit a degree of conformational flexibility, which may facilitate involvement at multiple stages within the HCV life cycle. Despite our current lack of knowledge concerning the full details of the HCV life cycle, it is likely that NS5A functions as a key nonenzymatic modulator of this process.
The modes of action (MOA) of NS5A inhibitors are a focus of great scientific interest but are currently unknown. Conceivably, these inhibitors could target any number of the functions attributed to NS5A. Therefore, the present study aimed to use NS5A-targeting molecules, representative of the NS5A inhibitor class, as tools to investigate the molecular mechanisms through which these molecules exert their inhibitory effects upon HCV RNA replication. We demonstrate that NS5A-targeting molecules promote specific phenotypic alterations in NS5A behavior, namely, a relocalization of the NS5A protein from the ER to LDs. We further reveal that the relocalization of NS5A is concomitant with the inhibition of HCV RNA replication and that an NS5A species encoding a Y93H substitution is refractory to compound-mediated redistribution. Collectively, our findings describe the first report of the biological consequences of NS5A inhibition, provide increased confidence for NS5A as a target for small-molecule-mediated inhibition, and begin to provide insight into the MOA for this class of HCV inhibitors.
MATERIALS AND METHODS
Cells.
Huh-7.5 cells (licensed from Apath LLC) were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, 1× nonessential amino acids, and 1× penicillin-streptomycin. The Huh-7-derived cell line, termed “Replicon 1b cells,” supports the replication of subgenomic RNA from the con1 strain of HCV genotype 1b, which is engineered to encode the firefly luciferase reporter gene and the neomycin phosphotransferase gene (licensed from RebLikon GmbH). This replicon contained tissue culture-adapted mutations in the NS3 gene (HCV polyprotein residues E1202G and T1280I) and the NS4B gene (HCV polyprotein residue K1846T) (35). Replicon 1b cells were maintained in Huh-7.5 medium supplemented with G418 at 500 μg/ml.
Compounds.
Inhibitors targeting the RNA replication of HCV genotypes 1a and 1b were identified by the screening of tissue culture cells containing HCV subgenomic replicons with a bespoke library of compounds that either were made based on literature examples or were designed at Pfizer, Sandwich, United Kingdom. Cytotoxicity assays were employed to eliminate compounds which displayed comparable concentrations that reduce cellular viability (through toxicity) by 50% (CC50s) and IC50s in Replicon 1b cells from further analysis. Structures of the example NS5A-targeting molecules used in this study are detailed in Fig. S1 in the supplemental material. NS5Ai 1 to NS5Ai 3 were identified from the screening of HCV replicon-containing cells at Pfizer. NS5Ai 4 is the Bristol-Myers Squibb (BMS) NS5A inhibitor BMS-790052 (Fig. S1) (12). The biotin-tagged HCV inhibitor (S form) was described previously by Gao et al. (12), and the R-form control is an enantiomer based thereupon (Fig. S1). HCV-796 (28) and BILN 2061 (30) are inhibitors of the HCV-encoded NS5B polymerase and NS3 protease, respectively (Fig. S1).
Antibodies.
Sheep antiserum used to detect HCV NS5A and human adipocyte differentiation-related protein (ADRP) were described previously (39, 54). Rabbit antiserum against calnexin (Sigma) was used according to the manufacturer's instructions. Mouse monoclonal antibodies specific for NS5A, NS5B, and double-stranded RNA (dsRNA) (J2) were purchased from Meridian Life Science, Axxora, and Scicons, respectively.
Generation of compound-resistant Replicon 1b cells.
Replicon 1b cells were seeded into 6-well tissue culture plates and cultured in G418-supplemented media and fixed concentrations of compounds (either 8× or 20× the IC50) for approximately 6 weeks. Cells treated with dimethyl sulfoxide (DMSO) only were maintained for equivalent periods of time to serve as passage controls. Following expansion, compound-resistant cells were subjected to phenotypic (IC50 determination) and genotypic (nucleotide sequencing) analyses.
For phenotypic analysis, 1 × 104 cells were added to each well of a white 96-well tissue culture assay plate in G418-supplemented medium containing the serially diluted compound. After 48 h of incubation, luciferase activity was determined by using BritelitePlus reagent (Perkin-Elmer).
For genotypic analysis, total RNA was extracted from replicon cell pellets (approximately 1 × 106 cells) by using an RNeasy minikit (Qiagen). First-strand cDNA synthesis was performed by using Superscript III enzyme (Invitrogen) and a replicon-specific primer (5′-GGATGGCCTATTGGCCTGGA-3′). PCR amplification of the HCV nonstructural genes was achieved with AccuPrime Taq DNA polymerase high-fidelity enzyme (Invitrogen) and gene-specific primers. Nucleotide sequencing of the purified amplicons was performed by Lark Technologies (Cambridge, United Kingdom).
Generation of mutant and chimeric replicons.
A plasmid containing HCV genotype 1b (con1 strain) subgenomic replicon cDNA (designated pBB7; licensed from Apath LLC) served as a template for the creation of replicons encoding L31V or Y93H substitutions in the NS5A open reading frame (ORF). The BB7-based replicon used in the present study contained tissue culture-adapted mutations in the NS3 gene (HCV polyprotein residues E1202G and T1280I) and the NS5A gene (HCV polyprotein residue S2197P). Site-directed mutagenesis was accomplished by using the QuikChange II XL site-directed mutagenesis kit (Stratagene-Agilent Technologies Inc.) according to the manufacturer's instructions. Nucleotide sequencing confirmed that only the desired changes were present following mutagenesis. Chimeric replicons containing NS5A sequences derived from genotype 1a (strain H77) were created in pBB7 by generating the appropriate NS5A genotype 1a nucleotide sequences by gene synthesis (Geneart), which were subcloned into the pBB7 backbone. Nucleotide sequencing confirmed the desired constructs. Chimeric replicons created were FL (full-length 1a NS5A sequence, encoding the tissue culture-adapted mutation S2204I, in the genotype 1b parental replicon background), D1 (genotype 1b replicon encoding a genotype 1a/1b chimeric NS5A protein composed of the N-terminal 213 amino acids from the genotype 1a strain), and D1P (genotype 1b replicon encoding a genotype 1a/1b chimeric NS5A protein composed of the N-terminal 149 amino acids from the genotype 1a strain).
Transient replication assays.
pBB7 and derivatives thereof were linearized with SpeI and used as a template for the production of replicon RNA by in vitro transcription (T7 Megascript transcription kit; Ambion). Transcribed RNA was purified (Megaclear purification kit; Ambion); and the quality and quantity of RNA were determined by gel electrophoresis and spectrophotometry, respectively. Huh-7.5 cells (5 × 106) were electroporated with 10 μg of replicon RNA by using an Amaxa Nucleofector device (Lonza Group Ltd.) according to the manufacturer's instructions. Following electroporation, 1 × 104 cells were added to each well of a white 96-well assay plate (CulturPlate; Perkin-Elmer) and incubated at 37°C overnight. Compounds serially diluted in DMSO were added to the cells the next day to give appropriate final assay concentrations in 0.5% DMSO. Following a further 72 h of incubation at 37°C, the luciferase activity was assayed by using a Renilla luciferase assay kit (Promega) according to the manufacturer's instructions.
Lentivirus construction and virus infections.
Recombinant lentiviruses expressing the NS5A wild type (WT) and Y93H variants with or without in-frame green fluorescent protein (GFP) fusions (45) were created by generating the appropriate ORFs by gene synthesis (Geneart), which were subcloned into the pLVX-Puro Lent-X expression vector (Clontech). Recombinant lentivirus was produced, purified, titrated, and used to infect cells according to the manufacturer's instructions (Clontech).
Quantitative RT-PCR.
The TaqMan Gene Expression Cells-to-CT kit (Ambion) was used to assay the abundance of HCV replicon RNA in a two-step reverse transcription (RT)-PCR directly from cultured-cell lysates according to the manufacturer's instructions. Primers specific for the HCV 5′ untranslated region (UTR) were 5′-TCCCGGGAGAGCCATAGTG-3′ (forward) and 5′-GGCATTGAGCGGGTTGATC-3′ (reverse), and 5′–6-carboxyfluorescein (6-FAM)–CCGGAATTGCCAGGACGACCG–MGB–3′ was used as the probe. Data were used to calculate arbitrary replicon RNA copy numbers.
Affinity isolation of the NS5A protein using biotin-tagged molecules.
