Abstract
The multifunctional helper component proteinase (HCpro) of potyviruses (genus Potyvirus; Potyviridae) shows self-interaction and interacts with other potyviral and host plant proteins. Host proteins that are pivotal to potyvirus infection include the eukaryotic translation initiation factor eIF4E and the isoform eIF(iso)4E, which interact with viral genome-linked protein (VPg). Here we show that HCpro of Potato virus A (PVA) interacts with both eIF4E and eIF(iso)4E, with interactions with eIF(iso)4E being stronger, as judged by the data of a yeast two-hybrid system assay. A bimolecular fluorescence complementation assay on leaves of Nicotiana benthamiana showed that HCpro from three potyviruses (PVA, Potato virus Y, and Tobacco etch virus) interacted with the eIF(iso)4E and eIF4E of tobacco (Nicotiana tabacum); interactions with eIF(iso)4E and eIF4E of potato (Solanum tuberosum) were weaker. In PVA-infected cells, interactions between HCpro and tobacco eIF(iso)4E were confined to round structures that colocalized with 6K2-induced vesicles. Point mutations introduced to a 4E binding motif identified in the C-terminal region of HCpro debilitated interactions of HCpro with translation initiation factors and were detrimental to the virulence of PVA in plants. The 4E binding motif conserved in HCpro of potyviruses and HCpro-initiation factor interactions suggest new roles for HCpro and/or translation factors in the potyvirus infection cycle.
INTRODUCTION
Picorna-like plant viruses of the genus Potyvirus (Potyviridae) produce polyprotein precursors processed into up to 11 mature proteins by three virus-encoded proteinases (17, 64). One of these virus-encoded proteinases is the multifunctional helper component proteinase (HCpro). For simplicity, HCpro can be considered to consist of N-proximal, central, and C-proximal domains, which correspond to ∼100, ∼200, and the remaining ∼150 amino acids (aa), respectively (62). The C-proximal domain confers proteolytic activity, which separates HCpro from the polyprotein at the HCpro C terminus (13). The N-proximal domain is structurally rather independent; for example, its deletion does not influence tetramerization of HCpro (62).
HCpro is involved in all essential steps of the potyviral infection cycle. The conserved KITC motif, which corresponds to aa 52 to 55 in the HCpro of Potato virus A (PVA) (65), and PTK motif (aa 310 to 312 in PVA) are needed for the aphid transmissibility of potyviruses (9, 10, 61). HCpro is an important contributor to viral genome amplification, as it suppresses RNA silencing that is the basal antiviral defense mechanism, in plants, induced by and targeting double-stranded RNA (3, 11, 38). RNA silencing also posttranscriptionally regulates gene expression via the microRNA (miRNA) pathway (47), and thus, suppression of RNA silencing interferes with gene regulation and causes disease symptoms. The nonspecific RNA binding activity of HCpro (50, 53) facilitates the binding of small interfering RNA (siRNA) and miRNA, which is suggested to be a mechanism for suppression of silencing by HCpro and requires the conserved FRNK box (aa 181 to 184 in PVA) in the central part of HCpro (81). However, all three domains of HCpro contain amino acids that, when mutated, affect RNA silencing (87). HCpro increases the size exclusion limit of plasmodesmata and facilitates viral cell-to-cell movement (71). In addition, the viral movement functions mediated by HCpro may be associated with the suppression of RNA silencing (39), because the conserved FRNK motif mentioned above and an IGN motif in the LAIGN box (PVA HCpro aa 248 to 252) in the central part of HCpro are involved in virus movement and amplification. Long-distance movement of Tobacco etch virus (TEV) also involves a conserved CCC motif (aa 292 to 294 in PVA) located in the central region of HCpro (19, 40).
Interactions of HCpro with host proteins and other viral proteins are probably essential for the viral functions mediated by HCpro. The HCpro of PVA interacts with the RING finger protein HIP1 and a putative microtubule-associated protein HIP2 in potato (Solanum tuberosum) and tobacco (Nicotiana tabacum) (32; T. Haikonen, unpublished data). HCpro of Lettuce mosaic virus (LMV) interferes and interacts with the 20S proteasome complex in cauliflower (Brassica oleracea) (6), whereas HCpro of Potato virus Y (PVY) interacts with several proteasome-subunits of Arabidopsis thaliana (35). The chloroplast division-related factor NtMinD of tobacco binds to HCpro of PVY (36), and a chloroplast precursor of ferredoxin-5 binds to HCpro of Sugar cane mosaic virus (SCMV) in maize (Zea mays) (15). A calmodulin-like cellular regulator of RNA silencing and the ethylene-inducible transcription factor RAV2 in tobacco (N. tabacum) interact with HCpro (2, 23).
HCpro forms dimers (30, 52, 88, 97), tetramers, and hexamers (62, 77) and interacts with the coat protein (CP) (10, 72), viral RNA helicase (CI) (16, 31), and the first protein (P1) of the potyviral polyprotein (52). It interacts also with the viral genome-linked protein (VPg) (31, 73, 97) and its precursor, nuclear inclusion protein a (NIa) (31), which, in turn, interact with the cap-binding translation initiation factor eIF4E and its isoform eIF(iso)4E (7, 45, 54, 96) that control important steps of the potyvirus infection cycle (reviewed in reference 70). Indeed, many host recessive genes that confer resistance to various potyviruses have been found to encode eIF4E or eIF(iso)4E, e.g., in A. thaliana (Brassicaceae) (22, 44, 80), Capsicum and Solanum (Solanaceae) (34, 74, 75, 76), lettuce (Lactuca sativa; Cichoriaceae) (60), and pea (Pisum sativum; Fabaceae) (27). However, the biochemical and physiological processes controlled by these host-virus protein interactions remain to be elucidated.
Because HCpro interacts with VPg, which in turn interacts with eIF4E and eIF(iso)4E, the aim of this study was to test whether the multifunctional HCpro interacts directly with eIF4E and/or eIF(iso)4E. This was indeed found with the HCpro of three potyviruses tested in vivo in yeast and in planta. The interaction was biologically significant because a 4E binding motif was identified in HCpro, and mutations in the motif debilitated the protein-protein interactions between eIF(iso)4E and HCpro in yeast and the virulence of PVA in plants. The 4E-HCpro interaction was observed in association with viral 6K2-induced membranous vesicles. These novel findings reveal a missing link in potyvirus-host interactions involving translation initiation factors and should be helpful for elucidating the roles of eIF4E and eIF(iso)4E in the potyvirus infection cycle, conclusions that have proven difficult to reach based on the interactions of translation initiation factors with VPg/NIa only.
MATERIALS AND METHODS
Cloning of the genes for eIF(iso)4E and eIF4E and the HCpro coding sequences for YTHS assays.
Total RNA was extracted from the leaves of tobacco (N. tabacum cv. Samsun NN) and the leaves of a diploid potato clone, 2x(v2)7, and its progeny lines (90, 93) using TRIzol reagent (Invitrogen, Carlsbad, CA). mRNA was isolated from total RNA using an Oligotex mRNA midi-kit (Qiagen, Hilden, Germany). cDNA was synthesized from 600 ng of total RNA or mRNA using Moloney murine leukemia virus reverse transcriptase (200 U/μl; Promega, Madison, WI) and Oligo(T)16 primers (Roche, Basel, Switzerland). Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) and primers based on the eIF(iso)4E and eIF4E gene sequences of tobacco, potato, or tomato (Solanum esculentum; see Table S1A in the supplemental material) were used to amplify the eIF4E and eIF(iso)4E coding regions using cDNA as a template. Restriction sites needed for cloning were added to the PCR fragments using primers (see Table S1A). PCR fragments were cloned into the yeast two-hybrid system (YTHS) vectors pGBKT7 and pGADT7 for fusion with the DNA binding domain (BD) or transcription activation domain (AD), respectively (Clontech, Mountain View, CA) (79). The PCR products and all constructs were verified by sequencing, which was carried out with an Applied Biosystems BigDye Terminator (version 3.1) kit and a 3100 capillary sequence analyzer at the sequencing core facility of the Haartman Institute, University of Helsinki. Sequence alignments were made using ClustalW2 (43).
The HCpro coding region was amplified with primers that included the appropriate restriction sites for cloning (see Table S1A in the supplemental material) from the infectious cDNA clone of PVA strain B11 (PVA-B11) (31, 63) and were cloned using the YTHS vectors as described above. PCR products encoding the full-length forms of HCpro were cloned into the BD and AD fusion vectors (Clontech) using BamHI and SalI restriction sites introduced into the PCR fragments with primers. The reading frame and orientation of inserts were verified with restriction analysis and sequencing.
