Abstract
Rift Valley fever virus (RVFV; family Bunyaviridae, genus Phlebovirus) is an important emerging pathogen of humans and ruminants. Its NSs protein has previously been identified as a major virulence factor that suppresses host defense through three distinct mechanisms: it directly inhibits beta interferon (IFN-β) promoter activity, it promotes the degradation of double-stranded RNA-dependent protein kinase (PKR), and it suppresses host transcription by disrupting the assembly of the basal transcription factor TFIIH through sequestration of its p44 subunit. Here, we report that in addition to PKR, NSs also promotes the degradation of the TFIIH subunit p62. Infection of cells with the RVFV MP-12 vaccine strain reduced p62 protein levels to below the detection limit early in the course of infection. This NSs-mediated downregulation of p62 was posttranslational, as it was unaffected by pharmacological inhibition of transcription or translation and MP-12 infection had no effect on p62 mRNA levels. Treatment of cells with proteasome inhibitors but not inhibition of lysosomal acidification or nuclear export resulted in a stabilization of p62 in the presence of NSs. Furthermore, p62 could be coprecipitated with NSs from lysates of infected cells. These data suggest that the RVFV NSs protein is able to interact with the TFIIH subunit p62 inside infected cells and promotes its degradation, which can occur directly in the nucleus.
INTRODUCTION
Rift Valley fever virus (RVFV) is a mosquito-borne Phlebovirus which is endemic to sub-Saharan Africa and has caused large outbreaks among humans and ruminants in several other African countries, including Egypt and Madagascar, as well as Saudi Arabia and Yemen (3, 38, 39). Human infection with RVFV usually manifests as an acute febrile illness; however, a small number of cases progress to encephalitis, retinal vasculitis, hepatitis, or hemorrhagic fever-like illness with a potentially fatal outcome (47). In ruminants, RVFV infection causes frequent abortions and has a newborn mortality rate of up to 95%, which leads to significant economic loss during an outbreak of RVFV (47). RVFV is a member of the family of Bunyaviridae and contains a tripartite, negative-stranded genome which encodes four structural proteins, the RNA-dependent RNA polymerase (RdRp) L, the envelope glycoproteins Gn and Gc, and the nucleocapsid N (45), two nonstructural proteins, NSs and NSm, and a less well-characterized 78-kDa protein (38). NSm and the 78-kDa protein are both encoded by the M segment (46) and are dispensable for viral replication and infection (11, 51). The NSs protein, on the other hand, is encoded in an ambisense manner on the S segment (13) and has been identified as the major virulence factor of RVFV (4, 49). NSs is a multifunctional protein, and it is able to manipulate the host cell through at least three independent mechanisms. It promotes the degradation of double-stranded RNA-dependent protein kinase (PKR) (14, 19), it specifically suppresses the induction of type I interferons by forming a repressor complex on the beta interferon (IFN-β) promoter (28), and it induces a general transcriptional shutoff of the infected cell (2, 27).
The general inhibition of host transcription is a strategy shared by a number of RNA viruses to evade cellular antiviral responses (32). These viruses have evolved to target different components of the host transcriptional machinery, and RVFV has been shown to effect transcriptional shutoff by disrupting the assembly of the basal transcription factor TFIIH (27). In eukaryotic cells, the transcription of protein-coding genes requires RNA polymerase II (Pol II) and the six general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (36). The last factor to be recruited into the preinitiation complex (PIC) is TFIIH, which is required both for promoter melting (48) and phosphorylation of the C-terminal domain (CTD) of the largest Pol II subunit during the transition from initiation to elongation (31). TFIIH is a multisubunit complex composed of 10 different proteins which form two distinct subcomplexes linked by XPD (gene defective in xeroderma pigmentosum patient complementation group D): the core complex, which consists of the subunits p8 (12), p34, p44, p52, p62, and XPB, and the cyclin-activating kinase (CAK) complex, which consists of ménage-à-trois 1 (MAT1), cyclin H, and cdk7 (44). In addition to its essential role in Pol II-driven transcription, TFIIH is also required for rRNA synthesis by RNA polymerase I (18) and for transcription-coupled DNA repair (9).
Although the replication cycle of RVFV is entirely cytoplasmic, its NSs protein localizes to the nucleus (52), where it prevents the assembly of a functional TFIIH complex by sequestering its p44 subunit. Furthermore, it has been reported that RVFV infection reduces the nuclear abundance of the TFIIH subunits p62 and XPD (27), although the mechanism for this reduction remains uncharacterized. Possible explanations for this observation include differences in the nuclear import/export of these subunits or a reduction in the abundance of their mRNAs, which might occur if host transcription is suppressed by p44 sequestration. Recently, NSs was found to promote the degradation of PKR, and we hypothesized that NSs could also degrade the TFIIH subunit p62 in order to suppress host transcription. The p62 subunit of TFIIH is essential for its function (10, 15), is required for the recruitment of TFIIH to the PIC through TFIIE (35), and mediates the interaction of TFIIH with transcriptional transactivators, such as p53 (7), VP16 (8), and the thyroid hormone receptor (30). We now demonstrate that NSs expression does not decrease the abundance of p62 mRNA but decreases the abundance of p62 protein through a posttranslational mechanism.
MATERIALS AND METHODS
Cells and viruses.
293 and VeroE6 cells were maintained in Dulbecco's modified minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin-streptomycin (all from Invitrogen). BHK/T7-9 cells (22), which stably express T7 RNA polymerase, were grown in MEM-alpha supplemented with 10% FBS, 100 μg/ml penicillin-streptomycin (all from Invitrogen), and 600 μg/ml hygromycin (Cellgro). The RVFV vaccine candidate MP-12 (6) was amplified in VeroE6 cells, and infectivity was determined by plaque assay in the same cells.
Virus rescue.
A recombinant MP-12 virus carrying an in-frame insertion of a Strep-Tag II–FLAG tag C terminal from the NSs open reading frame (ORF) (rMP12-NSs-SF; see below) was recovered as previously described (20). A recombinant MP-12 virus carrying a deletion of amino acids 16 to 198 in its NSs gene (clone 13-type [C13type]) was also recovered as previously described (20). Briefly, subconfluent monolayers of BHK/T7-9 cells were cotransfected with a mixture of pProT7-S(+), pPro-T7-M(+), pPro-T7-L(+), pT7-IRES-vN, pCAGGS-vG, and pT7-IRES-vL using TransIT-LT1 (Mirus Bio Corporation) according to the manufacturer's instructions. The culture medium was replaced with fresh medium 24 h later. At 5 days posttransfection, the culture supernatants were collected, clarified, and then inoculated into fresh VeroE6 cells. The culture supernatant from VeroE6 cells was collected at 3 days postinfection, and the virus titer was determined by plaque assay.
Antibodies and reagents.