Tissue culture plates (6-well) were seeded with 8 × 105 Replicon 1b or Huh-7.5 cells per well. Once the cells had adhered to the culture vessel (approximately 4 h at 37°C), biotin-tagged compounds were added to a final assay mixture concentration of 500 nM in 1% DMSO, and incubation at 37°C was continued for approximately 17 h. For lentivirus infections, cells were infected at 2 h postplating, treated with compounds 4 h later, and incubated for a further 17 h. Cells were subsequently washed twice in phosphate-buffered saline (PBS) and harvested into 140 μl of lysis buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.2), 120 mM KCl, 30 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 10% glycerol, 1× EDTA-free protease inhibitor cocktail] per well. The lysates were incubated on ice for 5 min prior to centrifugation at 19,357 × g for 5 min, and the supernatant was retained for analysis. Reacti-Bind streptavidin-coated 96-well enzyme-linked immunosorbent assay (ELISA) plates were washed twice with wash buffer (PBS, 0.1% Triton X-100) prior to use. Cell lysates were diluted 1:2 with lysis buffer, and 100 μl was transferred into each well of the Reacti-Bind streptavidin-coated plate. Plates were incubated at room temperature for 2 h with gentle agitation and then washed three times with wash buffer. Mouse monoclonal primary antibody specific for NS5A (Meridian Life Sciences) was diluted 1:500 in blocking buffer (PBS, 0.1% Triton X-100, 1% [wt/vol] bovine serum albumin [BSA]), and 100 μl was added to each well. Plates were incubated for 1 h with gentle agitation and then washed as described above. Horseradish peroxidase (HRP)-conjugated goat anti-mouse secondary antibody was diluted 1:800 in blocking buffer, and 150 μl was added to each well. Plates were incubated for a further hour with gentle agitation and washed as described above. Peroxidase reagent (DY999; R&D) was added (100 μl) to each well and incubated with gentle agitation until a color change was apparent (typically 5 to 10 min). The reaction was then stopped with 100 μl 2 M H2SO4; 170 μl from each well was transferred into a 96-well black-sided, clear-bottomed assay plate; and the absorbance at 450 nm was determined by using an Envision plate reader (Perkin-Elmer).
Indirect immunofluorescence.
Cells grown on 13-mm glass coverslips were processed for observation as described previously (53), and images were captured by using a Leica TCS SP5 inverted confocal microscope and associated software. Compounds were added to the indicated concentrations and incubated with the cells for appropriate periods of time. Control cells received the DMSO vehicle only (1% final assay concentration). Careful consideration was employed to capture images of a single cell or a few cells that were representative of the population of cells under scrutiny. Experiments were performed with a minimum of three independent repeats. Additionally, in an effort to ensure that images were representative of the cultures from which they were captured and to avoid operator bias, initial experiments were performed “blinded,” with the investigator unaware of which inhibitor (NS5Ai or control) was provided. To quantitate the number of cells exhibiting an altered NS5A distribution, a minimum of 3 multiples of 100 cells were counted from separate coverslips, and each cell was scored for the NS5A phenotype. Data are presented as average values over the multiple counts for each phenotype ± standard errors of the means.
Western blot analysis.
The preparation of cell extracts, polyacrylamide gel electrophoresis, and Western blot analysis were performed by using NuPage gels in combination with an iBlot apparatus (Invitrogen) according to the manufacturer's instructions. Membranes were probed with a mouse monoclonal primary antibody specific for NS5A (Meridian Life Science), followed by an HRP-conjugated secondary antibody. Bound antibody was visualized by using a GeneGnome bioimaging machine (Syngene) following the addition of Lumigen TMA-6 solution (GE Healthcare) to the nitrocellulose membrane. The densitometric quantitation of protein levels was performed with ImageJ software (http://rsb.info.nih.gov/ij/index.html).
IC50 determinations and t50 calculations.
IC50s for the activities of compounds in HCV genotype 1b replication assays were calculated from experimental data by using a Pfizer proprietary statistical analysis plug-in for Microsoft Excel 2007, which fitted the four-parameter sigmoidal maximum effect model. For experiments to determine the kinetics of inhibition, the change in the percent activity over time for each replicate was modeled using either an exponential decay model or a critical exponential model, as appropriate, and the t50s were estimated for both models as the time at which the percent activity was reduced to 50%. The significances of comparisons between the average compound t50s were made by using t tests with the appropriate degrees of freedom.
RESULTS
Targeting of NS5A-related functions by small-molecule inhibitors.
An internal discovery program identified several novel series of compounds (termed NS5Ai molecules) that demonstrated balanced IC50s in the picomolar range for cell lines constitutively replicating subgenomic RNA derived from both HCV genotypes 1a and 1b (strains H77 and con1, respectively). To gain insight into the potential MOA, the ability of the HCV replicon to adapt following an extended exposure of replicon-containing cell lines to representative molecules was investigated to determine whether the compound activity mapped to particular HCV-specific genetic loci. Accordingly, Replicon 1b cells were cultured in the presence of compounds at 8× and 20× the IC50 for 6 weeks. Resistant cell colonies were pooled, expanded, and then subjected to phenotypic analysis to reveal reduced susceptibility to the inhibitor; example data are presented in Fig. 1A. Replicon 1b cells cultured with NS5Ai 1 (a series example) at 20× the IC50 for 6 weeks exhibited decreased sensitivity to the compound (Fig. 1A). Specifically, the IC50 of NS5Ai 1 was increased 644-fold, from 32 pM in passage control Replicon 1b cells to 20.6 nM in NS5Ai 1-treated Replicon 1b cells (Fig. 1A). Replicon 1b cell populations that demonstrated reduced susceptibility to molecules with which they had been cultured were selected for genotypic analysis. Replicon RNA was isolated and amplified by RT-PCR, and DNA amplicons were sequenced to identify adaptive changes in the HCV nucleotide sequence associated with resistance to corresponding inhibitors. Parallel sequencing reactions were performed by using DNA amplicons derived from passage control Replicon 1b cells to identify nucleotide alterations not associated with resistance to compounds. Nucleotide sequencing data were obtained from DNA amplicons generated from cell populations treated separately with 8 different example compounds (see Table S1 in the supplemental material). Although a variety of sequence alterations were evident following the treatment of Replicon 1b cells with the NS5Ai molecules (Table S1), the NS5A genes from all amplicons contained nucleotide alterations that resulted in changes to the primary amino acid sequence at residues L31 and/or Y93; L31V and/or Y93H substitutions were predominantly observed (Table S1). To confirm that NS5A L31V and Y93H mutations conferred resistance to the NS5Ai series of small molecules, the corresponding nucleotide sequence alterations were engineered into the pBB7 HCV con1 replicon construct, and the activities of the compounds against the mutated subgenomic RNA were determined by transient replication assays. For NS5Ai 1, the activities in transient replication assays were reduced by 738- and 1,362-fold for replicons encoding L31V and Y93H mutations in the NS5A ORF, respectively, compared to the parental WT con1 replicon. NS5Ai 1 IC50s were 14.9 pM (n = 10), 11,000 pM (n = 2), and 20,300 pM (n = 4) for WT con1, L31V, and Y93H replicons, respectively. These data highlighted a genetic association between the MOA of the compound and the HCV NS5A gene and tentatively suggested that the molecules exerted their inhibitory effects by targeting one or more NS5A-related functions. To confirm that the tissue culture-adaptive mutation in the NS5A ORF present in the BB7 replicon sequence (HCV polyprotein residue S2197P) did not affect the potency of the compound, transient replication assays were also performed with example compounds by using a modified BB7 sequence that did not contain alterations in the NS5A ORF (see Table S2 in the supplemental material). Under these circumstances, the compound efficacies were comparable for replicon sequences that carried WT genotype 1b and 1a NS5A genes (Table S2).
Fig. 1.
Pfizer-identified small molecules target NS5A. (A) Replicon 1b cells were serially passaged in the presence of NS5Ai 1 at 20× the IC50 (labeled “resistant”); control cells were passaged in 1% DMSO only. After 6 weeks, cells were expanded, and the IC50 of NS5Ai 1 was determined for both cell populations. (B) Transient replication assays using chimeric replicons encoding genotype 1a NS5A sequences (dark shading) in a genotype 1b background were used to determine the IC50s for BILN 2061 and NS5Ai 2. (C) Biotin-tagged S- and R-form NS5A-targeting molecules were exposed to Replicon 1b cells or Huh-7.5 cells (500 nM final assay concentration), and bound NS5A was detected following the capture of protein-compound complexes, within cell lysates, on streptavidin-coated ELISA plates (Abs, NS5A antibodies). (D) Huh-7.5 cells were infected with a recombinant lentivirus (LV) expressing a polyhistidine-tagged NS5A species and then treated with the biotin-tagged NS5A-targeting compounds. The NS5A bound to the compound was assessed as described above (C). Cont., control.