A small-scale lithium acetate yeast transformation procedure was used (Clontech) to cotransform yeast cells (AH109) with the BD and AD fusion vectors. A concentration of the BD vector 2-fold higher than that of the AD vector was used for transformation as recommended. Synthetic minimal medium lacking leucine and tryptophan was used as the selective medium for growing the transformed yeast. Protein-protein interactions were detected by growth on medium lacking adenine, histidine, leucine, and tryptophan. Yeast growth at 30°C was observed for up to 14 days. Positive- and negative-control vectors supplied by the manufacturer were used for comparison. Experiments were carried out three times.
Protein-protein interactions were quantified in the yeast strain Y187 (Clontech) by measurement of β-galactosidase activity using the pellet X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (PXG) assay and processing images with Image J (National Institutes of Health) as described previously (56). The positive control (vector pair supplied by Clontech) was set to 100% β-galactosidase activity to determine the relative strength of interactions in other samples.
Bimolecular fluorescence complementation (BiFC).
The HCpro coding region of PVA-B11 cloned into the YTHS BD fusion vector was amplified with primers PVA-HCpro-Fwd and PVA-HCpro-Rev (see Table S1B in the supplemental material) for cloning into the pRT-YN and pRT-YC cassettes constructed and described by Zamyatnin et al. (98). The HCpro coding region of TEV (HCH10) was amplified from the plasmid pTRANS5′TEV (28), whereas the HCpro coding region of PVY (isolate Nevski) was amplified using cDNA of an infected tobacco plant. The primers are described in Table S1B. The primers included an NcoI restriction site that was required to express the yellow fluorescent protein (YFP) N-proximal region (YN) fused to the HCpro N terminus. The constructs of eIF4E and eIF(iso)4E were prepared to express the C-proximal part (YC) of YFP fused to the N or C terminus of the initiation factors and were obtained by PCR from the YTHS BD fusion vectors using the indicated primers (see Table S1B). NcoI was used to ligate the YC tag to genes for expression as N-terminal fusion proteins, whereas XhoI was used to obtain a C-terminal tag. The PCR products encoding HCpro from PVA, TEV, and PVY and genes for the translation initiation factors were first cloned into the pRT expression cassette that contains suitable HindIII sites that can be used to subclone the pRT expression cassette into the binary vector pLH7000 for expression under Cauliflower mosaic virus 35S promoter (35S) by agroinfiltration (98).
The binary vector plasmid pCAMBIA0390 (Cambia Labs) was modified by engineering a novel KpnI site using the QuikChange II XL site-directed mutagenesis system (Stratagene) and appropriate nucleotide primers. The infectious clone of PVA-B11 was modified for the fusion of YN to the HCpro N terminus by introducing the YN tag into cloning site 2 (CS2) (41) using BfrI and MluI. Subsequently, the clone was digested with KpnI and SalI, and the infectious PVA cDNA was transferred, including the 5′-proximal 35S and the 3′-proximal poly(A) tail, into the modified pCAMBIA0390 vector. Binary pLH-RFP was created by digestion with XhoI and NcoI to replace the YN fragment with the mono-red fluorescent protein (mRFP; AJ851291) in pLH-YN. PVA-6K2 was amplified from an infectious cDNA clone of PVA-B11 (31, 63) and cloned to pLH-RFP using XhoI sites. The resulting plasmid was named pLH6K2-RFP. The constructs were verified by restriction analysis and sequencing.
The binary vectors were introduced to Agrobacterium tumefaciens (pGV2260) cells by electroporation using a Bio-Rad Gene Pulser. Agrobacterium cultures were diluted with infiltration medium to a final optical density at 600 nm (OD600) of 0.5 and combined in equal volumes for coexpression experiments. Infiltration of Nicotiana benthamiana leaves was carried out as described previously (46). Experiments were carried out three times.
Visualization of protein-protein interactions in the living plant cells based on YFP fluorescence was done with a Leitz Laborlux S microscope with an epifluorescence extension, Leitz Ploemopak (Leica, Wetzlar, Germany), 2, 3, and 4 days postinfiltration. A green fluorescent protein (GFP) filter (Leica) with a bandpass of 470/40 nm for excitation and a bandpass of 525/50 nm for emission was used. For documentation of results, images were captured 3 days postinfiltration using the charge-coupled-device (CCD) camera DP-50 (Olympus, Hamburg, Germany), which was controlled by Viewfinder Lite, version 1.0 software (Olympus). Confocal microscopy was carried out 3 days postinfiltration with a Leica TCS SP2 AOBS scanner using a 63× 1.2 water immersion objective at the Institute of Biotechnology, University of Helsinki. Emission from YFP (bimolecular fluorescence complementation [BiFC]) and RFP were scanned sequentially as follows: YFP (BiFC) was excited with a 514-nm argon laser and captured at 520 to 581 nm, and RFP was excited with a 561-nm DPSS laser and captured at 600 to 640 nm. Chloroplast autofluorescence was excited with a 514-nm argon laser and captured at 650 to 700 nm. Images were processed using Leica Application Suite Advanced Fluorescence Lite (LAS AF Lite) and Adobe Photoshop 3.0.
Western blot analysis.
To verify fusion protein expression in yeast, total proteins were extracted as described previously (92), with some modifications. Overnight cultures (5 ml) of yeast cotransformants were harvested by centrifugation at 1,000 × g for 5 min. The pellet was frozen in liquid nitrogen and thawed on ice. One volume (100 μl) of fresh, ice-cold 1.85 M NaOH containing 7% β-mercaptoethanol was added, and the mixture was incubated on ice for 10 min. Precipitation was carried out by adding an equal volume (100 μl) of 50% trichloroacetic acid (TCA) and incubating for 5 min on ice. The precipitate was spun for 10 min at 3,000 × g, and the pellet was washed with 400 μl 1 M Tris-HCl (pH 8.0), dissolved in 100 μl 2× Laemmli buffer (20% glycerol, 100 mM Tris-HCl, 10% [vol/vol] β-mercaptoethanol, 0.2% bromophenol blue, 4% [wt/vol] sodium dodecyl sulfate [SDS]), and boiled for 5 min. The proteins were analyzed on a 12% SDS polyacrylamide gel by electrophoresis and electroblotted onto a Hybond-P polyvinylidene fluoride membrane (GE Healthcare, Buckinghamshire, United Kingdom). After blocking in 1× phosphate-buffered saline (PBS) with 0.1% Tween 20 and 5% milk powder, the membranes were probed with monoclonal AD and BD antibodies (1:50,000; Clontech) at 4°C overnight. Following washing with 1× PBS containing 0.1% Tween 20, the membrane was incubated with horseradish peroxidase (HRP)-conjugated anti-mouse sheep antibodies (GE Healthcare). Proteins were detected with the enhanced chemiluminescence method using Super Signal West Femto chemiluminescent substrate (Thermo Scientific, Rockford, IL) and visualized by autoradiography. For BiFC samples, leaf tissue (40 mg) was collected 3 postinfiltration and frozen in liquid nitrogen. The frozen leaf tissue was homogenized in a microcentrifuge tube on ice using a plastic pestle, and 100 μl 2× Laemmli buffer was added. Homogenized samples were boiled for 5 min and clarified by centrifugation. An aliquot of 10 μl of the homogenate was analyzed by SDS-PAGE, and proteins were transferred to a polyvinylidene difluoride membrane as described above. The membranes were probed with GFP polyclonal rabbit antiserum (Invitrogen), HCpro polyclonal antibodies (courtesy of F. Rabenstein, BAFZ, Aschersleben, Germany), or eIF4E polyclonal antibodies (courtesy of K. Browning, University of Texas, Austin, TX) and incubated with HRP-conjugated anti-rabbit donkey antibodies (GE Healthcare). Signals were detected as described above.
Mutation of PVA HCpro and inoculation of plants with the mutant virus.