Anti-p62 (Q-19), anti-β-actin (I-19), antiubiquitin (P4D1), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; V-18), anti-lamin B (C-20), anti-RanBP1 (C-19), and horseradish peroxidase (HRP)-labeled anti-mouse, anti-goat, and anti-rabbit secondary antibodies were purchased from Santa Cruz. Anti-FLAG M2 (F3165) and anti-green fluorescent protein (GFP; G1544) antibodies were purchased from Sigma. The anti-RVFV mouse polyclonal antibody was a gift of R. B. Tesh at the University of Texas Medical Branch. Goat anti-mouse IgG (H+L, Alexa Fluor 488-coupled), goat anti-rabbit (H+L, Alexa Fluor 488-coupled), and donkey anti-goat (H+L, Alexa Fluor 594-coupled) antibodies were purchased from Invitrogen. Leptomycin B and chloroquine were purchased from Santa Cruz, and lactacystin from Enzo Life Sciences. Actinomycin D (ActD) and MG132 were purchased from Sigma. Cycloheximide (CHX) was purchased from Acros Organics.
Plasmids.
Plasmids pT7-IRES-vNSs (20) and pT7-IRES-GFP (54) were described previously. pProT7-S(+)-NSs-SF was generated as follows: a PCR fragment encoding an NSs ORF which contains 2 Strep-Tag II epitopes and a Flag tag at the C terminus was amplified by 2 sequential reverse transcription (RT)-PCRs with the primer pair HpavNSF (AGT TGT TAA CAT GGA TTA CTT TCC TGT GAT ATC TGT TGA TTT GCA G) and NS-SF-1R (GGT GGC TCC AGG AGC CGC CTC CGG AGC CGC CTC CGG AGC CGC CTC CCT TCT CGA ACT GAG GGT GGC TCC AAT CAA CCT CAA CAA ATC CAT C), followed by amplification with primer pair HpavNSF and SpeSF-2R (CGC TAC TAG TCT ACT TGT CAT CGT CGT CCT TGT AGT CCT CAC CGC TGG CGC CCT TCT CGA ACT GAG GGT GGC TCC AGG AGC CGC CT). The PCR fragment was cloned between HpaI and SpeI of pProT7-S(+) (20).
RNA synthesis and transfection.
Capped and polyadenylated RNA transcripts were synthesized in vitro from pT7-IRES-NSs and pT7-IRES-GFP by using mMESSAGE mMACHINE T7 ultra (Ambion) according to the manufacturer's instructions. One microgram of in vitro-synthesized RNA was transfected into 293 cells in a 12-well plate using a TransIT mRNA transfection kit (Mirus Bio Corporation) according to the manufacturer's instructions.
Quantitative real-time PCR.
293 cells were infected with MP-12 at a multiplicity of infection (MOI) of 3, mock infected, or treated with 5 μg/ml ActD. After 8 h, cells were harvested with a rubber policeman and washed once in phosphate-buffered saline (PBS), and total RNA was purified from 6 × 105 cells using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. First-strand cDNA synthesis was performed from 1 μg RNA using a high capacity cDNA reverse transcription kit (Applied Biosciences). Real-time PCR was performed on a Mastercycler ep realplex2 (Eppendorf) using a QuantiFast SYBR green PCR kit and QuantiTect primer assays QT01681463 (p62) and QT01192646 (GAPDH) (all from Qiagen) according to the manufacturer's instructions.
Western blot analysis.
Western blot analysis was performed as previously described (19). Briefly, samples were normalized by cell number and disrupted by boiling in sample buffer for 10 min, followed by SDS-PAGE. Proteins were then transferred onto Immobilon P polyvinylidene fluoride membranes (Millipore) and probed with antibodies as indicated.
Fractionation of nuclear and cytoplasmic proteins.
Cells were infected with MP-12 at an MOI of 3 and treated with leptomycin B (LMB) at 0 h postinfection (h.p.i.) and MG132 at 3 h.p.i. as indicated below. At 8 h.p.i., cells were harvested by scraping with a rubber policeman, washed once in PBS, and resuspended in hypotonic lysis buffer containing 10 mM Tris, pH 7.8, 10 mM KCl, 1.5 mM MgCl2, 0.5% Triton X-100, 10 μM MG132, and 1× Complete protease inhibitor cocktail (Roche). Cells were incubated on ice for 10 min, followed by centrifugation for 2 min at 2,000 × g to sediment the nuclei. The supernatants were then transferred to a new tube, and the nuclei were washed once with lysis buffer containing no detergent. The nuclei were then disrupted by boiling in SDS-PAGE sample buffer. Both the cytoplasmic and nuclear fractions were adjusted to the same volume.
Click chemistry and immunofluorescence.
Labeling of nascent RNA was performed as previously described (23). Briefly, cells grown on 12-mm coverslips were incubated in medium containing 1 mM 5-ethynyluridine (EU; Berry & Associates) for 1 h prior to fixation in 4% paraformaldehyde for 30 min. Cells were then permeabilized in 0.2% Triton X-100 in PBS for 10 min, and nascent RNA with incorporated EU was detected by click chemistry using 20 μM Alexa Fluor 594-coupled azide (Invitrogen) in 100 mM Tris, pH 8.5, containing 1 mM CuSO4 and 100 mM ascorbic acid. For immunofluorescence, unspecific binding was blocked using 5% bovine serum albumin (BSA) in PBS for 30 min. The incubations with primary and Alexa Fluor-labeled secondary antibodies in blocking buffer were each carried out for 1 h. All steps were done at room temperature, and cells were washed with PBS in between steps. Cells were mounted on coverslips using Fluoromount-G (Southern Biotech), and images were acquired on an Olympus IX71 fluorescence microscope using a 20× LCPlanFl (numeric aperture, 0.4) objective.
FACS analysis.
For fluorescence-activated cell sorting (FACS) analysis, 293 cells were mock infected or infected with MP-12 at an MOI of 3 and treated with 0.5 mM EU (Berry & Associates) for 1 h prior to harvesting. Some samples were treated with 5 μg/ml ActD concurrently with the EU treatment to suppress cellular RNA synthesis. After harvesting, cells were fixed in 4% paraformaldehyde for 30 min and permeabilized in 0.2% Triton X-100 in PBS for 10 min, and nascent RNA with incorporated 5-ethynyluridine was detected by click chemistry using 200 nM Alexa Fluor 647-azide (Invitrogen) in 100 mM Tris, pH 8.5, containing 1 mM CuSO4 and 100 mM ascorbic acid. Cells were then stained with a mouse polyclonal antibody against RVFV, followed by an Alexa Fluor 488-labeled secondary antibody, to detect the expression of viral proteins. Cells were then analyzed by flow cytometry on an LSRII Fortessa instrument (Becton Dickinson).
Coaffinity precipitation.