The biological determinants required for the antiviral activities of compounds susceptible to L31V and Y93H mutations in NS5A were further mapped to increase the confidence that these molecules operated through an NS5A-related MOA. This was accomplished by using a tool compound (NS5Ai 2, which exhibited a 29-fold drop in potency for the Y93H mutant versus WT con1) and chimeric HCV subgenomic replicons. In cells constitutively replicating HCV subgenomic RNA, NS5Ai 2 was 506-fold less active against the genotype 1a strain (H77) than against the genotype 1b strain (con1) (Fig. 1B). Chimeric virus subgenomes based on genotype 1b were generated, in which genotype 1b NS5A, or parts of it, was replaced with the genotype 1a NS5A sequence to determine whether the tool compound exhibited reduced potency against the chimeras. Chimeric replicons constructed were designated FL, D1, and D1P (see Materials and Methods). The chimeric replicons were competent for replication in transient replication assays, and IC50s for BILN 2061, an inhibitor of the HCV-encoded NS3 protease (30), indicated that potency was maintained for compounds that exhibited broadly balanced genotype 1a/1b activity (Fig. 1B). However, the IC50s for NS5Ai 2 revealed a loss of activity when tested against the chimeric replicons: 680-, 870-, and 160-fold reductions in IC50s were observed with FL, D1, and D1P, respectively, while the sensitivity of the unmodified parental genotype 1b replicon to NS5Ai 2 was maintained (Fig. 1B). These data indicated that the biological determinants responsible for conferring sensitivity to this series of molecules were contained within the N-terminal sequences of NS5A, which corresponded to domain I of the protein. The increased susceptibility of the D1P chimera to NS5Ai 2 compared to the D1 chimera (IC50s of 16 nM and 87 nM, respectively) suggested that regions of NS5A that contributed determinants of compound sensitivity included amino acids 150 to 212 (Fig. 1B).
A variety of biophysical methods was employed to reveal the interaction of NS5A-targeting small molecules with the purified NS5A protein in vitro; however, specific binding was not detected (data not shown). Therefore, chemical tools were generated for use in experiments designed to highlight the interaction of compounds with NS5A in cells replicating HCV subgenomic RNA. Specifically, a biotin-tagged example of an NS5A-targeting compound was synthesized [previously referred to as the biotin-tagged HCV inhibitor 1:(S,S)-isomer by Gao et al. (12)] (see Fig. S1 in the supplemental material), together with its R stereoisoform to serve as a control, the activity of which was reduced approximately 7-fold compared to that with the S form in Replicon 1b cells (Fig. 1C). Following 17 h of incubation of the biotin-tagged inhibitors with Replicon 1b cells, cell lysates were prepared, and complexes composed of the protein-bound compound were captured on streptavidin-coated plates. The amount of the NS5A protein affinity isolated by the biotin-tagged compounds was quantitated by using an ELISA-based protocol (Fig. 1C). In Huh-7.5 cells or Replicon 1b cells treated with DMSO alone, no NS5A protein was retained on the streptavidin-coated plates (Fig. 1C). In contrast, NS5A bound in biotin-tagged compound complexes was clearly detected in lysates derived from Replicon 1b cells treated with the biotin-tagged S form of the molecule (Fig. 1C). Moreover, consistently less NS5A protein (from 3 analyses) was affinity isolated by using the less active biotin-tagged R form of the molecule, indicating that the activity of the compound correlated with the capacity to associate with the NS5A protein in cells (Fig. 1C). Notably, compounds required preincubation with Replicon 1b cells in order for the association with NS5A to be detected; it was not possible to use biotin-tagged molecules to isolate NS5A from lysates prepared from Replicon 1b cells that had not been preincubated with the compound (data not shown). Furthermore, in the absence of functional HCV RNA replication machinery, the association of the biotin-tagged compound with NS5A was inefficient, as evidenced by the affinity isolation of NS5A following an infection of cells with recombinant lentiviruses expressing only NS5A (Fig. 1D) or following the lentivirus-mediated expression of an NS3-NS5B polyprotein (see Fig. S2 in the supplemental material).
Taken together, these data provide genetic, biological, and biochemical evidence to link the NS5Ai compound series MOA to an NS5A-related mechanism. Since the virological profile of the NS5Ai molecules (e.g., susceptibility to substitution at L31 and Y93) was similar to those of previously reported NS5A-targeting inhibitors (11, 12, 32), we postulate that they exert their antiviral effects through an analogous MOA. Therefore, we believe that the NS5Ai molecules are representative of the NS5A inhibitor class and were subsequently utilized as tools to explore the MOA for this new class of HCV inhibitor.
NS5A-targeting compounds exhibit a delayed onset of inhibition.
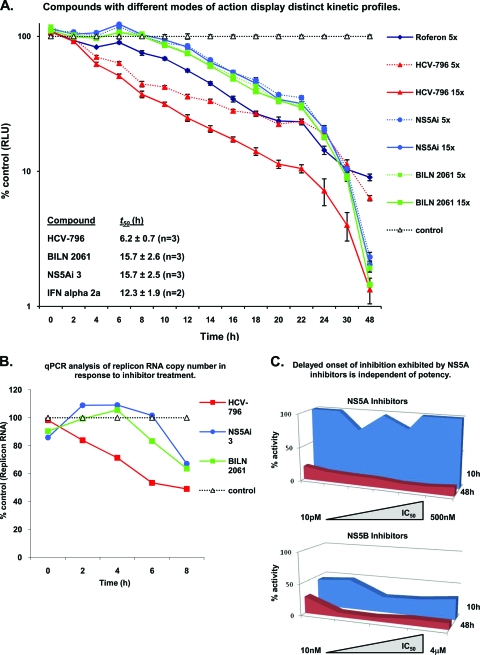
No enzymatic activity has been attributed to NS5A; therefore, compounds targeting NS5A are mechanistically distinct from molecules that inhibit the HCV-encoded protease and polymerase enzymes. To determine whether replicon assays can be utilized to differentiate separate DAA mechanistic classes, a panel of anti-HCV agents was examined for the capacity to inhibit HCV RNA replication shortly after administration to Replicon 1b cells. We hypothesized that in short-term replicon assays, NS5B-targeting molecules would exhibit rapid a inhibition of HCV RNA replication by virtue of the direct and immediate cessation of polymerase function within preformed RCs. Accordingly, Replicon 1b cells were treated with HCV-796 (a nonnucleoside inhibitor of NS5B) (see Fig. S1 in the supplemental material) (28), and luciferase activity was used to determine the relative levels of HCV RNA replication at 2-h intervals (Fig. 2A). A reduction in luciferase activity was evident from 2 h following the addition of compound (at both 5× and 15× the IC50), and the t50 (defined as the time at which luciferase activity was halved toward its baseline [see Materials and Methods]) was 6.2 h (Fig. 2A). Conversely, Replicon 1b cells treated with the NS3 protease inhibitor BILN 2061 exhibited a delay in the onset of replication inhibition; luciferase activity did not drop below control values until at least 8 h of compound treatment had elapsed. Furthermore, in comparison to HCV-796, the t50 of BILN 2061-treated Replicon 1b cells was increased to 15.7 h (Fig. 2A). The observed delay in inhibition presumably resulted from an inability of BILN 2061 to inhibit HCV genome synthesis within preformed RCs, since only the formation of de novo RCs would require NS3 protease activity. The treatment of Replicon 1b cells with an NS5A-targeting molecule (NS5Ai 3) resulted in replicon inhibition kinetics that were indistinguishable from those of BILN 2061-treated Replicon 1b cells, and the calculated t50 of 15.7 h was also identical (Fig. 2C). The treatment of Replicon 1b cells with alpha interferon 2a revealed an intermediate profile of inhibition; that is, a relatively rapid onset of inhibition was observed (a reduction in luciferase activity was apparent 2 h following administration to cells), but the decline in inhibition was lower than that observed for HCV-796 (Fig. 2A). Consequently, the t50 was calculated to be 12.3 h (Fig. 2A). However, after 48 h of treatment (at 5× and 15× the IC50 for alpha interferon 2a and the small molecules, respectively), all luciferase values were below 10% of those seen for control (DMSO-treated) samples. Statistically, the profile for HCV-796 had a significantly lower t50 (P < 0.001) than those of the other HCV inhibitors, but there were no significant differences in the t50 values between the other agents.
Fig. 2.
NS5A-targeting compounds exhibit delayed onset of inhibition. (A) Replicon 1b cells were treated with different HCV inhibitors at 5× or 15× their respective IC50s, and samples were analyzed for luciferase activity (RLU, relative light units) at the indicated time points. (B) Replicon 1b cells were treated with different HCV inhibitors at 15× their respective IC50s, and samples were analyzed for HCV RNA using RT-PCR at the indicated time points. For both A and B, data were normalized to data for DMSO controls, and average values from 8 replicates are plotted for each data point. qPCR, quantitative PCR. (C) Replicon 1b cells were exposed to a range of NS5A-targeting compounds (n = 8) and NS5B inhibitors (n = 7) at a fixed concentration of 40 μM. Individual compounds had IC50 values ranging from 10 pM to 500 nM (NS5A-targeting molecules) and 10 nM to 4 μM (NS5B inhibitors). Samples were harvested at 10 h and 48 h posttreatment, and luciferase activities within cell extracts were determined.