Two point mutations (Y345A and L350A) were introduced into the predicted 4E binding site of PVA HCpro using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations. The 5′ end of the PVA genome, including the HCpro encoding sequence, was cloned from the infectious PVA cDNA (31, 63) into pGEM-T (Promega), and the two mutations were introduced using the primer HCFwdY345A_L350A (5′-CCAAATCAGGATACTGCGCCATAAATATATTCGCGGCTATGCTTGTG-3′; mutated bases underlined) and its reverse complement HCRevComY345A_L350A. The ClaI and NruI restriction sites flanking the HCpro encoding sequence in pGEM-T were used to introduce the mutated HCpro into the infectious PVA cDNA driven by the Cauliflower mosaic virus 35S promoter (63), and verification of the mutations and cloning sites by sequencing followed. The mutated PVA cDNA was designated PVA-HC4Emut. The full-length point-mutated form of the HCpro-encoding sequence (HC4Emut) was inserted into the AD fusion vector (Clontech) using BamHI and SalI restriction sites introduced into the PCR fragment with primers (see Table S1A in the supplemental material).
Plants of Nicotiana tabacum cv. Samsun NN and N. benthamiana were grown from seeds in a controlled growth chamber (temperature, 18 to 22°C; relative humidity, 70%; photoperiod of 16 h, 200 μE m−2 · s−1 with illumination by fluorescent lamps) and watered with a solution containing 1.0% (N:P:K = 16:9:22) fertilizer. PVA-HC4Emut and the nonmutated (wild-type [wt]) PVA cDNA were inoculated biolistically to two full-grown leaves of N. tabacum and N. benthamiana (5 and 4 plants per experiment, respectively) using HandyGun in two experiments, as described previously (82). Gold particles (diameter of 1.0 μm; Bio-Rad Laboratories) were coated with the PVA plasmid, and 0.5 μg of the DNA was used for each bombardment. PVA was tested in the upper noninoculated leaves at 7, 11, and 21 days postinoculation (dpi), and in inoculated leaves at 14 dpi, by double-antibody sandwich (DAS) enzyme-linked immunosorbent assay (ELISA) using a PVA-CP-specific monoclonal antibody (MAb 58/0; Adgen) and alkaline phosphatase-conjugated MAb 58/0 (Adgen), as described previously (65). Systemic infection was tested by sampling expanding top leaves, whereas the bombarded leaf areas were excised for testing local infection. Leaf samples were weighed, ground in ELISA sample buffer at 1 g/10 ml, and aliquots of 100 μl were transferred to a microtiter plate coated with PVA antibodies. Due to very high titers of PVA, the sap extracted from leaves of N. benthamiana was diluted 100-fold with ELISA buffer prior to the transfer of aliquots (100 μl) to wells of the microtiter plate. Known amounts of purified PVA virions were included for comparison to estimate virus concentration.
For detection of PVA by reverse transcription-PCR (RT-PCR), virions of PVA were immunocaptured with MAb 58/0 from the same leaf samples that were subjected to DAS-ELISA and reverse transcribed as described above, and PCR amplification was carried out using PVA-specific primers flanking the mutated region in the HCpro-encoding sequence. PCR products were sequenced directly without cloning.
Nucleotide sequence accession numbers.
The sequences of the translation initiation factors T4Eb (FN666433), Tiso4Ea (FN666434), P4Ea (FN666435), P4Eb (FN666436), Piso4Ea (FN666437), and Piso4Eb (FN666438) were deposited in the EMBL sequence database. The sequences of T4Ea and Tiso4Eb determined in this study were identical to previously published sequences (AY702653 and AY699609, respectively). The HCpro-encoding sequences of PVA-B11, PVY-Nevski, and TEV-HCH10 are available as accession no. AJ296311, FR837956, and DQ986288, respectively.
RESULTS
Characterization of eIF(iso)4E and eIF4E sequences.
The sequences encoding eIF(iso)4E and eIF4E in tobacco (N. tabacum cv. Samsun NN) and potato (S. tuberosum hybrids) (90, 93), both of which belong to the family Solanaceae, were amplified from cDNA by PCR, and five clones of each were sequenced. Two variants, designated a and b, were detected for both eIF4E and eIF(iso)4E in the two species, and they were named, according to their species, T (tobacco) or P (potato). Hence, four different eIF(iso)4E proteins (Tiso4Ea, Tiso4Eb, Piso4ea, and Piso4Eb) and four different eIF4E proteins (T4Ea, T4Eb, P4Ea, and P4Eb) were studied for protein interactions. The deduced eIF(iso)4E amino acid sequences were 90 to 92% identical in the two species and were similar in length (200 aa), except for Tiso4Ea, which lacked 5 aa (Fig. 1A). Piso4Ea and Piso4Eb differed only at a single amino acid, whereas Tiso4Ea and Tiso4Eb differed at 12 aa and were different in length (Fig. 1A). The amino acid sequences of P4Ea and P4Eb were 231 aa and differed at 7 aa residues, whereas T4Ea and T4Eb were 222 aa and differed at 10 aa residues (Fig. 1B). Identities of the potato and tobacco eIF4E amino acid sequences were only 70 to 72%.
Fig. 1.
Alignment of amino acid sequences of the translation initiation factors. (A) Alignment of the sequence variants (a and b) of eIF(iso)4E from tobacco (Tiso4E) and potato (Piso4E). (B) Alignment of the sequence variants (a and b) of eIF4E from tobacco (T4E) and potato (P4E). Identical amino acids are indicated by dots, and deletions are indicated by hyphens.
HCpro interacts with eIF(iso)4E and eIF4E.
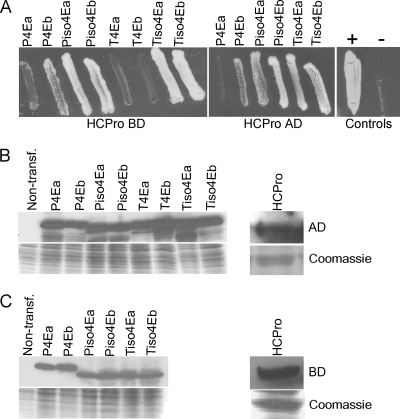
Interactions between HCpro of PVA and the translation initiation factors mentioned above were tested in vivo using a YTHS. Protein-protein interactions were readily detected between the four different eIF(iso)4E proteins and HCpro, as indicated by the fast growth of yeast on stringent selection medium (Fig. 2A). These interactions were observed irrespective of whether the proteins were fused to BD or AD and despite the amino acid sequence differences in the initiation factors. However, the growth of yeast was slower when HCpro was coexpressed with P4Eb, and even slower growth was observed when P4Ea, T4Ea, or T4Eb was coexpressed with HCpro (Fig. 2A). The interactions with eIF4E proteins of tobacco could be tested only in one orientation (fused to AD) because T4Ea and T4Eb fused to the BD autoactivated reporter gene expression. No yeast growth was observed when HCpro was coexpressed with human lamin C (pGBKT7-lam) provided by the manufacturer (Clontech) and used as a negative control (see Fig. S1A and B in the supplemental material). Expression of all tested fusion proteins was readily detected in yeast cells by Western blot analysis (Fig. 2B and C).
Fig. 2.
Interactions between HCpro and the translation initiation factors of tobacco and potato using the GAL4 YTHS. Two variants (a and b) of the translation initiation factors eIF(iso)4E and eIF4E (4E) from tobacco (Tiso4Ea and Tiso4Eb; T3Ea and T4Eb) and potato (Piso4Ea and Piso4Eb; P3Ea and P4Eb) were tested for interactions with the HCpro of PVA. HCpro and initiation factors were fused to the AD or BD and were expressed following cotransformation of yeast cells with the respective vector plasmids. (A) Growth of yeast (Saccharomyces cerevisiae AH109) 10 days after plating on stringent selective medium lacking adenine, histidine, leucine, and tryptophan indicates an interaction between the tested proteins. T4Ea and T4Eb autoactivated the reporter genes when fused to the BD vector and were not included in the assay. Western blot analysis with monoclonal antibodies specific to the AD (B) or the BD (C) was used to detect the expressed proteins. Coomassie blue-stained SDS-PAGE gel served as a protein loading control.
Interactions of HCpro with eIF(iso)4E and eIF4E in planta.
One variant each of eIF(iso)4E and eIF4E from tobacco and potato was tested for interactions with the HCpro proteins of three potyviruses (PVA, PVY, and TEV) in N. benthamiana using BiFC (98). BiFC is based on the reconstitution of the complementary nonfluorescent N-proximal (YFP1–154, YN) and C-proximal (YFP155–239, YC) halves of YFP fused to the tested protein pairs. YN and YC are brought together by interaction of the tester proteins, which is detected by the restored fluorescence of YFP. In this study, YN was fused to the N terminus of HCpro (YN-HCpro), because protein fusions at the HCpro N terminus interfere less with the structure and function of HCpro according to studies of other potyviruses (8, 20) and our unpublished data on PVA.