Cells were infected with rMP-12-NSs-SF at an MOI of 3 and harvested at 5 h postinfection. Lysis was performed in 20 mM Tris, pH 7.6, 0.1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1× Complete protease inhibitors (Roche), and 100 U/ml Benzonase using 3 cycles of freeze-thaw. Cleared lysates (15 min at 13,000 rpm and 4°C) were incubated with 50 μl Strep-Tactin magnetic beads (Qiagen) for 2 h at 4°C in an end-over-end rotor. Precipitates were washed 3 times in lysis buffer, boiled in SDS-PAGE sample buffer, and analyzed by Western blotting.
RESULTS
RVFV NSs downregulates p62.
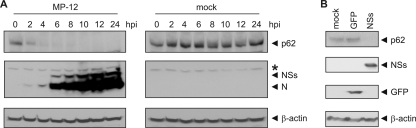
The NSs protein of RVFV has previously been shown to induce the degradation of PKR (14, 19). Furthermore, it has been reported that the nuclear abundance of TFIIH subunit p62 was decreased in cells infected with the virulent ZH548 strain of RVFV (27). We studied this phenomenon further because it was possible that NSs could promote posttranslational downregulation of two different substrates, i.e., PKR and TFIIH p62. To determine whether RVFV infection was able to affect the total cellular content of p62, we infected 293 cells with the MP-12 candidate vaccine strain (6) at an MOI of 3 and analyzed the abundance of p62 in whole-cell lysates by Western blotting. The rationale for the use of MP-12 instead of wild-type RVFV is as follows: (i) MP-12 is avirulent to humans, a non-select agent and risk group 2 pathogen, which is safer than wild-type RVFV, and (ii) although it is attenuated through mutations in its M and L segments, its S segment remains unattenuated and the NSs protein is fully functional (1). The results in Fig. 1A show that in cells infected with MP-12, the abundance of p62 was quickly reduced to below detection levels early in the course of infection. No p62 could be detected at 8 h.p.i. or later, and this absence persisted until 24 h.p.i., when cells started to display signs of cytopathic effect. To determine whether the NSs protein was responsible for this downregulation, we transfected 293 cells with in vitro-synthesized RNA for NSs. To exclude the possibility that the observed p62 downregulation was an unspecific effect of the transfection of T7 polymerase-synthesized RNA, we also transfected cells with RNA for GFP as a negative control. In cells transfected with NSs RNA, p62 was undetectable at 8 h posttransfection, whereas the p62 levels were unaffected in cells transfected with RNA for GFP (Fig. 1B). The results suggest that NSs is responsible for the downregulation of p62.
Fig. 1.
NSs downregulates p62 in whole-cell lysates. (A) 293 cells were mock infected or infected with MP-12 at an MOI of 3 and harvested at the indicated time points postinfection. Whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, and anti-β-actin antibodies (from top to bottom). The asterisk denotes a nonspecific band. (B) 293 cells were transfected with in vitro-synthesized RNA for GFP or NSs and harvested 8 h posttransfection. Whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, anti-GFP, and anti-β-actin antibodies (from top to bottom). Representative examples of at least 3 independent experiments are shown.
NSs suppresses host transcription.
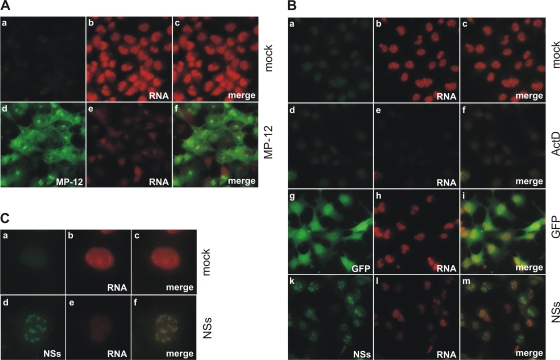
It has been shown that infection with wild-type RVFV resulted in a reduction of cellular RNA synthesis, and the interaction of NSs with TFIIH p44 was suggested to be responsible for this transcriptional suppression (27). To confirm that MP-12 NSs was indeed able to inhibit host transcription, we analyzed the ongoing RNA synthesis in cells infected with MP-12 or transfected with NSs RNA. To determine whether MP-12 infection was able to induce transcriptional suppression in the host cell, 293 cells were infected at an MOI of 3 and treated with the uridine analog 5-ethynyluridine (EU) directly before fixation. Newly synthesized RNA was visualized by detecting the incorporated EU via click chemistry with an Alexa Fluor-coupled azide. Subsequently, we performed an immunostaining with anti-RVFV antiserum to visualize viral proteins. MP-12-infected cells displayed a markedly reduced amount of newly synthesized RNA compared to the levels in mock-infected cells (Fig. 2A). We also transfected 293 cells with in vitro-synthesized RNA for NSs to ensure that the transcriptional suppression observed in MP-12-infected cells was indeed mediated by NSs. Cells transfected with NSs RNA, which expresses NSs proteins that can be detected by anti-RVFV antibodies, showed marked suppression of EU incorporation compared to the levels of EU incorporation in mock-transfected cells (Fig. 2B, k to m), whereas cells transfected with GFP RNA still actively incorporated EU (Fig. 2B, g to i). As a positive control for transcriptional suppression, cells were treated with 5 μg/ml actinomycin D (ActD) concurrently with the EU treatment (Fig. 2B, d to f). The suppression of EU incorporation by treatment with ActD ensured that the incorporated EU represents the results of ongoing cellular transcription (23). TFIIH is a required cofactor for both Pol II and Pol I, which is responsible for ribosomal DNA transcription that takes place in the nucleolus (56). Figure 2C shows magnified images of mock- and NSs-transfected cells (Fig. 2B, a to c and k to m, respectively) that demonstrate that NSs expression is able to silence transcription equally well in the nucleoplasm (where Pol II-mediated transcription occurs) and the nucleoli.
Fig. 2.
NSs induces host transcriptional suppression. (A) 293 cells were mock infected (a to c) or infected with MP-12 (d to f) at an MOI of 3. To label newly synthesized RNA, cells were treated with 1 mM ethynyluridine (EU) from 15 to 16 h.p.i., followed by fixation and permeabilization. Labeled RNA was subsequently detected by click chemistry with an Alexa Fluor 594-coupled azide. The expression of viral proteins was visualized by immunostaining with a polyclonal anti-RVFV antiserum followed by an Alexa Fluor 488-labeled secondary antibody. (B) 293 cells were mock transfected (a to f) or transfected with in vitro-synthesized RNA for GFP (g to i) or NSs (k to m) and treated with 1 mM EU for 1 h before fixation at 16 h posttransfection. Cells shown in panels d to f were treated with 5 μg/ml actinomycin D (ActD) for 1 h concurrently with the EU treatment. Labeled RNA was subsequently detected by click chemistry with an Alexa Fluor 594-coupled azide. The expression of GFP and NSs was visualized by immunostaining with anti-GFP (g and i) or anti-RVFV (k and m) antibodies followed by an Alexa Fluor 488-labeled secondary antibody. (C) Magnification of a mock-transfected cell (a to c) and an NSs-transfected cell (k to m) from the experiment whose results are shown in panel B to visualize RNA synthesis in the nucleoli. Representative examples of at least 3 independent experiments are shown.