To confirm that the inhibition profiles measured using luciferase activity translated to a concomitant reduction in replicon RNA levels, quantitative RT-PCR was employed to assess the decline in relative levels of HCV RNA following the administration of the compound (Fig. 2B). This approach revealed similar findings: a delay in the decline of RNA levels was evident using BILN 2061 and NS5Ai 3 compared to HCV-796 (Fig. 2B). Specifically, whereas less HCV RNA was detected 2 h following treatment with HCV-796 (compared to the control), a reduction in HCV RNA levels was detected only 4 and 6 h following the exposure of Replicon 1b cells to BILN 2061 and NS5Ai 3, respectively (Fig. 2B). The different absolute values obtained with the two methods for assaying HCV RNA replication following the inhibition of HCV genome synthesis (luciferase and TaqMan) presumably resulted from dissimilar half-lives of the luciferase protein compared to HCV RNA in response to compound treatments. Next, we sought to determine whether the differentiated inhibition profiles exhibited by NS5A- and NS5B-targeting molecules were consistent for each respective mechanistic class and to identify if the compound IC50 affected the profile of NS5A inhibitors. Accordingly, the inhibition profiles of a panel of NS5A- and NS5B-targeting small molecules, with IC50 ranges of 10 pM to 500 nM and 10 nM to 4 μM, respectively, were analyzed with Replicon 1b cells (Fig. 2C). Results were consistent with previous data: the levels of luciferase activity in cell extracts derived from Replicon 1b cells treated with NS5B inhibitors were less than 50% of those exhibited by control (DMSO-treated) extracts at 10 h after exposure to the compound (Fig. 2C). In contrast, the panel of NS5A-targeting molecules exhibited reduced inhibition within the same time frame (Fig. 2C). After 48 h of treatment with the compound, luciferase activities in cell lysates from all compound-treated cells were less than 20% of control values (Fig. 2C).
Collectively, these data revealed that short-term replicon inhibition assays could distinguish between distinct classes of HCV inhibitors. Furthermore, the similarity in the activity profiles exhibited by both NS3- and NS5A-targeting molecules suggested a consistent mechanism; i.e., inhibitory effects may be exerted upon the formation and/or function of de novo, rather than preexisting, RCs.
NS5A-targeting compounds redistribute the NS5A protein.
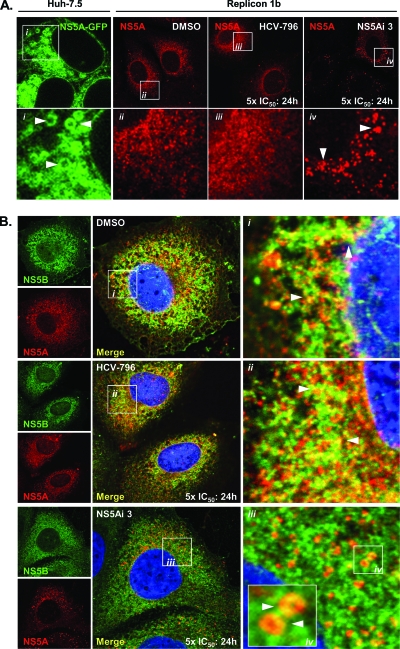
To investigate whether NS5A-targeting compounds cause detectable phenotypic alterations in the NS5A protein within cells, confocal microscopy was used to assess the distribution of NS5A following treatments with compounds. In Huh-7.5 cells expressing an NS5A-GFP fusion protein (in the absence of other HCV-encoded proteins), NS5A-GFP exhibited a cytoplasmic distribution, with apparent LD localization (Fig. 3Ai), as previously reported (51). Conversely, in Replicon 1b cells, no apparent LD localization was evident for NS5A; the protein was distributed in numerous small, brightly stained, clustered foci with a clear nuclear membrane and, presumably, ER staining (Fig. 3Aii), consistent with findings reported previously (53, 56). The observed change in the localization of NS5A in Replicon 1b cells presumably reflected the hierarchy of interactions formed between the HCV nonstructural proteins and host factors, which sequestered NS5A to RCs (53). No further differences in NS5A distribution were noted for Replicon 1b cells treated with HCV-796 (Fig. 3Aiii). However, following the treatment of Replicon 1b cells with low concentrations of NS5Ai 3 (50 pM) for 24 h, NS5A adopted an altered pattern of distribution (Fig. 3Aiv). Specifically, NS5A was observed as fewer foci, which were larger and spacially more discrete; a reduced staining of nuclear membranes was evident (Fig. 3Aiv). Of note, the distribution of NS5A observed for lentivirus-infected cells expressing NS5A-GFP (as in Fig. 3Ai) was not altered in the presence of NS5Ai 3 (data not shown).
Fig. 3.
NS5A-targeting inhibitors disrupt the association between NS5A and NS5B in replicon-containing cells. (A) Huh-7.5 cells were infected with a recombinant lentivirus expressing genotype 1b NS5A encoding an in-frame GFP fusion, and live cells were examined by using confocal microscopy (i). Replicon 1b cells were treated with 1% DMSO (ii), 5× the IC50 of HCV-796 (iii), or 5× the IC50 of NS5Ai 3 (iv) for 24 h and then analyzed for the distribution of NS5A. Boxed regions in the top panels are magnified in the corresponding bottom panels. Arrowheads indicate the NS5A distribution. (B) Replicon 1b cells were treated with DMSO (i), HCV-796 (ii), or NS5Ai 3 (iii) as described above (A), and the distributions of NS5A and NS5B were assessed by using confocal microscopy. Boxed regions i to iv are magnified in the corresponding panels. Arrowheads indicate regions of NS5A-NS5B colocalization in i and ii and NS5A-coated spherical structures in iv.
In the absence of inhibition, NS5A and NS5B are both sequestered in RCs derived from the ER membrane in Replicon 1b cells and, hence, exhibit overlapping patterns of distribution in DMSO-treated cells (Fig. 3Bi). The treatment of Replicon 1b cells with HCV-796 did not affect the distribution of either NS5B or NS5A compared to that in DMSO-treated samples (Fig. 3Bii), nor did the localization of NS5B change in the presence of NS5Ai 3 (Fig. 3Biii). However, the compound had a marked effect on the NS5A distribution, which served to reduce levels of colocalization with NS5B (Fig. 3Biv). Moreover, when viewed at a higher magnification, NS5A was relocalized to circular structures in the cytoplasm of Replicon 1b cells treated with NS5Ai 3 (Fig. 3Biv), reminiscent of structures seen when NS5A was expressed in the absence of other HCV-encoded proteins (Fig. 3Ai).
To further explore the NS5A distribution following exposure to NS5A-targeting molecules, Replicon 1b cells were treated with DMSO, BILN 2061, or NS5Ai 3, and the distribution of NS5A, calnexin (an ER membrane protein), and dsRNA (a marker for sites of HCV RNA replication [53]) was observed by using confocal microscopy (Fig. 4). Consistent with findings reported previously (53), RCs containing replicating HCV RNA and NS5A were observed in association with the ER membrane of DMSO-treated control cells, as judged by the colocalization of dsRNA, NS5A, and calnexin (Fig. 4Bi, arrowheads). In Replicon 1b cells exposed to BILN 2061, the intensity of dsRNA staining appeared reduced, consistent with an inhibitory effect on HCV RNA replication (Fig. 4B). Furthermore, the distribution of NS5A appeared more punctuate, with less colocalization evident with the ER and dsRNA, which presumably resulted from an accumulation of uncleaved HCV polyprotein precursors (Fig. 4Bi, arrowheads). Similarly, in the presence of NS5Ai 3, less dsRNA was evident; however, NS5A exhibited a dramatic relocalization from the ER to numerous spherical cytoplasmic organelles (Fig. 4Ci, arrowheads) and closely resembled the distribution evident in Fig. 3Ai. Taken together, these findings indicated that the redistribution of the NS5A protein was a specific biological phenomenon resulting from the exposure of Replicon 1b cells to NS5A-targeting molecules. Furthermore, these data provided additional confidence that the NS5Ai series of small molecules exerted its inhibitory properties by targeting one or more functions of the HCV-encoded NS5A protein.
Fig. 4.
NS5A-targeting molecules cause a redistribution of NS5A to spherical structures in replicon-containing cells. Replicon 1b cells were treated with 1% DMSO (A), 5× the IC50 of BILN 2061 (B), or 5× the IC50 of NS5Ai 3 (C) for 24 h. Cells were examined for the presence of dsRNA, NS5A, and calnexin (labeled as ER) by using confocal microscopy. Boxed regions are magnified in panels i for each case. Arrowheads highlight the distribution of NS5A.
NS5A-targeting molecules redistribute NS5A to lipid droplets.