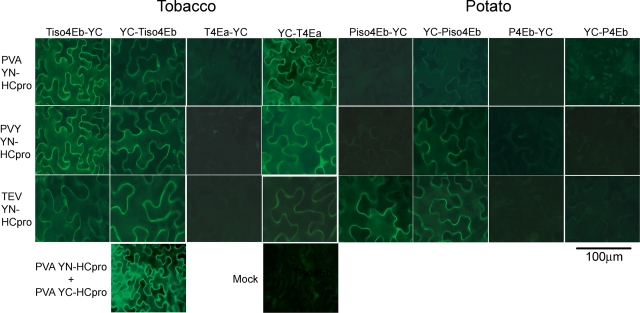
Visualization of protein-protein interactions in living plant cells revealed that the HCpro proteins from the three potyviruses interacted with YC-Tiso4Eb and Tiso4Eb-YC (Fig. 3). Interactions between the HCpro proteins of PVA and PVY with YC-T4Ea were also detected, but interactions of HCpro of TEV with T4Ea were weaker as judged by the low intensity of fluorescence (Fig. 3). Results with T4Ea-YC were negative. All three HCpro proteins showed interactions with YC-Piso4Eb, but only HCpro of TEV showed some interaction with Piso4Eb-YC (Fig. 3). In contrast, no unambiguous fluorescence signals or protein interactions were observed between HCpro and P4Eb (Fig. 3). These results were not due to poor expression of the fusion proteins, because they were detected by Western blot analysis (see Fig. S2 in the supplemental material). YFP fluorescence resulting from protein interactions was distributed throughout the cells and showed no specific subcellular localization (Fig. 3).
Fig. 3.
Detection of protein-protein interactions in leaves of N. benthamiana using BiFC. HCpro proteins of three potyviruses, PVA, PVY, and TEV, were expressed with YN fused to the HCpro N terminus. Translation initiation factors eIF(iso)4E (Tiso4Eb and Piso4Eb) and eIF4E (T4Ea and P4Eb) of tobacco (T) and potato (P) were expressed with YC fused to the N or C terminus. Leaves were infiltrated with pairs of Agrobacterium strains expressing tester proteins tagged with the opposite halves of YFP (the corresponding bacterial cultures were mixed in a 1:1 ratio for infiltration). YFP fluorescence indicating protein-protein interactions was detected in epidermal cells using an epifluorescence microscope 3 days postinfiltration. Images were taken using the same exposure time. For detection of the expressed recombinant proteins using protein-specific antibodies, see Fig. S1 in the supplemental material.
Subsequently, interactions of HCpro and Tiso4Eb were tested in PVA-infected plant cells by expressing the YN-HCpro fusion protein from a chimeric, infectious PVA clone (PVA-YN-HC) (Fig. 4A and B) during virus infection and introducing Tiso4Eb-YC (Fig. 4A and B) to the same tissues by agroinfiltration. PVA-YN-HC infects N. benthamiana systemically, although this recombinant virus spreads at a lower rate and virus titers accumulate to 25 to 40% (0.3 to 0.5 μg virus/g fresh leaf tissue [gflt]) of the infectious PVA clone lacking YN (1 to 2 μg virus/gflt) in the upper systemically infected leaves (17 to 28 dpi).
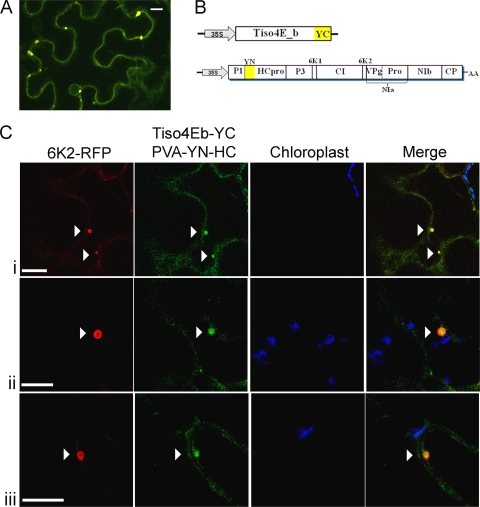
Fig. 4.
Detection of YFP fluorescence resulting from interaction between PVA HCpro and Tiso4Eb, and its association with 6K2-induced viral vesicles in N. benthamiana. (A) YN-HCpro was expressed from an engineered infectious clone of PVA during the course of virus infection, and Tiso4Eb-YC was coexpressed from a binary vector under 35S promoter. The fluorescence resulting from the interaction between HCpro and Tiso4Eb fluorescence was partly confined to round structures or bodies. Scale bars represent 10 μm. (B) Schematic presentations of the constructs used for protein coexpression. The mature viral proteins: P1, the first protein; HC-Pro; P3, third protein; 6K1 and 6K2, 6-kDa proteins; CI, cylindrical inclusion protein; VPg; NIaPro, the main viral proteinase; NIb, replicase; and CP, coat protein. AA indicates the poly(A) tail. The 5′ and 3′ untranslated regions are not depicted. (Ci to iii) Localization of interacting HCpro and Tiso4E with 6K2-induced viral vesicles. Arrows highlight fluorescence signals and colocalization. Confocal images are single optical sections selected from z-series for viewing with LAS AF Lite.
Interactions between YN-HCpro and Tiso4Eb-YC were evident by YFP fluorescence (Fig. 4A). Fluorescence was distributed throughout the cell and was also concentrated in round structures in the cytoplasm (Fig. 4A). When PVA-YN-HC and Tiso4Eb-YC were expressed together with 6K2-RFP, round YFP fluorescent structures encircled by peripheral red fluorescence were observed (Fig. 4Ci to iii). Due to the need of virus polyprotein processing and the reconstitution of YFP, the fluorescence derived from YFP was generating a weaker signal and observed in fewer cells compared to the RFP-tagged recombinant proteins that generally were found in all cells.
4E binding site located in the C-proximal part of HCpro.
Since the interaction of HCpro with eIF4E and eIF(iso)4E was detected with many independent methods, it was of interest to analyze the PVA HCpro amino acid sequence for putative 4E binding sites. The consensus motif for 4E binding is YXXXXLΦ, where X is a variable amino acid and Φ is a hydrophobic residue (reviewed in reference 68). A putative 4E binding motif (YINIFLA, amino acids 345 to 351) was found in the C-proximal part of PVA HCpro (the protein consists of 458 amino acids). Comparison of the HCpro sequences of 47 potyviruses available from a curated plant virus sequence database (www.dpvweb.net) (see Fig. S3 in the supplemental material) indicated that the Tyr and Leu residues of the YINIFLA motif are highly conserved and substituted by a His and a Phe residue, respectively, in only two and six viruses, respectively; the hydrophobic Ala residue was fully conserved (see Fig. S3). One of the deviant viruses was TEV, for which all 28 isolates available in the database at www.dpvweb.net contained Phe instead of Leu in the motif. In addition, the C-proximal part of HCpro in all TEV isolates contained another motif, YLLSILY (residues 391 to 397), also corresponding to the consensus 4E binding motif YXXXXLΦ, but this motif was not conserved in other potyviruses.
According to secondary structure predictions, the region of HCpro containing the 4E binding motif may adapt to an alpha helix conformation (see Fig. S3 in the supplemental material), similar to the structural changes induced in human eIF4GII and 4E-BP1 upon the binding of eIF4E (51).