For a more quantitative evaluation of MP-12-mediated transcriptional suppression, we analyzed the amount of nascent RNA in MP-12-infected or mock-infected cells by flow cytometry (Fig. 3A). ActD treatment again served as our control for efficient transcriptional suppression. Nascent RNA was labeled with EU for 1 h at various time points postinfection, and cells were harvested and stained immediately following the labeling. Transcriptional suppression could be observed as early as 4 h.p.i. and progressed continuously with increasing times of viral infection. Infection with MP-12 for 24 h reduced transcriptional activity as potently as pharmacological inhibition with ActD. To better visualize the progression of transcriptional suppression during the course of infection, Fig. 3B displays the RNA fluorescence intensities of MP-12-infected cells. Cells were gated on anti-RVFV reactivity for this graph. Figure 3C displays the RNA fluorescence intensities of mock-infected and ActD-treated cells. The gate was placed on anti-RVFV-negative cells for this graph.
Fig. 3.
NSs induces host transcriptional suppression. (A) 293 cells were mock infected (mock) or infected with MP-12 at an MOI of 3 and treated with 0.5 mM EU for 1 h at the indicated time points of infection. As a control for transcriptional suppression, cells were treated with 5 μg/ml ActD concurrently with the EU labeling (ActD). Cells were harvested, fixed, and permeabilized immediately after the labeling step. EU-labeled RNA was then detected by click chemistry with an Alexa Fluor 647-coupled azide. The expression of viral proteins was visualized by immunostaining with a polyclonal anti-RVFV antiserum followed by an Alexa Fluor 488-labeled secondary antibody. Cells were then analyzed by flow cytometry. (B) To better demonstrate the progressive loss of transcriptional activity in MP-12-infected cells, data from the experiment whose results are shown in panel A were gated on anti-RVFV-positive cells, and the fluorescence of labeled RNA is displayed as a histogram. (C) Data from the experiment whose results are shown in panel A were gated on anti-RVFV-negative cells, and the fluorescence of labeled RNA in mock-infected and ActD-treated cells is displayed as a histogram. (D) 293 cells were mock infected or infected with either MP-12 or an MP-12 mutant carrying a large deletion in its NSs gene that renders the NSs protein nonfunctional (C13type). Cells were harvested at 12 h.p.i., and whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, and anti-β-actin antibodies (from top to bottom). (E) 293 cells were mock infected (mock) or infected with either MP-12 or C13type at an MOI of 3 and were treated with 0.5 mM EU from 12 to 13 h.p.i. Cells were harvested, fixed, and permeabilized, and EU-labeled RNA was detected by click chemistry with an Alexa Fluor 647-coupled azide. The expression of viral proteins was visualized by immunostaining with a polyclonal anti-RVFV antiserum followed by an Alexa Fluor 488-labeled secondary antibody. Cells were then analyzed by flow cytometry.
To confirm that NSs was required for both transcriptional suppression and p62 degradation, we infected 293 cells with a recombinant MP-12 virus (C13type) carrying a nonfunctional NSs gene (20). Clone 13 is a naturally occurring attenuated isolate of RVFV which carries a large in-frame deletion (amino acids 16 to 198, 69% of the gene) in its NSs gene. The resulting truncated NSs protein is nonfunctional. C13type was unable to downregulate p62 (Fig. 3D) and did not suppress transcription (Fig. 3E), even though it was able to replicate well, as evidenced by the large amount of N protein present in C13type-infected cells (Fig. 3D). The results demonstrate that MP-12 NSs indeed inhibits host RNA synthesis.
p62 downregulation is posttranslational.
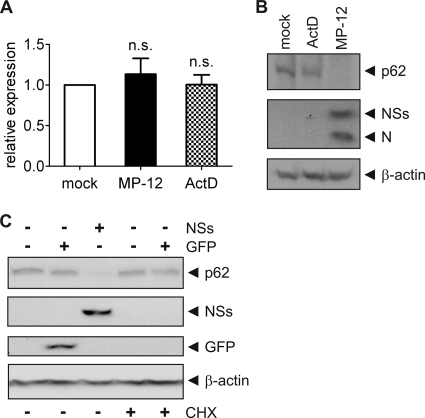
Our results show that p62 is downregulated by 8 h after MP-12 infection or NSs RNA transfection. In order to determine whether this downregulation of p62 by NSs was an indirect effect of the suppression of host mRNA synthesis or increased instability of p62 mRNA caused by MP-12 infection, we analyzed the relative amounts of p62 mRNA in mock- or MP-12-infected cells by quantitative real-time PCR using GAPDH as a reference gene. We observed no significant difference between the p62 mRNA levels in infected and mock-infected cells (Fig. 4A). To confirm that the observed reduction in p62 levels was not caused by the general transcription suppression, cells were treated with 5 μg/ml of ActD for 9 h, while control cells were either left untreated or were infected with MP-12 at an MOI of 3 for 8 h. Downregulation of p62 was induced in cells infected with MP-12, whereas in cells treated with ActD, the abundance of p62 was only marginally reduced compared to the levels in mock-treated cells (Fig. 4B). Interestingly, ActD treatment for 9 h had no effect on the relative levels of expression of p62, suggesting that the turnover of p62 mRNA can be longer than 9 h. Because NSs induced a rapid downregulation of the p62 protein (Fig. 1), we suspected that the reduction of p62 could occur during or after the translation of p62.
Fig. 4.
Shutoff of host transcription or translation does not induce p62 downregulation. (A) 293 cells were mock infected or infected with MP-12 at an MOI of 3 or treated with 5 μg/ml of actinomycin D (ActD) for 9 h and harvested at 8 h.p.i. p62 mRNA expression levels were analyzed by RT-PCR using relative quantification normalized to the level of GAPDH. Error bars represent the standard deviations (SD) of the results of three independent experiments. Statistical analysis was carried out using Student's t test (n.s., not significant). (B) 293 cells were mock treated, treated with 5 μg/ml ActD for 9 h, or infected with MP-12 at an MOI of 3 for 8 h. Whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, and anti-β-actin antibodies (from top to bottom). (C) 293 cells were mock transfected or transfected with in vitro-synthesized RNA for GFP or NSs. Immediately after transfection, cells were treated with 100 μg/ml cycloheximide (CHX) where indicated. Cells were harvested at 8 h posttransfection, and whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, anti-GFP, and anti-β-actin antibodies (from top to bottom). Representative examples of at least 3 independent experiments are shown.