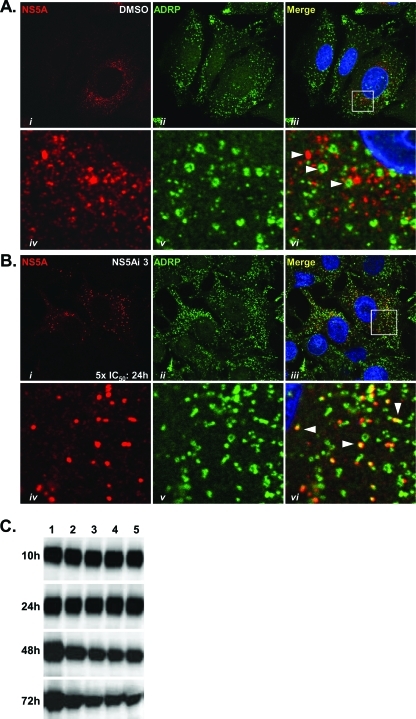
We hypothesized the spherical structures coated by NS5A in the cytoplasm of Replicon 1b cells treated with NS5Ai 3 were LDs, since NS5A has an intrinsic capacity to localize to LDs (51), and the spherical structures closely resembled the previously described morphology of LDs in live (54) and fixed (20, 53) cells. To first determine the relationship between LDs and NS5A in the absence of inhibition, the distribution of NS5A was compared to that of ADRP, a cell-encoded LD marker protein (4, 44, 53–55), in Replicon 1b cells (Fig. 5A). The results revealed that no colocalization was evident between NS5A and ADRP under control conditions; the distributions of ADRP-coated LDs and NS5A did not overlap (Fig. 5Ai to vi). In the presence of NS5Ai 3, no difference in the size, number, or distribution of ADRP-coated LDs was noted (compare Fig. 5Aii and Bii). In response to NS5Ai 3 treatment, NS5A was redistributed in a manner previously observed (Fig. 3Aiv), and NS5A foci now colocalized with ADRP-coated LDs (Fig. 5Bvi). These data suggested that the small-molecule-mediated inhibition of NS5A resulted in a redistribution of the protein from RCs (derived from ER membranes) to LDs. This phenomenon was specific to NS5A-targeting inhibitors and restricted to the NS5A protein.
Fig. 5.
NS5A-targeting compounds redistribute NS5A to LDs in Replicon 1b cells. (A and B) Replicon 1b cells were treated with 1% DMSO (A) or 5× the IC50 of NS5Ai 3 (B) for 24 h, and the distributions of NS5A (i and iv) and ADRP (ii and v) were assessed by using confocal microscopy. Merged images are presented in iii and iv. The boxed regions in panels iii are shown magnified in panels iv to vi. Arrowheads in vi indicate NS5A that does not (A) or does (B) colocalize with ADRP-coated LDs. (C) Replicon 1b cells were treated with DMSO (lane 1) or a range of HCV inhibitors at 10× the IC50 (lane 2, HCV-796; lane 3, BILN-2061; lane 4, NS5Ai 3; lane 5, NS5Ai 4). Samples were harvested at the indicated time points and assessed for NS5A contents by Western blot analysis. Protein levels were quantitated by using densitometry, and results contributed to data presented in Table 1.
To reveal whether the compound-mediated inhibition of HCV RNA replication was associated with the degradation of NS5A, Replicon 1b cells were treated with HCV-796, BILN 2061, and two NS5A inhibitors (NS5Ai 3 and NS5Ai 4 [BMS-790052]), and levels of the NS5A protein were assessed by using Western blotting (Fig. 5C and Table 1). Results revealed that the NS5A protein levels in Replicon 1b cell extracts treated with HCV inhibitors were 91.5 to 97.5% of those in DMSO-treated cells following 24 h of exposure to the compound (Fig. 5C and Table 1). These findings indicated that the degradation of NS5A was not associated with inhibitor-mediated NS5A redistribution. A loss of the NS5A protein was detected at 48 and 72 h following the addition of compounds (Fig. 5C and Table 1), presumably a result of a blocking of de novo HCV protein synthesis as a consequence of the inhibition of HCV RNA replication (Fig. 2A). Notably, the loss of the NS5A protein from Replicon 1b cells was not dramatically enhanced in response to NS5A inhibitors compared to NS3- or NS5A-targeting molecules at these later time points (Fig. 5C and Table 1). The concentrations of the compounds used were 10 times their respective IC50s for Replicon 1b cells, selected to stimulate a redistribution of the NS5A protein but not abrogate replicon replication. Hence, the half-life of the NS5A protein observed (Table 1) was greater than the previously reported value of 16 h (48), because RNA replication was not inhibited to 100% following the exposure of Replicon 1b cells to compounds at 10× the IC50.
Table 1.
Abundance of the NS5A protein in Replicon 1b cells following treatment with HCV inhibitorsa
| Time (h) | Mean % NS5A protein level relative to control ± SD |
||||
|---|---|---|---|---|---|
| DMSO | HCV-796 | BILN-2061 | NS5Ai 3 | NS5Ai 4 | |
| 10 | 100 | 96.5 ± 1.5 | 95 ± 2 | 100.5 ± 2.5 | 92.5 ± 1.5 |
| 24 | 100 | 91.5 ± 0.5 | 94.5 ± 0.5 | 97 ± 1 | 97.5 ± 0.5 |
| 48 | 100 | 84.5 ± 1.5 | 75.5 ± 1.5 | 74 ± 2 | 74 ± 2 |
| 72 | 100 | 87 ± 2 | 74 ± 2 | 73 ± 2 | 65.5 ± 1.5 |
Densitometry was used to analyze the relative levels of the NS5A protein revealed following Western blot analysis of cell extracts prepared at different times after exposure to the inhibitor at 10× the IC50. Data were normalized to DMSO values for each time point and are expressed as averages of data from 2 experiments ± standard errors of the means (data in Fig. 5C contributed 1 set of data; Western blot data for 2 experiments not shown).
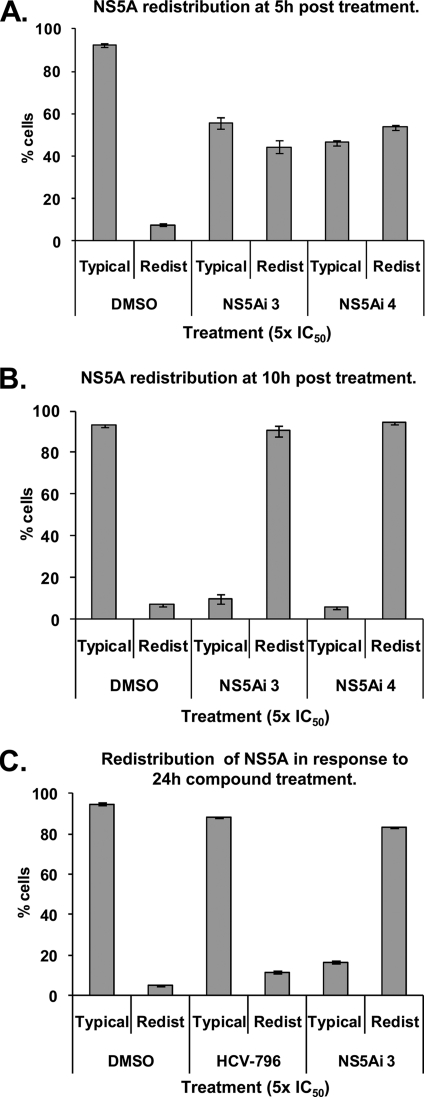
To determine whether the NS5A redistribution (Fig. 3 to 5) was concomitant with the onset of inhibition (Fig. 2A), the percentages of cells exhibiting a relocalized NS5A phenotype were quantitated at various time points following exposure to the compound (Fig. 6). After 5 h, 8% of control Replicon 1b cells (treated with DMSO only) exhibited a relocalized NS5A distribution reminiscent of that illustrated in Fig. 3Aiv (designated “redistributed”); 92% of cells stained for NS5A appeared as shown in Fig. 3Aii (designated “typical”) (Fig. 6A). However, following the treatment of Replicon 1b cells with NS5Ai 3 or NS5Ai 4 for 5 h, 44% and 54% of cells exhibited a redistribution of the NS5A protein, respectively (Fig. 6A). These values rose to 90% and 94% for NS5Ai 3- and NS5Ai 4-treated Replicon 1b cells following 10 h of compound exposure, respectively (Fig. 6B). No further NS5A redistribution was apparent at time points beyond 10 h (Fig. 6C). The NS5A distribution in DMSO- or HCV-796-treated Replicon 1b cells remained equivalent at all time points (Fig. 6). These findings correlated the time-dependent inhibition of HCV RNA replication observed following the exposure of Replicon 1b cells to NS5A inhibitors (Fig. 2A) with the phenotypic behavior of the NS5A protein in response to inhibitor treatment (Fig. 3Aiv and 6B).
Fig. 6.