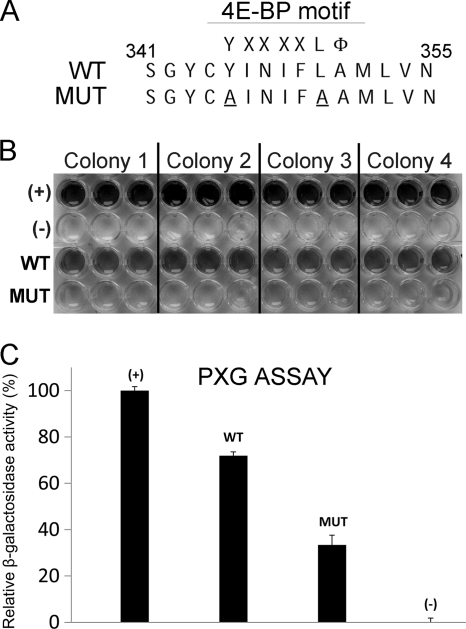
The YINIFLA motif of PVA HCpro was mutated by the introduction of two amino acid substitutions at the conserved positions (Y345A and L350A) (Fig. 5A). Interaction of the mutated HCpro (HC4Emut) with Tiso4Eb was clearly weaker than that of wt HCpro in cotransformed yeast, as indicated by the difference in β-galactosidase activity (Fig. 5B and C). The strength of the interaction between wt HCpro and Tiso4Eb was ca. 75% of that measured for the positive control provided by the manufacturer (Clontech), whereas the interaction between HC4Emut and Tiso4Eb was ca. 35% (Fig. 5C). No activity was detected in the negative control supplied by the manufacturer (Clontech) (Fig. 5C). The application of 1.0 mM or 10 mM 3-amino-1,2,4-triazole (3-AT) to the yeast culture selection medium revealed clear differences in the growth rate of yeast cotransformed with wt HCpro or HC4Emut and Tiso4Ea, Tiso4Eb, P4Ea, P4Eb, Piso4ea, or Piso4Eb (see Fig. S1A and B in the supplemental material), consistent with the role of the YINIFLA motif in interaction with eIF(iso)4E and eIF4E.
Fig. 5.
The putative 4E binding protein (4E-BP) motif of HCpro. (A) The amino acids 345 and 351 of PVA HCpro correspond to the conserved residues in the 4E-BP consensus motif YXXXXLΦ, where X is a variable amino acid and Φ is a hydrophobic residue, and were substituted for alanine (underlined) in the PVA HCpro [HC4Emut (MUT)]. (B) Interactions of the wild-type (WT) HCpro of PVA and HC4Emut (MUT) with the eIF(iso)4E of tobacco (Nicotiana tabacum) (Tiso4Eb) illustrated by the β-galactosidase enzyme activity in cotransformed yeast (96-well microtiter plate format) following overnight incubation. The results from three technical replicates of four independent yeast colonies are shown. (+) and (-), the positive and negative control provided by Clontech, respectively. (C) Mean relative β-galactosidase activities corresponding to the samples shown in panel B and quantified by Image J (National Institutes of Health). Standard deviation is indicted with error bars.
The N-proximal parts of PVA HCpro contained additional 4E binding-like motifs (amino acids 1 to 7, 116 to 122, 214 to 220, 223 to 229, 236 to 242, 288 to 294, 304 to 310, and 343 to 349), and one additional 4E binding-like motif is found at the C terminus of HCpro (amino acids 396 to 402). However, they are not conserved among potyviruses and are less likely to be functional 4E binding sites, while their contribution to the interaction is possible. Since truncated forms of PVA HCpro (see Fig. S4A in the supplemental material) were available from another study (T. Haikonen, unpublished data), they were included in the YTHS analyses to verify the area of HCpro responsible for the interaction with Tiso4Eb by another approach. Yeast cotransformed with Tiso4Eb and HCpro(Δ1–229) or HCpro(Δ1–108), which lack the entire or partial N-proximal domain of HCpro, respectively, grew similarly well as yeast cotransformed with Tiso4Eb and the full-length wt HCpro, indicating that the N terminus of HCpro is redundant for interaction with eIF(iso)4E. Consistent with this result, HCpro(Δ169–458), which expresses only the N-proximal domain and the first third of the central domain, showed no detectable interaction with Tiso4Eb. HCpro(Δ325–458), which lacks the C-proximal portion of HCpro, showed weaker interactions with Tiso4Eb than did HCpro(Δ1–229), HCpro(Δ1–108), and full-length HCpro. HCpro(Δ245–354), which expresses most parts of HCpro except the last third of the central domain and the first third of the C-proximal domain, also interacted more weakly with Tiso4Eb than did proteins containing the entire C-proximal domain (see Fig. S4A). HCpro(Δ1–108 Δ325–458), which is a segment of HCpro lacking the entire N-proximal domain and nearly the entire C-proximal domain, showed only weak interaction with Tiso4Eb. The further reduced fragment of HCpro [HCpro(Δ1–229 Δ325–458)] showed no interaction with Tiso4Eb. All truncated proteins were expressed in yeast cells, as shown by Western blot analysis (Fig. S4B). Taken together, these results provided additional evidence that the C-proximal part of HCpro containing the 4E binding motif (residues 345 to 351) is pivotal for interaction of PVA HCpro with Tiso4Eb.
Mutations in the 4E binding site of HCpro reduce virulence of PVA.
The two amino acid substitutions (Y345A and L350A) which reduced the interaction of HCpro with Tiso4Eb were introduced into an infectious clone of PVA by site-directed mutagenesis, and the mutant virus (PVA-HC4Emut) was inoculated into N. benthamiana and N. tabacum in three experiments. Titers of the PVA CP antigen were estimated in inoculated leaf tissue by DAS-ELISA at 14 dpi, including known amounts of purified PVA virions for comparison. The detection threshold was 0.3 to 1.0 ng PVA CP/gflt. Two leaves of N. tabacum (40 to 50 ng CP/gflt) and a single leaf of N. benthamiana (5.0 μg CP/gflt) were PVA positive following inoculation with PVA-HC4Emut in the 14 and 12 plants tested, respectively, in three experiments. In contrast, all leaves inoculated with wt PVA were virus positive (4.0 to 12 μg CP/gflt) in both host species.
All plants of N. tabacum and N. benthamiana were infected systemically with wt PVA, as tested by DAS-ELISA at 11 dpi (Table 1). At 21 dpi, all plants of N. benthamiana infected with wt PVA displayed severe leaf malformation and mosaic symptoms in upper leaves and contained 200 to 300 μg of PVA CP/gflt, whereas the upper leaves of N. tabacum were symptomless and contained 0.1 to 0.7 μg CP/gflt. In contrast, following inoculation with PVA-HC4Emut, no plant of N. benthamiana was found to be infected systemically (Table 1). The top leaves of only 1 out of the 14 N. tabacum plants inoculated with PVA-HC4Emut generated somewhat elevated ELISA absorbance values at 21 dpi (Table 1) and were estimated to contain 0.6 to 0.9 ng CP/gflt. Immunocapture-RT-PCR analysis with primers specific to the PVA HCpro-encoding sequence and sequencing of the PCR product showed that the mutations introduced into the 4E binding site of HCpro were retained in the progeny viruses in the systemically infected leaves of the tobacco plants.
Table 1.
Systemic infections of plants of Nicotiana tabacum and N. benthamiana with PVA-HC4Emut that carries two amino acid substitutions (Y345A and L350A) in the putative 4E-binding site of HCpro, or with the nonmutated Potato virus A (wt PVA)a
| Strain | Expt 1 |
Expt 2 |
Expt 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 7 dpi | 11 dpi | 21 dpi | 7 dpi | 11 dpi | 21 dpi | 7 dpi | 11 dpi | 21 dpi | |
| N. tabacum | |||||||||
| PVA-HC4Emut | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 1b/5 | 0/4 | 0/4 | 0/4 |
| wt PVA | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | 4/4 | 4/4 | 4/4 |
| N. benthamiana | |||||||||
| PVA-HC4Emut | 0/3 | 0/3 | 0/3 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| wt PVA | 3/3 | 3/3 | 3/3 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 |
The number of systemically infected plants out of the number of inoculated plants. The upper noninoculated leaves were tested for PVA by DAS-ELISA at 7, 11, and 21 days dpi.
The HCpro-encoding sequence in the progeny viruses was amplified by immunocapture-RT-PCR, and the introduced mutations were verified by sequencing.
DISCUSSION
The results of this study show that HCpro proteins of PVA, PVY, and TEV interact with eIF(iso)4E and eIF4E characterized from potato and tobacco, which are hosts for the three potyviruses (89). Interactions between HCpro and eIF4E were, however, mostly weaker or absent compared with those with eIF(iso)4E. These observed differences were not associated with differences in protein expression levels, as shown by Western blot analysis, and instead indicated differential interactions of HCpro with the two types of translation initiation factors. The three tested potyvirus species are more closely related to other potyvirus species than to each other in the genus Potyvirus (83), which suggests that the novel interactions between viral and host proteins reported in this study may occur widely among potyviruses that infect solanaceous hosts and other plant species.