RVFV infection has been shown to have a suppressive effect on host translation (19). Therefore, we tested whether downregulation of p62 could be induced by the suppression of host translation by MP-12 infection. We treated 293 cells with 100 μg/ml cycloheximide (CHX) for 8 h to inhibit translation. The pharmacological inhibition of protein synthesis in mock- or GFP RNA-transfected cells had no significant effect on p62 levels, whereas p62 was undetectable in cells transfected with NSs RNA. The efficacy of our CHX treatment was confirmed by the lack of GFP expression after GFP RNA transfection and CHX treatment (Fig. 4C). The results suggest that neither host transcription suppression nor translation suppression could induce p62 downregulation within 8 h.
The proteasome is involved in p62 downregulation.
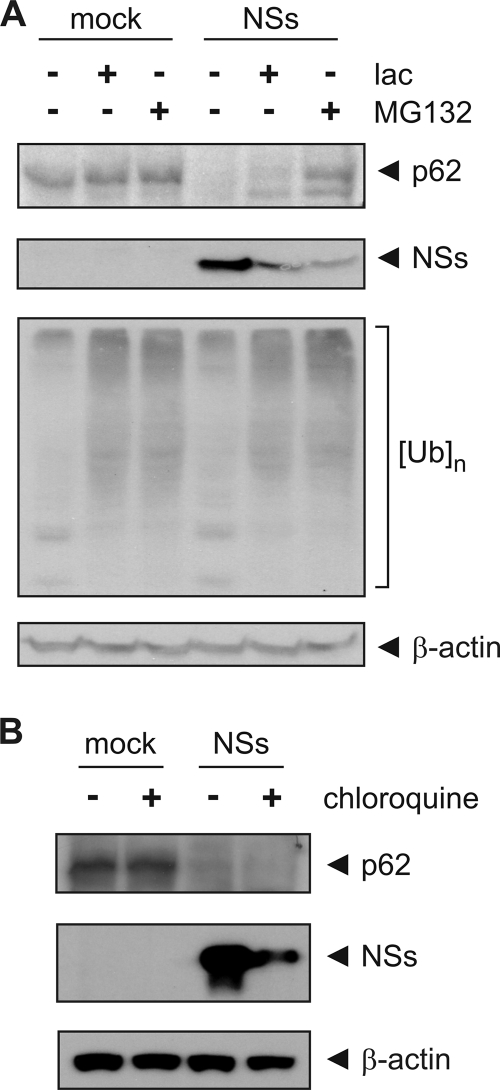
Since NSs displays no sequence similarities with known proteases (data not shown), we hypothesized that it promotes the downregulation of p62 by targeting it for either proteasomal or lysosomal degradation. Since the proteasome has been implicated in the degradation of PKR (14, 19), we treated cells with the proteasome inhibitors MG132 or lactacystin. Thirty minutes after starting the inhibitor treatment, cells were transfected with in vitro-synthesized RNA for NSs, and they were harvested 8 h posttransfection. The abundance of p62, NSs, and ubiquitin in whole-cell lysates was then determined by Western blotting. The accumulation of high-molecular-weight ubiquitin conjugates was used as a marker for effective proteasomal inhibition. Treatment with the peptide aldehyde inhibitor MG132, which displays some unspecific inhibitory activity against lysosomal cathepsins (26), was able to stabilize p62 in the presence of NSs, whereas treatment with the more specific proteasome inhibitor lactacystin stabilized p62 less efficiently than MG132 (Fig. 5A). We observed an additional band with anti-p62 reactivity but a lower molecular weight than full-length p62 in cells expressing NSs only when treated with lactacystin or MG132. This band was also seen in mock-transfected cells, yet its abundance was lower than in NSs-transfected cells. We also observed that treatment with MG132 or lactacystin decreased the amount of NSs protein (Fig. 5A). The reason why a large portion of p62 was still downregulated by NSs in the presence of lactacystin remains unknown. We suspected that NSs could alternatively promote degradation of p62 through the lysosomal pathway when proteasomal degradation was suppressed by lactacystin. Since downregulation of p62 still occurred, albeit to a lesser extent, in lactacystin-treated cells, we treated cells with 100 μM chloroquine to inhibit lysosomal degradation and transfected them with RNA for NSs 30 min later. Treatment with chloroquine also reduced NSs protein levels but was unable to stabilize p62 protein levels in the presence of NSs (Fig. 5B). These results indicate that NSs does not mediate degradation of p62 through the lysosomal pathway and suggest involvement of the proteasome.
Fig. 5.
The proteasome is involved in p62 downregulation. (A) 293 cells were pretreated with 50 μM lactacystin (lac) or 5 μM MG132 for 30 min to inhibit proteasomal degradation before being mock transfected or transfected with in vitro-synthesized RNA for NSs. Cells were harvested at 8 h posttransfection, and whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, anti-ubiquitin, and anti-β-actin antibodies (from top to bottom). [Ub]n denotes high-molecular-weight polyubiquitin conjugates. (B) 293 cells were mock transfected or transfected with in vitro-synthesized RNA for NSs. Immediately following transfection, cells were treated with 100 μM chloroquine to inhibit lysosomal degradation. Cells were harvested at 8 h posttransfection, and whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, and anti-β-actin antibodies (from top to bottom).
NSs interacts with p62 in infected cells.
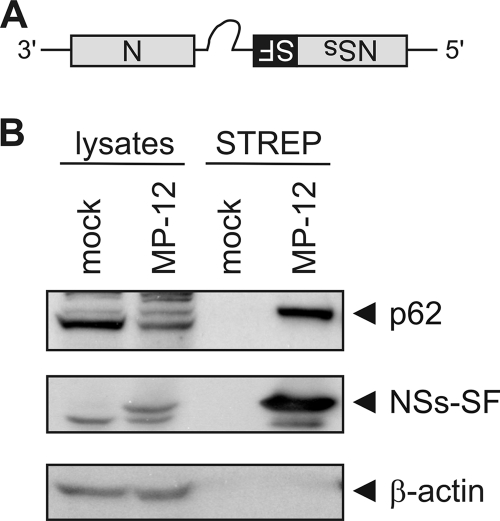
RVFV NSs is known to interact with the TFIIH p44 subunit in its free form but not when it is incorporated into the assembled TFIIH complex (27). The p44 subunit is part of the TFIIH ring structure, and its binding partners are p34, XPB, and XPD (44). We suspected that NSs forms a complex with p62 either directly or indirectly through p44 to target p62 for degradation. To determine whether NSs was able to interact with p62, we investigated whether both proteins could be coprecipitated from MP-12-infected cells. To aid in the detection and precipitation of NSs, we created a recombinant virus (rMP-12-NSs-SF) carrying a tandem Strep-Tag II–FLAG tag at the C terminus of the NSs gene (Fig. 6A). The recovered virus replicated to the same titers and with the same kinetics as MP-12 and induced the degradation of p62 (data not shown). Cells were infected with rMP-12-NSs-SF at an MOI of 3 and harvested at 5 h.p.i. We selected this time point to ensure that viral replication had produced sufficient levels of NSs inside infected cells but p62 had not been completely degraded. NSs was affinity precipitated from cleared lysates with a Strep-Tactin matrix, and the precipitated proteins were analyzed by Western blotting. Even though p62 was present in infected cells in considerably smaller amounts than in the uninfected controls, p62 could only be efficiently coprecipitated with the Strep-Tactin matrix in infected cells (Fig. 6B). These results suggest that NSs forms a complex with p62.