The redistribution of NS5A to LDs is concomitant with the onset of inhibition exhibited by NS5A-targeting inhibitors. Replicon 1b cells were treated with 1% DMSO or HCV inhibitors at the indicated concentrations and times. Cells were probed for the NS5A protein and examined by using confocal microscopy. Individual counts of at least 100 cells were performed, and the distribution of NS5A was scored as typical (as in Fig. 3Aii) or redistributed (as in Fig. 3Aiv). A minimum of 3 counts were performed under each condition; data were averaged and are presented with standard errors of the means.
NS5A encoding a Y93H mutation is refractory to redistribution.
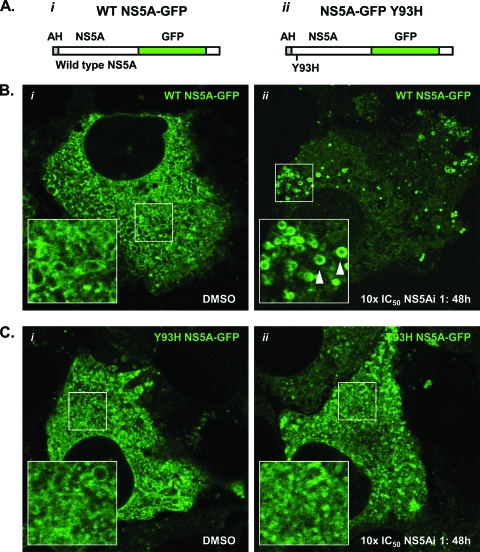
The replacement of tyrosine with histidine at residue 93 of the NS5A primary amino acid sequence is associated with a reduced susceptibility to NS5A-targeting molecules (Fig. 1A) (32). To determine whether an NS5A species encoding a Y93H substitution was refractory to redistribution under conditions that stimulated a redistribution of WT NS5A, we constructed recombinant lentiviruses designed to deliver NS5A proteins (WT and Y93H) encoding in-frame GFP fusions to Replicon 1b cells (Fig. 7A). This approach was selected because replicons encoding Y93H substitutions in NS5A replicate poorly in transient replication assays (data not shown). The NS5A-GFP species were designed to be competent components of the HCV RNA replication machinery, as reported previously (25, 45). The infection of Replicon 1b cells with lentiviruses expressing either WT or Y93H NS5A-GFP revealed similar patterns of NS5A-GFP localization (Fig. 7B and C, respectively). Specifically, NS5A-GFP was localized in the cytoplasm with an ER-like distribution and some evidence of an LD association (magnified insets in Fig. 7B and C). However, the degree of the LD association was not as great as that seen when NS5A-GFP was expressed in the absence of the HCV replication machinery (Fig. 3Ai). This presumably resulted from the sequestration of some NS5A-GFP molecules to RCs in replicon-containing cells, as reported previously (25, 45). In the presence of NS5Ai 1 at 10× the IC50 for WT con1 (150 pM) (Fig. 1A), WT NS5A-GFP was clearly relocalized to LDs (Fig. 7Bii, arrowheads). Conversely, using the same concentration of NS5Ai 1, NS5A Y93H-GFP was refractory to relocalization and exhibited a pattern of distribution similar to that within DMSO-treated Replicon 1b cells (compare Fig. 7Ci and ii). Importantly, NS5Ai 1 is not inhibitory for the resistant Y93H NS5A species at 150 pM; this concentration is 135-fold lower than the IC50 for a genotype 1b subgenomic replicon encoding a Y93H substitution in NS5A (Fig. 1A). An increase of the concentration of NS5Ai 1 to 200 nM (10× the IC50 for the NS5A Y93H-containing replicon) resulted in Y93H NS5A-GFP adopting a pattern of distribution comparable to that of WT NS5A-GFP in Replicon 1b cells treated with 150 pM NS5Ai 1 (Fig. 7Bii and data not shown). Therefore, these findings highlighted the link between the pharmacology of NS5A-targeting compounds and the biological consequences of the inhibition of NS5A.
Fig. 7.
An NS5A species encoding a Y93H substitution is refractory to relocalization. Recombinant lentiviruses were designed to express WT NS5A-GFP (i) or Y93H NS5A-GFP (ii) (AH, amphipathic helix) (A) and used to infect Replicon 1b cells (B and C, respectively). Following infection, cells were treated with 1% DMSO or 10× the IC50 of NS5Ai 1 for 48 h and examined for GFP fluorescence using live-cell confocal microscopy (i and ii, respectively, in B and C). Boxed regions in i and ii (B and C) are magnified in the bottom left corner of their respective images. Arrowheads indicate LDs.
DISCUSSION
This study describes findings related to the NS5Ai series of small molecules, which exert their antiviral activity by targeting the HCV-encoded NS5A protein. In the absence of biological screening assays for definitive NS5A functions amenable to small-molecule-mediated inhibition, attributing NS5A as the target of specific small molecules requires a weight of indirect evidence rather than a direct indication. To this end, we have demonstrated that (i) the NS5Ai series of compounds promotes the selection of resistance substitutions in the NS5A ORF, (ii) the biological determinants required for compound activity map to the N terminus of NS5A, (iii) biotin-tagged NS5Ai molecules affinity isolate protein complexes containing NS5A, and (iv) NS5A-targeting molecules cause specific phenotypic alterations in the behavior of NS5A. Collectively, these findings provided a substantial body of evidence to indicate that the NS5Ai series of small molecules targeted the HCV-encoded NS5A protein. Additionally, novel biological consequences that resulted from the inhibition of the NS5A protein were revealed. Since the NS5Ai molecules are susceptible to resistance substitutions at residues previously reported for other NS5A-targeting compounds (11, 12, 32), we believe that our findings are pertinent to the NS5A inhibitor class.
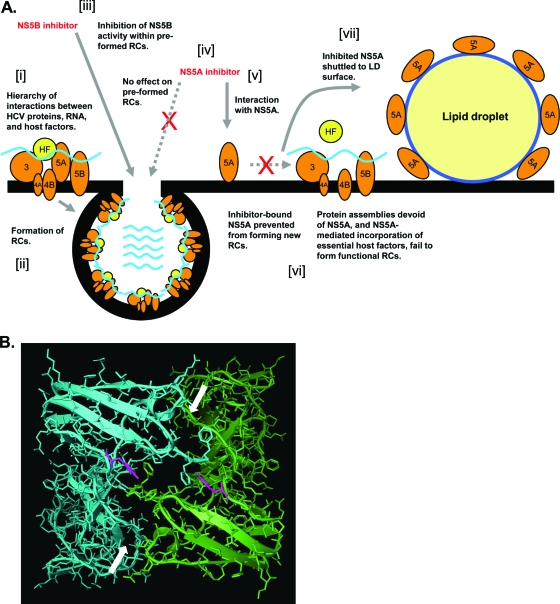
Resistance data and potency mapping experiments demonstrated that domain I of NS5A encoded functions that were sensitive to small-molecule-mediated inhibition. Domain I of NS5A is conserved among HCV genotypes, contains the amphipathic membrane-anchoring helix (5) and a Zn-binding motif (59), and exhibits affinity for HCV RNA (10, 21, 24). The crystal structure of NS5A domain I from genotype 1b has been solved to reveal a dimeric molecule (38, 60). Exactly how NS5A-targeting molecules perturb domain I-associated functions remains unknown, but the NS5Ai series of inhibitors does not affect the capacity of NS5A to bind HCV RNA in vitro (see Fig. S3 in the supplemental material). The present study and previous studies (11, 12, 32) identified that a substitution at residue Y93 was associated with a reduced susceptibility to the NS5A-targeting compound-mediated inhibition of HCV RNA replication. Each domain I monomer contributes its respective Y93 residue to opposing surfaces of the dimer interface (Fig. 8B). The propensity for substitutions at Y93 after prolonged exposure to an inhibitor suggests that a subtle modification of the nature of the monomeric contacts within the dimer molecule is capable of decreasing the sensitivity of the protein to NS5A-targeting compounds. It is therefore possible that the domain I dimer interface may also constitute the compound interaction site on the NS5A protein. An intriguing property of NS5A-targeting molecules is their exquisite potency; low-picomolar IC50s for cell-based HCV RNA replication assays are unprecedented for anti-HCV small-molecule inhibitors. Using previously reported data relating to the quantitation of NS proteins in HCV replicon-containing cells (49), together with calculated values for the average cytoplasmic volume of hepatocytes, it is possible to estimate the approximate molarity of NS proteins within a replicon cell. This equates to 0.6 nM to 600 nM; the 1,000-fold window accounts for excess NS proteins not actively involved in HCV genome synthesis in replicon-containing cells (49). These values are consistent for inhibitors, such as BILN 2016, which are expected to inhibit their target protein (NS3) with 1:1 stoichiometry and exhibit single-figure nanomolar IC50s in HCV replicon assays. However, NS5A-targeting compounds are almost 1,000-fold more potent than BILN 2061; therefore, these calculations suggest a “potency paradox” in which few molecules of the NS5A-targeting inhibitor can abrogate the function of many more molecules of the NS5A protein. Thus, NS5A inhibitors may exert their inhibitory effects in a dominant negative manner. Conceivably, one could envisage a scenario whereby a single molecule of inhibitor affects the structural distortion of a single NS5A dimer that is communicated allosterically to many NS5A molecules in an oligomeric complex.