Epifluorescence and confocal microscopy detected BiFC in plant tissues coexpressing HCpro and the translation initiation factors, which was indicative of interactions between these proteins in the cytoplasm but not in the nucleus. These results are consistent with the absence of nuclear localization of HCpro in infected cells reported in previous studies (49, 55, 66, 69). Similarly, eIF(iso)4E of A. thaliana is associated primarily with the endoplasmic reticulum in the cytoplasm (7), although some of the protein is found also in the nucleus (12). However, the previously reported interactions with NIa, which contains a bipartite nuclear and nucleolar localization signal in the VPg domain (67), can translocate eIF(iso)4E to the nucleus (7).
Interactions between HCpro proteins and potato eIF(iso)4E seemed to be weaker than those with tobacco eIF(iso)4E in the cells of N. benthamiana, as observed with BiFC, although no differences were observed in the accumulation of the proteins in infiltrated leaf tissues. However, no differences in interactions of HCpro with tobacco and potato eIF(iso)4E were observed in the YTHS, a system in which protein interactions are tested in the nucleus. These results suggest that other unidentified proteins may participate in these interactions in plant cells and that the cells of N. benthamiana (BiFC experiments) provided a better matched protein interaction network for eIF(iso)4E derived from tobacco (N. tabacum) than for eIF(iso)4E derived from the more distantly related potato (S. tuberosum). Furthermore, interactions of the HCpro of PVY and TEV with the eIF(iso)4E of potato and tobacco were detected in the cells of N. benthamiana by BiFC, whereas the results of YTHS were negative (data not shown), underlining the importance of testing interactions in the natural cellular environment in planta.
The two initiation factors studied here, eIF(iso)4E and eIF4E, belong to different translation complexes [eIF(iso)4F and eIF4F, respectively]. eIF(iso)4F plays a role in the translation of normally capped mRNAs, whereas eIF4F is also involved in the translation of noncapped mRNAs and mRNAs with a structured 5′ leader (25). The eIF4F and eIF(iso)4F complexes are also involved in cap-independent translation of the potyviral polyprotein, which is mediated via the eIF4G/eIF(iso)4G-dependent binding to an internal ribosome entry site (IRES) of the 5′ nontranslated region in TEV (24). The affinity of the translation complexes to IRES in TEV is improved by the interaction of eIF4E and eIF(iso)4E with VPg (42). Results of the YTHS and BiFC were consistent with the notion that HCpro may preferentially interact with eIF(iso)4E. Therefore, preferential interactions between eIF(iso)4E and HCpro as shown here suggest a novel mechanism by which potyviruses might control interactions with translation initiation factors.
Previous studies of A. thaliana mutants indicate that potyviruses may be different in their requirements for eIF(iso)4E and eIF4E during the infection cycle (22, 80). For example, TEV, TuMV, and LMV require eIF(iso)4E for the infection of A. thaliana, whereas infection with ClYVV requires eIF4E (22, 44, 80). Many recessive host resistance genes that protect crop plants from infections with the picorna-like potyviruses or bymoviruses (Potyviridae) encode mutated forms of eIF4E or eIF(iso)4E (see, e.g., references 22, 27, 34, 37, 44, 60, 74, 75, 76, and 80). In some virus-host combinations, simultaneous mutations in eIF(iso)4E and eIF4E are required to confer recessive resistance, e.g., resistance to Pepper veinal mottle virus and Chili veinal mottle virus in pepper (34, 76), suggesting that these viruses may use both translation initiation factors for infection. Resistance conferred by the translation initiation factors can be overcome by amino acid changes in the VPg of TuMV, PVY, and TEV (5, 14, 26, 37, 57) but also the CI protein of LMV (1), the P3 protein of PSbMV (33), and the P1 protein of Clover yellow vein virus (58). The mechanisms by which the mutations in genes for translation initiation factors compromise virus infection and by which mutations in viral proteins overcome the resistance are not yet known. This may be because only interactions between the translation initiation factors and VPg/NIa have been described (70). It is anticipated that future studies considering the multifunctional HCpro as an interaction partner for translation initiation factors will uncover important novel mechanisms by which the translation initiation factors confer resistance to potyviruses.
Interaction between eIF4E and the scaffold protein eIF4G is important in cap-dependent translation initiation (78) and mediated by a binding site possessing a conserved amino acid sequence, YXXXXLΦ, where X is a variable amino acid and Φ is a hydrophobic residue (reviewed in reference 68). Similar 4E binding motifs have been identified in other 4E binding proteins which regulate translation in eukaryotes (68). In this study, we detected a putative 4E binding motif in the C-proximal part of HCpro in PVA and 46 additional potyviruses whose HCpro sequences were retrieved from databases and compared. The substitution of two conserved amino acids (Tyr and Leu) for Ala in the motif (YINIFLA) of PVA HCpro weakened the HCpro-initiation factor interactions significantly and impaired the virulence of PVA drastically. These findings show that the interaction of HCpro with eIF(iso)4E and/or eIF4E is important in the infection cycle of the virus. However, the mutations introduced to the 4E binding motif of HCpro did not abolish the infectivity of PVA or prevent systemic movement of the virus completely, although virus accumulation in the inoculated and upper noninoculated leaves was reduced heavily. It is noteworthy that the 4E binding motif YINIFLA in PVA HCpro overlaps with the motif YCYINIFL that confers interactions with an eIF4E homolog protein (4EHP) in the mouse (91). Therefore, the 4E binding motif in HCpro may control interactions with many homologs of eIF4E and eIF(iso)4E in plants, which requires further study.
Our results are consistent with those obtained on encephalomyocarditis virus (EMCV; genus Cardiovirus) (29). A 4E binding motif was identified recently in the 2A proteinase of EMCV. Mutations introduced to the motif decreased eIF4E binding by the 2A proteinase and reduced virus multiplication in human HeLa cells (29). The PVA HCpro proteinase and EMCV 2A proteinase seem to be the first viral proteins reported to bind translation initiation factors via a specific 4E binding motif.
Both capped host cell mRNA and uncapped viral RNA are present in virally infected plant cells, with the consequence that the viral RNA has to compete for the translation factors. Positive-strand RNA viruses are known to manipulate translation initiation factors, their activity, and subcellular localization to get access to the host translation machinery (21, 84, 85). When the interaction of HCpro and eIF(iso)4E was tested in tissues infected with PVA, fluorescence was partly confined to round structures or bodies in the cytoplasm. These round cytoplasmic structures were associated with 6K2-induced vesicles. Viral replication factors and viral RNA have been detected in 6K2-induced vesicles, and in addition, previous studies have detected the presence of translation initiation factors in viral replication vesicles, implying that potyviral replication may be intimately linked to translation (7, 18, 86, 95). Our study shows that the HCpro-Tiso4Eb interaction is associated with the viral replication/translation vesicles, suggesting that HCpro might contribute to viral genome amplification by additional means other than suppressing RNA silencing. HCpro might enhance the association of eIF4E or its isoform to the virus replication/translation complex and thus increase translation of viral RNA on the cost of cellular mRNAs. Alternatively, by binding to eIF4E, HCpro may prevent the formation of functional cap-dependent translation complexes required for host protein production and thereby inhibit selectively host cell protein synthesis, which is observed with potyvirus-infected plant cells (4, 94). It is also not excluded that HCpro, a cysteine proteinase, might cleave eIF4G, the poly(A) binding protein, or other components in the host mRNA translation complexes, as reported for proteinases of poliovirus, foot-and-mouth disease virus, human immunodeficiency virus type 1, and other entero- and caliciviruses (reviewed in reference 48).
Taken together, this study's findings demonstrated interactions between HCpro of three potyviruses and the translation initiation factor 4E, particularly the isoform eIF(iso)4E, from two plant species in planta. These interactions were controlled by an eIF4E binding motif detected in HCpro and were important for viral virulence. These results reveal novel, biologically significant potyvirus-host interactions and their control mechanism involving two host proteins known to play a crucial role in the success of potyviral infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank K. Browning, I. Fridborg, F. Rabenstein, A. Zamyatnin, and Y. Tian for providing materials and the anonymous reviewers for their valuable criticism and suggestions.
This work was supported financially by the Academy of Finland (grants 1102003, 1118766, and 1134759).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 27 April 2011.