Fig. 6.
NSs interacts with p62. (A) Schematic representation of the S segment of the recombinant virus (rMP-12-NSs-SF) used in the experiments whose results are shown in panel B. SF, Strep-Tag II–FLAG tag. (B) 293 cells were mock infected or infected with rMP12-NSs-SF at an MOI of 3. At 5 h.p.i., NSs-SF was affinity precipitated with Strep-Tactin magnetic beads. Whole-cell lysates and the precipitated proteins (STREP) were analyzed by Western blotting with anti-p62, anti-FLAG, and anti-β-actin antibodies (from top to bottom). Representative examples of at least 3 independent experiments are shown.
p62 degradation can occur directly in the nucleus.
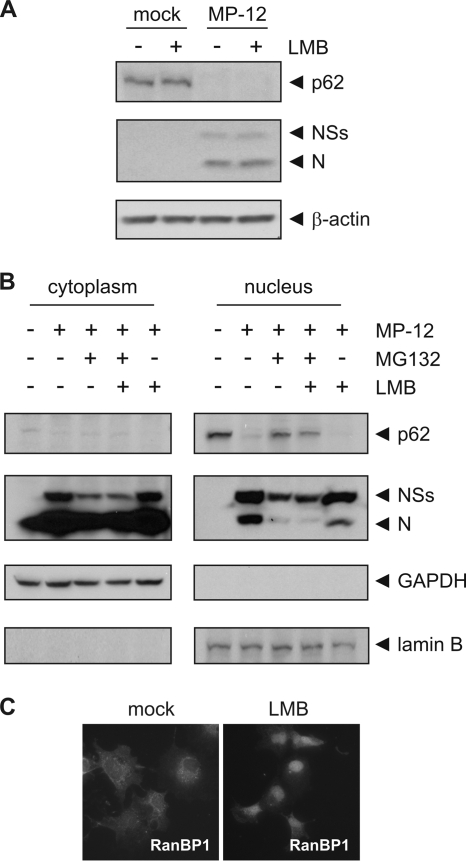
NSs is distributed throughout the cell during RVFV infection (52); however, its ability to suppress transcription has been suggested to require nuclear localization and the formation of filamentous structures inside the nucleus (2). The localization of p62 is predominantly nuclear (24, 42), and it is thought that, immediately after translation, it is translocated into the nucleus, where it assembles with the other subunits of TFIIH to form the functional holocomplex (43). Le May et al. suggested that RVFV NSs sequesters p44 subunits before the assembly of TFIIH but does not interact with p44 when already associated with XPD within TFIIH (27). Our results suggest that NSs can posttranslationally downregulate p62 relatively early after infection (2 to 6 h.p.i.) (Fig. 1A). Thus, we hypothesized that NSs could degrade p62 in fully assembled TFIIH complexes, which are only present in the nucleus (43). In order to determine whether the NSs-mediated degradation of p62 could take place directly inside the nucleus, we treated MP-12-infected cells (MOI of 3) with leptomycin B (LMB) over the entire course of infection to inhibit nuclear export (33, 50). We hypothesized that if p62 needed to be transported back into the cytoplasm to be degraded, blocking the CRM1/exportin-dependent nuclear export of proteins with LMB would stabilize the p62 levels inside infected cells. On the other hand, if p62 could be degraded directly inside the nucleus, blocking nuclear export would have no effect on its degradation. The results shown in Fig. 7A demonstrate that we were unable to stabilize p62 levels by treating RVFV-infected cells with LMB, whereas nuclear export was clearly inhibited, as shown by the nuclear accumulation of RanBP1 in LMB-treated cells (Fig. 7C). Since LMB only inhibits the CRM1/exportin-dependent nuclear export of proteins (33, 50), it is possible that p62 could be exported through an as-yet-uncharacterized pathway and subsequently degraded by cytoplasmic proteasomes. We therefore fractionated mock-infected or MP-12-infected cells and observed the stabilization of p62 by MG132 treatment in both the cytoplasmic and nuclear fractions. We reasoned that if p62 was exported out of the nucleus prior to its degradation, it would accumulate in the cytoplasm after MG132 treatment. On the other hand, if NSs was able to promote the degradation of p62 directly inside the nucleus, p62 should remain nuclear after MG132 treatment. The results in Fig. 7B demonstrate that p62, which is localized almost exclusively in the nucleus in uninfected cells, is degraded by MP-12 infection and that it still remains in the nucleus after MG132 treatment. Again, treatment with LMB had no effect on the localization or degradation of p62. Our results suggest that NSs can promote the degradation of p62 directly inside the nucleus.
Fig. 7.
p62 is degraded in the nucleus of infected cells. (A) 293 cells were mock infected or infected with MP-12 at an MOI of 3. Immediately after infection, cells were treated with 20 nM leptomycin B (LMB) where indicated. Cells were harvested at 8 h.p.i., and whole-cell lysates were analyzed by Western blotting with anti-p62, anti-RVFV, and anti-β-actin antibodies (from top to bottom). (B) 293 cells were mock infected or infected with MP-12 at an MOI of 3 and treated with 5 μM MG132 at 3 h.p.i. or 20 nM LMB at 0 h.p.i. where indicated. Cells were harvested at 8 h.p.i. and fractioned into cytoplasmic and nuclear compartments, followed by analysis by Western blotting with anti-p62, anti-RVFV, anti-GAPDH, and anti-lamin B antibodies (from top to bottom). (C) 293 cells were mock treated or treated with 20 nM LMB for 7 h before fixation and permeabilization. RanBP1 localization was visualized by immunostaining. Representative examples of at least 3 independent experiments are shown.
DISCUSSION
Our results presented here imply that RVFV NSs promotes the degradation of two different substrates, PKR and p62, in order to ensure efficient viral translation and the suppression of the host antiviral response, respectively. Le May et al. (27) have shown that NSs is able to interfere with TFIIH assembly by sequestering its p44 subunit. Prompted by the observation that nuclear extracts of RVFV-infected cells contain reduced amounts of the TFIIH subunit p62 in comparison to the amounts in uninfected cells (27) and by the recent discovery that NSs is able to promote the proteasomal degradation of PKR (14, 19), we identified p62 as a second substrate of NSs-mediated degradation.