Fig. 8.
Model for NS5A inhibition in replicon-containing cells. (A) A hierarchy of interactions formed between HCV-encoded proteins, HCV RNA, and host factors (HF) (i) results in the formation of RCs on the ER membrane (ii). NS5B-targeting molecules inhibit the HCV polymerase within preformed RCs (iii). However, the activity of NS5A sequestered within preformed RCs is refractory to inhibition by NS5A-targeting molecules (iv), perhaps as a consequence of protein-protein interactions masking the compound-binding site of NS5A in RCs. NS5A is accessible for inhibition prior to the formation of de novo RCs (v); the interaction of the NS5A inhibitor with the NS5A protein alters/locks NS5A in a conformation not permissive for incorporation into RCs (vi). The failure of NS5A to be sequestered into RCs results in the protein adopting a distribution reminiscent of that when expressed in the absence of other HCV-encoded proteins; i.e., NS5A is shuttled to the surface of lipid droplets (vii). (B) Y93 residues (pink) are located on either side of the dimer interface in the published crystal structure of domain I of NS5A from HCV genotype 1b (60). Separate domain I monomers are blue and green, respectively, and arrows indicate the dimer interface. The orientation of molecule is as seen “through” the ER membrane from the lumen to the cytoplasm; i.e., the viewed molecule area faces the cytosolic surface of the ER membrane.
Structural information gained from the cocrystallization of the inhibitor and protein can inform a detailed analysis of structure-activity relationships (47, 62). Unfortunately, as yet, it has not been possible to demonstrate a specific binding of NS5A inhibitors to purified protein in vitro by using biophysical methods (data not shown). This probably resulted from an inability to recapitulate the exact conformation of the NS5A protein required for compound interactions within cells. Additionally, it was recently shown that uracil-rich RNA can promote the formation of NS5A dimers (24), suggesting that the inclusion of uracil-rich RNA in future biophysical analyses may yield more promising results. However, the affinity isolation of NS5A-containing complexes was achieved by using biotin-tagged compounds when preincubated with Replicon 1b cells. Notably, the affinity isolation of NS5A-containing complexes was possible only following a preincubation of Replicon 1b cells with biotin-tagged compounds; no binding to the compound was detected when Replicon 1b cell lysates were used as a source of the NS5A protein, nor was specific binding to the compound observed when purified NS5A was used alone (data not shown). Furthermore, a minimal binding of NS5A to the compound was detected upon the intracellular expression of NS5A only or following the lentivirus-mediated expression of an NS3-NS5B polyprotein. These findings indicated that the interaction of the compound with NS5A required the presence of functional RNA replication machinery. The absolute requirement for the preincubation of Replicon 1b cells with the compound in order to detect inhibitor-bound complexes containing NS5A suggested that the inhibitor interaction site on NS5A is accessible only at specific stages during the involvement of NS5A in HCV genome synthesis. This may result from the masking of the compound-binding site by certain protein-protein interactions formed in RCs. Additionally, conformational flexibility, associated with NS5A as the protein performs defined roles during RNA replication, may reveal the interaction site only intermittently. The half-life of HCV NS proteins in replicon-containing cells is 11 to 16 h (48); presumably, HCV RNA replication is not synchronized in replicon-containing cells, and at any given moment in time, rates of RC formation, activity, and degradation are equilibrated. Hence, exposure to an inhibitor that interacts with NS5A only at a defined stage during the protein's functional cycle would require a period of time necessary for a sufficient number of compound molecules to bind enough protein for subsequent detection. In agreement with this notion, we found that maximal levels of NS5A association with the biotin-tagged compound were evident following at least 17 h of incubation of Replicon 1b cells with the biotin-tagged compound.
Consistent with our hypothesis that an interaction surface on the NS5A protein may not be perpetually accessible to NS5A-targeting molecules, kinetic profiling experiments revealed that NS3 and NS5A inhibitors exhibited a delayed onset of inhibition compared to NS5B-targeting molecules. For NS3 inhibitors, this phenotype correlates with the requirement for NS3 function to initiate de novo RC formation. This may also be the case for NS5A inhibitors, which exhibited kinetics of inhibition identical to those displayed by NS3-targeting molecules. Other causes for the observed delay in NS5A inhibitor activity may include reduced cellular permeability or a requirement for conversion from prodrug to active species. However, a preclinical bioanalysis of NS5A-targeting molecules revealed that this was unlikely (data not shown). In the absence of further evidence, we speculate that the kinetic profiles observed for the separate classes of HCV DAAs are likely related to their MOA. Therefore, our findings suggested that preformed RCs were refractory to inhibition using NS5A-targeting molecules and that compound-mediated antiviral effects resulted from an inhibition of de novo RC function or formation.
NS5A performs a pivotal role at the LD surface (2, 44, 57), and in virus-infected cells there probably exists regulated mechanisms that dictate the spatial properties of NS5A in a temporal fashion, directing NS5A to regions of the cell where it is needed at specific stages of the HCV life cycle. Consistent with this hypothesis, subpopulations of intracellular NS5A form highly mobile structures (66), indicating that the trafficking of NS5A-containing complexes does occur during HCV genome replication. A key finding of the present study was the discovery that NS5A-targeting compounds caused distinctive phenotypic alterations in the behavior of the NS5A protein. Specifically, NS5Ai molecules deregulated the distribution of NS5A and promoted the relocalization of the NS5A protein from the ER to LDs in Replicon 1b cells. The redistribution of NS5A was a phenomenon associated with NS5Ai molecules only; no evidence for NS5A relocalization was evident following the exposure of Replicon 1b cells to HCV-796 or BILN 2061. Furthermore, the redistribution of NS5A was concomitant with the onset of inhibition, as judged by kinetic profiling experiments, suggesting that the two events may be linked; i.e., the relocalization of NS5A was a cause, or a consequence, of the compound-mediated inhibition of HCV RNA replication. Importantly, we demonstrated that an NS5A species encoding a Y93H substitution was refractory to compound-mediated relocalization. This finding reinforced a link between the pharmacological activity of NS5A-inhibiting compounds and the biological consequences of NS5A inhibition. In virus-infected cells, NS5A is already localized to LD-associated sites (44, 53); hence, the treatment of JFH1-infected cells with NS5Ai molecules did not reveal an appreciable deviation from the pattern of NS5A localization evident in control cells infected with HCV (data not shown). Therefore, the relocalization of NS5A to LDs served as a marker for the MOA of NS5A-targeting molecules in Replicon 1b cells. Accordingly, high-content imagining techniques could be developed to specifically screen for additional chemical material that promotes similar effects on the NS5A distribution. Indeed, such analyses were previously employed to examine the behaviors of LDs in response to various stimuli (15, 41, 65).
Exactly how do NS5A-targeting molecules promote the redistribution of NS5A to LDs in replicon-containing cells, and how does this phenomenon relate to their MOA? Although still unclear, we propose a model to describe the MOA of NS5A-targeting molecules based upon findings from the present study (Fig. 8A). During de novo RC formation, a compound interaction site becomes accessible on the surface of the NS5A protein. The binding of the inhibitor to NS5A serves to lock, or alter, the conformation of the protein, rendering it incapable of RC incorporation. The location of the Y93H resistance substitution in domain I of NS5A suggests that the NS5A dimer interface may constitute the molecular surface recognized by NS5A-targeting compounds (Fig. 8B). The binding of the inhibitor to NS5A may result in an abrogation of specific protein-protein interactions involving NS5A (including homo-oligomerization) or a disruption of specific contacts formed with the ER membrane. Additionally, although we did not detect any gross inhibitor-mediated effects on NS5A phosphorylation (data not shown), it is possible that a subtle deregulation of NS5A phosphorylation may also contribute to the antiviral properties of these molecules. Inhibited NS5A, no longer sequestered to RCs, adopts a pattern of localization reminiscent of that when expressed in the absence of other HCV proteins; i.e., the protein is redistributed to the LD surface. Through the use of click chemistry to reveal the intracellular localization of NS5A-targeting molecules in Replicon 1b cells, we previously demonstrated that an NS5Ai compound and the NS5A protein were colocalized (27), indicating that redistributed NS5A is probably bound with the inhibitor at the LD surface. In virus-infected cells, the compound-mediated accumulation of NS5A on the LD surface is probably masked by the inherent capacity of preexisting RCs to localize to core-coated LDs. Despite the close proximity of LD-associated RCs, compound-bound NS5A on the LD surface presumably remains nonfunctional, locked in a configuration not permissive to functioning in HCV genome synthesis. Therefore, the redistribution of NS5A to the LD surface per se is likely not directly responsible for the antiviral properties of NS5A-targeting molecules. Based upon our findings, we believe that the specific deregulation of NS5A-related mechanisms, which serve to govern the precise microenvironment required for RC formation, is a credible MOA for this class of HCV inhibitor. Additional work will aim to refine this hypothesis.