REFERENCES
- 1. Abdul-Razzak A., et al. 2009. Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E-mediated resistance against lettuce mosaic potyvirus. Mol. Plant Pathol. 10:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anandalakshmi R., et al. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142–144 [DOI] [PubMed] [Google Scholar]
- 3. Anandalakshmi R., et al. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. U. S. A. 95:13079–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aranda M. A., Escaler M., Wang D. W., Maule A. J. 1996. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. U. S. A. 93:15289–15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ayme V., et al. 2006. Different mutations in the genome-linked protein VPg of Potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant Microbe Interact. 19:557–563 [DOI] [PubMed] [Google Scholar]
- 6. Ballut L., et al. 2005. HCPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymatic activities. J. Gen. Virol. 86:2595–2603 [DOI] [PubMed] [Google Scholar]
- 7. Beauchemin C., Boutet N., Laliberté J. F. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation initiation factor iso 4E in planta. J. Virol. 81:775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanc S., Dolja V. V., Llave C., Pirone T. P. 1999. Histidine-tagging and purification of tobacco etch potyvirus helper component protein. J. Virol. Methods 77:11–15 [DOI] [PubMed] [Google Scholar]
- 9. Blanc S., et al. 1998. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J. Gen. Virol. 79:3119–3122 [DOI] [PubMed] [Google Scholar]
- 10. Blanc S., et al. 1997. A specific interaction between coat protein and helper component correlates with aphid transmission of potyvirus. Virology 231:141–147 [DOI] [PubMed] [Google Scholar]
- 11. Brigneti G., et al. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Bush M. S., et al. 2009. Selective recruitment of proteins to 5′ cap complexes during the growth cycle in Arabidopsis. Plant J. 59:400–412 [DOI] [PubMed] [Google Scholar]
- 13. Carrington J. C., Herndon K. L. 1992. Characterization of the potyviral HC-Pro autoproteolytic cleavage site. Virology 187:308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Charron C., et al. 2008. Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54:56–68 [DOI] [PubMed] [Google Scholar]
- 15. Cheng Y.-Q., et al. 2008. HC-Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J. Gen. Virol. 89:2046–2054 [DOI] [PubMed] [Google Scholar]
- 16. Choi I.-R., Stenger D. C., French R. 2000. Multiple interactions among proteins encoded by the mite-transmitted wheat streak mosaic tritimovirus. Virology 267:185–198 [DOI] [PubMed] [Google Scholar]
- 17. Chung B. Y.-W., Miller W. A., Atkins J. F., Firth A. E. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105:5897–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cotton S., et al. 2009. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 20:10460–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cronin S., Verchot J., Haldeman-Cahill R., Schaad M. C., Carrington J. C. 1995. Long distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7:549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dolja V. V., Herndon K. L., Pirone T. P., Carrington J. C. 1993. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J. Virol. 67:5968–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dreher T. W., Miller W. A. 2006. Translational control in positive strand RNA plant viruses. Virology 344:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duprat A., et al. 2002. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32:927–934 [DOI] [PubMed] [Google Scholar]
- 23. Endres M. W., et al. 2010. Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog. 6(1):e1000729 doi:10.1371/journal.ppat.1000729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gallie D. R. 2001. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 75:12141–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gallie D. R., Browning K. S. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 276:36951–36960 [DOI] [PubMed] [Google Scholar]
- 26. Gallois J.-L., et al. 2010. Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF(iso)4E and eIF(iso)4G. J. Gen. Virol. 91:288–293 [DOI] [PubMed] [Google Scholar]
- 27. Gao Z., et al. 2004. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40:376–385 [DOI] [PubMed] [Google Scholar]
- 28. Goytia E., Fernándes-Calvino L., Martínez-García B., López-Abella D., López-Moya J. J. 2006. Production of plum pox virus HC-Pro functionally active for aphid transmission in a transient-expression system. J. Gen. Virol. 87:3413–3423 [DOI] [PubMed] [Google Scholar]
- 29. Groppo R., Brown B. A., Palmenberg A. C. 2011. Mutational analysis of the EMCV 2A protein identifies a nuclear localization signal and an eIF4E binding site. Virology 410:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo D., Merits A., Saarma M. 1999. Self-association and mapping of interaction domains of helper component-proteinase of potato A potyvirus. J. Gen. Virol. 80:1127–1131 [DOI] [PubMed] [Google Scholar]
- 31. Guo D. Y., Rajamäki M. L., Saarma M., Valkonen J. P. T. 2001. Towards a protein interaction map of potyviruses: protein interaction matrixes of two potyviruses based on the yeast two-hybrid system. J. Gen. Virol. 82:935–939 [DOI] [PubMed] [Google Scholar]
- 32. Guo D., Spetz C., Saarma M., Valkonen J. P. T. 2003. Two potato proteins, including a novel RING finger protein (HIP1), interact with the potyviral multifunctional protein HC-Pro. Mol. Plant Microbe Interact. 16:405–410 [DOI] [PubMed] [Google Scholar]
- 33. Hjulsager C. K., et al. 2006. Multiple determinants in the coding region of Pea seed-borne mosaic virus P3 are involved in virulence against sbm-2 resistance. Virology 355:52–61 [DOI] [PubMed] [Google Scholar]
- 34. Hwang J., et al. 2009. Double mutations in eIF4E and eIF(iso)4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells 27:329–336 [DOI] [PubMed] [Google Scholar]
- 35. Jin Y., et al. 2007. HC-Pro protein of Potato virus Y can interact with three Arabidopsis 20S proteasome subunits in planta. J. Virol. 81:12881–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin Y., et al. 2007. The HC-Pro protein of Potato Virus Y interacts with NtMinD of tobacco. Mol. Plant Microbe Interact. 20:1505–1511 [DOI] [PubMed] [Google Scholar]
- 37. Kang B. C., Yeam I., Frantz J. D., Murphy J. F., Jahn M. M. 2005. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42:392–405 [DOI] [PubMed] [Google Scholar]
- 38. Kasschau K. D., Carrington J. C. 1998. A counter defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470 [DOI] [PubMed] [Google Scholar]
- 39. Kasschau K. D., Carrington J. C. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71–81 [DOI] [PubMed] [Google Scholar]
- 40. Kasschau K. D., Cronin S., Carrington J. C. 1997. Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 288:251–262 [DOI] [PubMed] [Google Scholar]
- 41. Kelloniemi J., Mäkinen K., Valkonen J. P. T. 2008. Three heterologous proteins simultaneously expressed from a chimeric potyvirus: infectivity, stability, and the correlation of genome and virion length. Virus Res. 135:282–291 [DOI] [PubMed] [Google Scholar]
- 42. Khan M. A., Miyoshi H., Gallie D. R., Gross D. J. 2008. Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 283:1340–1349 [DOI] [PubMed] [Google Scholar]
- 43. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 44. Lellis A. D., Kasshau K. D., Whitman S. A., Carrington J. C. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12:1046–1051 [DOI] [PubMed] [Google Scholar]
- 45. Léonard S., et al. 2000. Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74:7730–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Llave C., Kasschau K. D., Carrington J. C. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. U. S. A. 97:13401–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Llave C., Xie Z. X., Kasschau K. D., Carrington J. C. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056 [DOI] [PubMed] [Google Scholar]
- 48. Lloyd R. E. 2006. Translational control by viral proteinases. Virus Res. 119:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lózsa R., Csorba T., Lakatos L., Burgyan J. 2008. Inhibition of 3′ modification of small RNAs in virus-infected plants require spatial and temporal co-expression of small RNAs and viral silencing-suppressor proteins. Nucleic Acids Res. 36:4099–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maia I. G., Bernardi F. 1996. Nucleic acid-binding properties of a bacterially expressed potato virus Y helper component-proteinase. J. Gen. Virol. 77:869–877 [DOI] [PubMed] [Google Scholar]
- 51. Marcotrigiano J., Gingras A. C., Sonenberg N., Burley S. K. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3:707–716 [DOI] [PubMed] [Google Scholar]
- 52. Merits A., Guo D., Järvekülg L., Saarma M. 1999. Biochemical and genetic evidence for interactions between Potato A potyvirus-encoded proteins P1 and P3 and proteins of the putative replication complex. Virology 263:15–22 [DOI] [PubMed] [Google Scholar]
- 53. Merits A., Guo D., Saarma M. 1998. VPg, coat protein and five non-structural proteins of potato A potyvirus bind RNA in sequence-unspecific manner. J. Gen. Virol. 79:3123–3127 [DOI] [PubMed] [Google Scholar]
- 54. Michon T., Estevez Y., Walter J., German-Retana S., Le Gall O. 2006. The potyviral virus genome-linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. FEBS J. 273:1312–1322 [DOI] [PubMed] [Google Scholar]
- 55. Mlotshwa S. W., et al. 2002. Subcellular location of the helper component-protease of Cowpea aphid-borne mosaic virus. Virus Genes 25:207–216 [DOI] [PubMed] [Google Scholar]
- 56. Möckli N., Auerbach D. 2004. Quantitative β-galactosidase assay suitable for high-throughput applications in the yeast two-hybrid system. Proteomics Tech. 36:872–876 [DOI] [PubMed] [Google Scholar]
- 57. Moury B., et al. 2004. Mutations in Potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant Microbe Interact. 17:322–329 [DOI] [PubMed] [Google Scholar]
- 58. Nakahara K. S., et al. 2010. Involvement of the P1 cistron in overcoming eIF4E-mediated recessive resistance against Clover yellow vein virus in pea. Mol. Plant Microbe Interact. 23:1460–1469 [DOI] [PubMed] [Google Scholar]
- 59. Reference deleted.