Infection of cells with the RVFV MP-12 vaccine strain resulted in a complete reduction of p62 protein levels at 6 h.p.i. and beyond. Transfection of cells with in vitro-synthesized RNA for NSs was equally able to downregulate p62, indicating that this effect was specific to NSs and not a general consequence of RVFV infection. NSs did not affect p62 transcription or alter the mRNA stability of p62, as p62 mRNA levels remained unchanged at 8 h.p.i. in MP-12-infected cells. This is further corroborated by the finding that pharmacological inhibition of transcription by treatment of cells with actinomycin D for 8 h only marginally decreased p62 levels. This finding also suggests that the transcriptional suppression observed in RVFV-infected cells is indeed a consequence and not the cause of p62 downregulation. To determine whether NSs downregulated p62 by inhibiting p62 protein neosynthesis or, rather, by turnover, we treated cells with cycloheximide for 8 h to inhibit translation. Pharmacological inhibition of protein neosynthesis for 8 h did not alter the p62 levels, indicating that NSs promoted p62 downregulation via a posttranslational mechanism.
Because the selective degradation of most cellular proteins is carried out by the ubiquitin-proteasome system (17), we investigated whether inhibitors of proteasomal (MG132 and lactacystin) or lysosomal (chloroquine) degradation were able to stabilize p62 in the presence of NSs. Interestingly, MG132 was able to inhibit p62 degradation, while treatment with the more specific proteasomal inhibitor lactacystin only resulted in a partial stabilization of p62. Conversely, treatment of cells with chloroquine had no effect on the degradation of p62. Treatment of cells with either a proteasomal or lysosomal inhibitor greatly reduced the amount of NSs transcribed from transfected RNA. We believe this might be caused by activation of the unfolded protein response (UPR) by pharmacological inhibition of protein degradation (34). Activation of the UPR in turn results in translational arrest (55) and, ultimately, in reduced NSs levels in comparison to the levels in untreated cells. Since treatment of cells with proteasomal inhibitors stabilizes p62 and reduces NSs levels at the same time, it is unclear whether this stabilization of p62 is mediated directly by inhibition of the proteasome rather than being due to the loss of NSs. However, as the amount of NSs is equally reduced in chloroquine-treated cells, although this inhibitor does not stabilize p62 levels, these results suggest involvement of the proteasome rather than the lysosome in the degradation of p62.
Recent findings demonstrate extensive cross talk between the pathways of lysosomal and proteasomal degradation; most notably, proteasomal substrates have been shown to be targeted to the lysosome via selective autophagy as a compensatory mechanism during proteasomal inhibition (25). We therefore speculate that by treating cells with MG132, which also has partial activity against lysosomal proteases (26), this compensatory mechanism is also affected, which ultimately results in a better stabilization of p62 than can be achieved by inhibition of the proteasome alone. Unfortunately, concurrent treatment of cells with both lactacystin and chloroquine resulted in massive cell death (unpublished results), so the effect of MG132 could not be recapitulated by combining these two inhibitors.
Both NSs (52) and the proteasome (37, 41) are present in cytoplasmic as well as nuclear compartments, and the nuclear localization of NSs has previously been shown to be required for efficient transcriptional suppression (2). The localization of p62, on the other hand, is predominantly nuclear, and pharmacological inhibition of nuclear export has no effect on the NSs-mediated degradation of p62. Furthermore, after MP-12 infection and in the presence of proteasomal inhibitors, p62 remained localized in the nucleus. This suggests that p62 does not need to be exported into the cytoplasm to be degraded but, rather, is degraded directly inside the nucleus.
Proteins are canonically targeted for proteasomal degradation by covalent attachment of a chain of ubiquitin moieties (17), and it will be important to determine whether NSs is able to promote the polyubiquitination of p62. Interestingly, two distinct ubiquitin ligases (NEDD4 and the elongin/Rbx1/Cul5 complex) have recently been identified as being responsible for the polyubiquitination of the large subunit of RNA polymerase II (16). The elongin/Rbx1/Cul5 complex is also recruited by the adenoviral E4orf6 protein (40) and HIV-1 Vif (53). Furthermore, although it displays no sequence similarity to the RVFV NSs protein, the NSs protein of Bunyamwera virus has been shown to interact with MED8 (29), which also serves as a substrate recognition factor of the elongin/Rbx1/Cul2 E3 complex (5). We hypothesize that NSs functions by recruiting a cellular ubiquitin ligase to promote the ubiquitination and subsequent degradation of p62, and it will be important to characterize the complex containing NSs and p62 for a more detailed understanding of NSs-mediated p62 degradation.
The question remains as to whether and, if so, why RVFV employs two redundant mechanisms to induce transcriptional suppression in its host cell, i.e., (i) sequestration of p44 by NSs and (ii) NSs-mediated p62 degradation. Le May et al. suggested that the sequestration of p44 can only prevent the assembly of new TFIIH complexes and leaves preexisting ones unaffected (27). Furthermore, NSs only interacts with free p44 and not with p44 which has been assembled into the TFIIH complex (27). If NSs acted only through the sequestration of p44, the onset of transcriptional suppression would be delayed until existing TFIIH had been turned over naturally. By promoting the degradation of p62, on the other hand, NSs might be able to disrupt and inactivate fully assembled complexes. Indeed, partial transcriptional suppression can be observed as early as 4 h.p.i. (Fig. 3A). Furthermore, substoichiometric amounts of NSs would be sufficient to completely inactivate all existing and newly synthesized TFIIH, which might be important since the synthesis of large amounts of NSs occurs only after secondary transcription of the ambisense S segment and relatively late during viral replication (21).
ACKNOWLEDGMENTS
We gratefully acknowledge R. B. Tesh for providing the anti-RVFV antiserum.