The discovery and clinical validation (12) of NS5A-targeting molecules herald a new and exciting era for HCV drug development, enhancing an already promising HCV DAA pipeline and contributing optimism to the millions of HCV-infected patients at risk of developing debilitating liver disease. Continued research will aim to elucidate the precise MOA of NS5A inhibitors to further our understanding of the function of NS5A and to develop specific screening assays to enable the discovery of novel NS5A inhibitors with a higher genetic barrier to resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ben Sidders for analysis of data, Caroline Smith-Burchnell for JFH-1 infections, and Owen Everson, Michael Paradowski, and Duncan Hay for compound preparation. We thank Toshana Foster and Mark Harris (University of Leeds, United Kingdom) for the NS5A RNA-binding analysis.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 20 April 2011.
REFERENCES
- 1. Alberti A., Chemello L., Benvegnu L. 1999. Natural history of hepatitis C. J. Hepatol. 31(Suppl. 1):17–24 [DOI] [PubMed] [Google Scholar]
- 2. Appel N., et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blight K. J., Kolykhalov A. A., Rice C. M. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 4. Boulant S., et al. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268–1282 [DOI] [PubMed] [Google Scholar]
- 5. Brass V., et al. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130–8139 [DOI] [PubMed] [Google Scholar]
- 6. Deuffic-Burban S., Poynard T., Sulkowski M. S., Wong J. B. 2007. Estimating the future health burden of chronic hepatitis C and human immunodeficiency virus infections in the United States. J. Viral Hepat. 14:107–115 [DOI] [PubMed] [Google Scholar]
- 7. Dimitrova M., Imbert I., Kieny M. P., Schuster C. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Egger D., et al. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evans M. J., Rice C. M., Goff S. P. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:13038–13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foster T. L., Belyaeva T., Stonehouse N. J., Pearson A. R., Harris M. 2010. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 84:9267–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fridell R. A., Qiu D., Wang C., Valera L., Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao M., et al. 2010. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Global Burden of Hepatitis C Working Group 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 44:20–29 [DOI] [PubMed] [Google Scholar]
- 14. Gosert R., et al. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grandl M., Schmitz G. 2010. Fluorescent high-content imaging allows the discrimination and quantitation of E-LDL-induced lipid droplets and Ox-LDL-generated phospholipidosis in human macrophages. Cytometry A 77:231–242 [DOI] [PubMed] [Google Scholar]
- 16. Hamamoto I., et al. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473–13482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanoulle X., Badillo A., Verdegem D., Penin F., Lippens G. 2010. The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept. Lett. 17:1012–1018 [DOI] [PubMed] [Google Scholar]
- 18. Hanoulle X., et al. 2009. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 381:634–638 [DOI] [PubMed] [Google Scholar]
- 19. Hezode C., et al. 2009. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N. Engl. J. Med. 360:1839–1850 [DOI] [PubMed] [Google Scholar]
- 20. Hope R. G., McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913–1925 [DOI] [PubMed] [Google Scholar]
- 21. Huang L., et al. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417–36428 [DOI] [PubMed] [Google Scholar]
- 22. Huang Y., Staschke K., De Francesco R., Tan S. L. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9 [DOI] [PubMed] [Google Scholar]
- 23. Hughes M., Griffin S., Harris M. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J. Gen. Virol. 90:1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hwang J., et al. 2010. Hepatitis C virus non-structural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteins. J. Virol. 84:12480–12491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones D. M., Gretton S. N., McLauchlan J., Targett-Adams P. 2007. Mobility analysis of an NS5A-GFP fusion protein in cells actively replicating hepatitis C virus subgenomic RNA. J. Gen. Virol. 88:470–475 [DOI] [PubMed] [Google Scholar]
- 26. Jones D. M., McLauchlan J. 2010. Hepatitis C virus: assembly and release of virus particles. J. Biol. Chem. 285:22733–22739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones L., et al. 2011. In-cell click labelling of small molecules to determine subcellular localisation. J. Chem. Biol. 4:49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kneteman N. M., et al. 2009. HCV796: a selective nonstructural protein 5B polymerase inhibitor with potent anti-hepatitis C virus activity in vitro, in mice with chimeric human livers, and in humans infected with hepatitis C virus. Hepatology 49:745–752 [DOI] [PubMed] [Google Scholar]
- 29. Kwo P. Y., et al. 2010. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet 376:705–716 [DOI] [PubMed] [Google Scholar]
- 30. Lamarre D., et al. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186–189 [DOI] [PubMed] [Google Scholar]
- 31. Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl. 1):74–81 [DOI] [PubMed] [Google Scholar]
- 32. Lemm J. A., et al. 2010. Identification of hepatitis C virus NS5A inhibitors. J. Virol. 84:482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang Y., Ye H., Kang C. B., Yoon H. S. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46:11550–11558 [DOI] [PubMed] [Google Scholar]
- 34. Lindenbach B. D., et al. 2007. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J. Virol. 81:8905–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lohmann V., Hoffmann S., Herian U., Penin F., Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lohmann V., Korner F., Dobierzewska A., Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 38. Love R. A., Brodsky O., Hickey M. J., Wells P. A., Cronin C. N. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macdonald A., et al. 2003. The hepatitis C virus non-structural NS5A protein inhibits activating protein-1 function by perturbing ras-ERK pathway signaling. J. Biol. Chem. 278:17775–17784 [DOI] [PubMed] [Google Scholar]
- 40. Masaki T., et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonough P. M., et al. 2009. Quantification of lipid droplets and associated proteins in cellular models of obesity via high-content/high-throughput microscopy and automated image analysis. Assay Drug Dev. Technol. 7:440–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McHutchison J. G., et al. 2009. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360:1827–1838 [DOI] [PubMed] [Google Scholar]
- 43. McHutchison J. G., et al. 2010. Telaprevir for previously treated chronic HCV infection. N. Engl. J. Med. 362:1292–1303 [DOI] [PubMed] [Google Scholar]
- 44. Miyanari Y., et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 45. Moradpour D., et al. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78:7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okamoto T., et al. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ortqvist P., et al. 2010. Structure-activity relationships of HCV NS3 protease inhibitors evaluated on the drug-resistant variants A156T and D168V. Antivir. Ther. 15:841–852 [DOI] [PubMed] [Google Scholar]
- 48. Pietschmann T., Lohmann V., Rutter G., Kurpanek K., Bartenschlager R. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinkert D., Bartenschlager R., Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594–13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rong L., Dahari H., Ribeiro R. M., Perelson A. S. 2010. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci. Transl. Med. 2:30ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi S. T., et al. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292:198–210 [DOI] [PubMed] [Google Scholar]
- 52. Shimakami T., et al. 2004. Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78:2738–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Targett-Adams P., Boulant S., McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Targett-Adams P., et al. 2003. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J. Biol. Chem. 278:15998–16007 [DOI] [PubMed] [Google Scholar]
- 55. Targett-Adams P., Hope G., Boulant S., McLauchlan J. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J. Biol. Chem. 283:16850–16859 [DOI] [PubMed] [Google Scholar]
- 56. Targett-Adams P., McLauchlan J. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 86:3075–3080 [DOI] [PubMed] [Google Scholar]
- 57. Tellinghuisen T. L., Foss K. L., Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tellinghuisen T. L., Foss K. L., Treadaway J. C., Rice C. M. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tellinghuisen T. L., Marcotrigiano J., Gorbalenya A. E., Rice C. M. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 60. Tellinghuisen T. L., Marcotrigiano J., Rice C. M. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van der Meer A. J., de Knegt R. J. 2009. Telaprevir for chronic HCV infection. N. Engl. J. Med. 361:533–534 [DOI] [PubMed] [Google Scholar]
- 62. Venkatraman S., Njoroge F. G. 2007. Macrocyclic inhibitors of HCV NS3-4A protease: design and structure activity relationship. Curr. Top. Med. Chem. 7:1290–1301 [DOI] [PubMed] [Google Scholar]
- 63. Waller H., Chatterji U., Gallay P., Parkinson T., Targett-Adams P. 2010. The use of AlphaLISA technology to detect interaction between hepatitis C virus-encoded NS5A and cyclophilin A. J. Virol. Methods 165:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang C., et al. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425–434 [DOI] [PubMed] [Google Scholar]
- 65. Whittaker R., et al. 2010. Identification of microRNAs that control lipid droplet formation and growth in hepatocytes via high-content screening. J. Biomol. Screen. 15:798–805 [DOI] [PubMed] [Google Scholar]
- 66. Wolk B., Buchele B., Moradpour D., Rice C. M. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.