- 60. Nicaise V., et al. 2003. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 132:1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Peng Y.-H., et al. 1998. Mutations in the HC-Pro gene of zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J. Gen. Virol. 79:897–904 [DOI] [PubMed] [Google Scholar]
- 62. Plisson C., et al. 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 278:23753–23761 [DOI] [PubMed] [Google Scholar]
- 63. Puurand U., Valkonen J. P. T., Mäkinen K., Rabenstein F., Saarma M. 1996. Infectious in vitro transcripts from cloned cDNA of the potato A potyvirus. Virus Res. 40:135–140 [DOI] [PubMed] [Google Scholar]
- 64. Rajamäki M. L., Mäki-Valkama T., Mäkinen K., Valkonen J. P. T. 2004. Infection with potyviruses, p. 68–91 In Talbot N. J. (ed.), Plant-pathogen interactions. Blackwell Publishing, Sheffield, United Kingdom [Google Scholar]
- 65. Rajamäki M. L., et al. 1998. Biological, serological, and molecular differences among isolates of potato A potyvirus. Phytopathology 88:311–321 [DOI] [PubMed] [Google Scholar]
- 66. Rajamäki M. L., Valkonen J. P. T. 2003. Localization of a potyvirus and the viral genome-linked protein in wild potato leaves at an early stage of systemic infection. Mol. Plant Microbe Interact. 16:25–34 [DOI] [PubMed] [Google Scholar]
- 67. Rajamäki M. L., Valkonen J. P. T. 2009. Control of nuclear and nucleolar localization of nuclear inclusion protein A of picorna-like Potato virus A in Nicotiana species. Plant Cell 21:2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rhoads R. 2009. eIF4E: new family members, new binding partners, new roles. J. Biol. Chem. 284:16711–16715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Riedel D., Lesemann D.-E., Maiss E. 1998. Ultrastructural localization of non-structural and coat proteins of 19 potyviruses using antisera to bacterially expressed proteins of plum pox potyvirus. Arch. Virol. 143:2133–2158 [DOI] [PubMed] [Google Scholar]
- 70. Robaglia C., Caranta C. 2006. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11:40–45 [DOI] [PubMed] [Google Scholar]
- 71. Rojas M., Zerbini F. M., Allison R. F., Gilbertson R. L., Lucas W. J. 1997. Capsid protein and helper component-proteinase function as potyvirus cell-to-cell movement proteins. Virology 237:283–295 [DOI] [PubMed] [Google Scholar]
- 72. Roudet-Tavert G., et al. 2002. Interaction between potyvirus helper component-proteinase and capsid protein in infected plants. J. Gen. Virol. 83:1765–1770 [DOI] [PubMed] [Google Scholar]
- 73. Roudet-Tavert G., et al. 2007. Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HCPro. J. Gen. Virol. 88:1029–1033 [DOI] [PubMed] [Google Scholar]
- 74. Ruffel S., et al. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32:1067–1075 [DOI] [PubMed] [Google Scholar]
- 75. Ruffel S., Gallois J. L., Lesage M. L., Caranta C. 2005. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Gen. Genomics 274:346–353 [DOI] [PubMed] [Google Scholar]
- 76. Ruffel S., et al. 2006. Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent Pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87:2089–2098 [DOI] [PubMed] [Google Scholar]
- 77. Ruiz-Ferrer V., et al. 2005. Structural analysis of tobacco etch potyvirus HC-Pro oligomers involved in aphid transmission. J. Virol. 79:3758–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sachs A. B., Varani G. 2000. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat. Struct. Biol. 7:356–361 [DOI] [PubMed] [Google Scholar]
- 79. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 80. Sato M., Nakahara K., Yoshii M., Ishikawa M., Uyeda I. 2005. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 579:1167–1171 [DOI] [PubMed] [Google Scholar]
- 81. Shiboleth Y. M., et al. 2007. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 81:13135–13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sikorskaite S., et al. 2010. HandyGun: an improved custom-designed, non-vacuum gene gun suitable for virus inoculation. J. Virol. Methods 165:320–324 [DOI] [PubMed] [Google Scholar]
- 83. Spetz C., et al. 2003. Molecular resolution of a complex of potyviruses infecting solanaceous crops at the centre of origin in Peru. J. Gen. Virol. 84:2565–2578 [DOI] [PubMed] [Google Scholar]
- 84. Sukarieh R., Sonenberg N., Pelletier J. 2010. Nuclear assortment of eIF4E coincides with shut-off of host protein synthesis upon poliovirus infection. J. Gen. Virol. 91:1224–1228 [DOI] [PubMed] [Google Scholar]
- 85. Svitkin Y. V., et al. 2005. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 25:10556–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thivierge K., et al. 2008. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377:216–225 [DOI] [PubMed] [Google Scholar]
- 87. Torres-Barceló C., Martín S., Darós J. A., Elena S. F. 2008. From hypo- to hypersuppression: effect of amino acid substitutions on the RNA-silencing suppressor activity of the Tobacco etch potyvirus HC-Pro. Genetics 180:1039–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Urcuqui-Inchima S., Maia I. G., Drugeon G., Haenni A. L., Bernardi F. 1999. Effect of mutations within the Cys-region of potyvirus helper component-proteinase on the self-interaction. J. Gen. Virol. 80:2809–2812 [DOI] [PubMed] [Google Scholar]
- 89. Valkonen J. P. T., Kyle M. M., Slack S. A. 1996. Comparison of resistance to potyviruses within Solanaceae: infection of potatoes with tobacco etch potyvirus and peppers with potato A and Y potyviruses. Ann. Appl. Biol. 129:25–38 [Google Scholar]
- 90. Valkonen J. P. T., Slack S. A., Plaisted R. L., Watanabe K. N. 1994. Extreme resistance is epistatic to hypersensitive resistance to potato virus Yo in a Solanum tuberosum subsp. andigena-derived potato genotype. Plant Dis. 78:1177–1180 [Google Scholar]
- 91. Villaescusa J. C., et al. 2009. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One 4(4):e5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Volland C., Urban-Grimal D., Géraud G., Haguenauer-Tapis R. 1994. Endocytosis and degradation of the yeast uracil permease under adverse condition. J. Biol. Chem. 269:9833–9841 [PubMed] [Google Scholar]
- 93. Vuorinen A. L., et al. 2010. Factors underpinning the responsiveness and higher levels of virus resistance realised in potato genotypes carrying virus-specific R genes. Ann. Appl. Biol. 157:229–241 [Google Scholar]
- 94. Wang D. W., Maule A. J. 1995. Inhibition of host gene-expression associated with plant-virus replication. Science 267:229–231 [DOI] [PubMed] [Google Scholar]
- 95. Wei T., et al. 2010. Sequential recruitment of the endoplasmatic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84:799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wittmann S., Chatel H., Fortin M. G., Laliberté J. F. 1997. Interaction of the viral protein genome linked of Turnip mosaic potyvirus with the translation eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast-two hybrid system. Virology 234:84–92 [DOI] [PubMed] [Google Scholar]
- 97. Yambao M. L. M., Masuta C., Nakahara K., Uyeda I. 2003. The central and C-terminal domains of VPg of Clover yellow vein virus are important for VPg-HCPro and VPg-VPg interactions. J. Gen. Virol. 84:2861–2869 [DOI] [PubMed] [Google Scholar]
- 98. Zamyatnin A. A., Jr., et al. 2006. Assessment of the integral membrane protein topology in living cells. Plant J. 46:145–154 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.