This work was supported by NIH grant 5U54AI057156-07 from NIAID through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases, by funding from the Sealy Center for Vaccine Development at the University of Texas Medical Branch (UTMB), and by the James W. McLaughlin Fellowship fund.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Billecocq A., et al. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Billecocq A., et al. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bird B. H., Ksiazek T. G., Nichol S. T., Maclachlan N. J. 2009. Rift Valley fever virus. J. Am. Vet. Med. Assoc. 234:883–893 [DOI] [PubMed] [Google Scholar]
- 4. Bouloy M., et al. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brower C. S., et al. 2002. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 99:10353–10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caplen H., Peters C. J., Bishop D. H. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J. Gen. Virol. 66:2271–2277 [DOI] [PubMed] [Google Scholar]
- 7. Di Lello P., et al. 2006. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 22:731–740 [DOI] [PubMed] [Google Scholar]
- 8. Di Lello P., et al. 2005. NMR structure of the amino-terminal domain from the Tfb1 subunit of TFIIH and characterization of its phosphoinositide and VP16 binding sites. Biochemistry 44:7678–7686 [DOI] [PubMed] [Google Scholar]
- 9. Drapkin R., et al. 1994. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368:769–772 [DOI] [PubMed] [Google Scholar]
- 10. Drapkin R., Reinberg D. 1994. The multifunctional TFIIH complex and transcriptional control. Trends Biochem. Sci. 19:504–508 [DOI] [PubMed] [Google Scholar]
- 11. Gerrard S. R., Bird B. H., Albariño C. G., Nichol S. T. 2007. The NSm proteins of Rift Valley fever virus are dispensable for maturation, replication and infection. Virology 359:459–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giglia-Mari G., et al. 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36:714–719 [DOI] [PubMed] [Google Scholar]
- 13. Giorgi C., et al. 1991. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology 180:738–753 [DOI] [PubMed] [Google Scholar]
- 14. Habjan M., et al. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hampsey M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harreman M., et al. 2009. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 106:20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 18. Iben S., et al. 2002. TFIIH plays an essential role in RNA polymerase I transcription. Cell 109:297–306 [DOI] [PubMed] [Google Scholar]
- 19. Ikegami T., et al. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ikegami T., Won S., Peters C. J., Makino S. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J. Virol. 80:2933–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ikegami T., Won S., Peters C. J., Makino S. 2005. Rift Valley fever virus NSs mRNA is transcribed from an incoming anti-viral-sense S RNA segment. J. Virol. 79:12106–12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ito N., et al. 2003. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol. Immunol. 47:613–617 [DOI] [PubMed] [Google Scholar]
- 23. Jao C. Y., Salic A. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U. S. A. 105:15779–15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jordan P., Cunha C., Carmo-Fonseca M. 1997. The cdk7-cyclin H-MAT1 complex associated with TFIIH is localized in coiled bodies. Mol. Biol. Cell 8:1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korolchuk V. I., Menzies F. M., Rubinsztein D. C. 2010. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 584:1393–1398 [DOI] [PubMed] [Google Scholar]
- 26. Lee D. H., Goldberg A. L. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397–403 [DOI] [PubMed] [Google Scholar]
- 27. Le May N., et al. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550 [DOI] [PubMed] [Google Scholar]
- 28. Le May N., et al. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leonard V. H., Kohl A., Hart T. J., Elliott R. M. 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J. Virol. 80:9667–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y., et al. 2005. p62, a TFIIH subunit, directly interacts with thyroid hormone receptor and enhances T3-mediated transcription. Mol. Endocrinol. 19:879–884 [DOI] [PubMed] [Google Scholar]
- 31. Lu H., Zawel L., Fisher L., Egly J. M., Reinberg D. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641–645 [DOI] [PubMed] [Google Scholar]
- 32. Lyles D. S. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64:709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishi K., et al. 1994. Leptomycin B targets a regulatory cascade of Crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269:6320–6324 [PubMed] [Google Scholar]
- 34. Obeng E. A., et al. 2006. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107:4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okuda M., et al. 2008. Structural insight into the TFIIE-TFIIH interaction: TFIIE and p53 share the binding region on TFIIH. EMBO J. 27:1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Orphanides G., Lagrange T., Reinberg D. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657–2683 [DOI] [PubMed] [Google Scholar]
- 37. Palmer A., et al. 1996. Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem. J. 316:401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pepin M., Bouloy M., Bird B. H., Kemp A., Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 41:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters C. J., Meegan J. M. 1981. Viral zoonoses, p. 403–420 In Steele J. H., CRC handbook series in zoonoses, vol. 1 CRC Press, Boca Raton, FL [Google Scholar]
- 40. Querido E., et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reits E. A., Benham A. M., Plougastel B., Neefjes J., Trowsdale J. 1997. Dynamics of proteasome distribution in living cells. EMBO J. 16:6087–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Santagati F., Botta E., Stefanini M., Pedrini A. M. 2001. Different dynamics in nuclear entry of subunits of the repair/transcription factor TFIIH. Nucleic Acids Res. 29:1574–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Satoh M. S., Hanawalt P. C. 1997. Competent transcription initiation by RNA polymerase II in cell-free extracts from xeroderma pigmentosum groups B and D in an optimized RNA transcription assay. Biochim. Biophys. Acta 1354:241–251 [DOI] [PubMed] [Google Scholar]
- 44. Schultz P., et al. 2000. Molecular structure of human TFIIH. Cell 102:599–607 [DOI] [PubMed] [Google Scholar]
- 45. Struthers J. K., Swanepoel R., Shepherd S. P. 1984. Protein synthesis in Rift Valley fever virus-infected cells. Virology 134:118–124 [DOI] [PubMed] [Google Scholar]
- 46. Suzich J. A., Kakach L. T., Collett M. S. 1990. Expression strategy of a phlebovirus: biogenesis of proteins from the Rift Valley fever virus M segment. J. Virol. 64:1549–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Swanepoel R., Coetzer J. A. W. 2004. Rift Valley fever, p. 1037–1070 In Coetzer J. A. W., Thompson G. R., Tustin R. D., et al. (ed.), Infectious diseases of livestock with special reference to Southern Africa, 2nd ed. Oxford University Press, Cape Town, South Africa [Google Scholar]
- 48. Tirode F., Busso D., Coin F., Egly J. M. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3:87–95 [DOI] [PubMed] [Google Scholar]
- 49. Vialat P., Billecocq A., Kohl A., Bouloy M. 2000. The S segment of Rift Valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J. Virol. 74:1538–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolff B., Sanglier J. J., Wang Y. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139–147 [DOI] [PubMed] [Google Scholar]
- 51. Won S., Ikegami T., Peters C. J., Makino S. 2006. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J. Virol. 80:8274–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yadani F. Z., Kohl A., Prehaud C., Billecocq A., Bouloy M. 1999. The carboxy-terminal acidic domain of Rift Valley fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J. Virol. 73:5018–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu X., et al. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060 [DOI] [PubMed] [Google Scholar]
- 54. Zamoto-Niikura A., Terasaki K., Ikegami T., Peters C. J., Makino S. 2009. Rift Valley fever virus L protein forms a biologically active oligomer. J. Virol. 83:12779–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhao L., Ackerman S. L. 2006. Endoplasmic reticulum stress in health and disease. Curr. Opin. Cell Biol. 18:444–452 [DOI] [PubMed] [Google Scholar]
- 56. Zurita M., Merino C. 2003. The transcriptional complexity of the TFIIH complex. Trends Genet. 19:578–584 [DOI] [PubMed] [Google Scholar]