Abstract
The activity of the DAF-2 insulin-like receptor is required for Caenorhabditis elegans reproductive growth and normal adult life span. Informatic analysis identified 37 C. elegans genes predicted to encode insulin-like peptides. Many of these genes are divergent insulin superfamily members, and many are clustered, indicating recent diversification of the family. The ins genes are primarily expressed in neurons, including sensory neurons, a subset of which are required for reproductive development. Structural predictions and likely C-peptide cleavage sites typical of mammalian insulins suggest that ins-1 is most closely related to insulin. Overexpression of ins-1, or expression of human insulin under the control of ins-1 regulatory sequences, causes partially penetrant arrest at the dauer stage and enhances dauer arrest in weak daf-2 mutants, suggesting that INS-1 and human insulin antagonize DAF-2 insulin-like signaling. A deletion of the ins-1 coding region does not enhance or suppress dauer arrest, indicating a functional redundancy among the 37 ins genes. Of five other ins genes tested, the only other one bearing a predicted C peptide also antagonizes daf-2 signaling, whereas four ins genes without a C peptide do not, indicating functional diversity within the ins family.
Keywords: Insulin signaling, Caenorhabditis elegans, dauer, life span, C-peptide, DAF-2
Insulin and its related proteins define a superfamily of secreted proteins that share a structural motif stabilized by a set of stereotypical disulfide bonds (Blundell and Humbel 1980; Murray-Rust et al. 1992). Insulin superfamily genes are ubiquitous in vertebrates, and have been identified in invertebrates, including insects, molluscs, and the nematode Caenorhabditis elegans (Duret et al. 1998; Gregoire et al. 1998; Smit et al. 1998; Kawano et al. 2000). Seven members of the insulin superfamily have been identified in humans, including insulin (Brown et al. 1955), insulin-like growth factors (IGFs) I and II (Rinderknecht and Humbel 1978a,b), relaxins HI and HII (Bedarkar et al. 1977; Schwabe and McDonald 1977), early placenta insulin-like peptide (EPIL) (Chassin et al. 1995; Koman et al. 1996), and relaxin-like factor (Bullesbach and Schwabe 1995). These hormones mediate diverse functions. Insulin is a metabolic hormone that acts on target tissues to increase glucose uptake and energy storage, IGFs are mitogenic stimulators that control cell survival and proliferation, and relaxin causes dilation of the symphysis pubis before parturition and vasodilation. No function is yet known for either EPIL or relaxin-like factor. Bombyxin in silk moths (Satake et al. 1997), and the neurons that secrete locust and molluscan insulin-related proteins (Smit et al. 1988; Lagueux et al. 1990) regulate metabolism, implicating insulin-like proteins in metabolic control broadly in animal phylogeny. The insulin-like proteins that regulate metabolism, insulin in vertebrates, bombyxin from silk moths, molluscan MIP, and locust LIRP, appear to be processed proteolytically to remove an internal C peptide (Smit et al. 1988; Lagueux et al. 1990; Kondo et al. 1996). This processing is associated with loading of insulin into secretory vesicles (Orci et al. 1987). Insulin-related proteins are secreted from neurosecretory cells of both vertebrates and invertebrates and may regulate survival and differentiation of neurons and/or function as neuromodulators (Smit et al. 1998).
The receptors for insulin and IGFs are closely related cell surface receptor tyrosine kinases (RTKs) that, unlike most RTKs, are processed from a larger precursor (McDonald et al. 1989). No receptors have been identified yet for the remaining members of the vertebrate insulin superfamily. Bombyxin binds to an insulin receptor-like protein on target tissues (Fullbright et al. 1997).
A C. elegans insulin-like signaling pathway also controls metabolism, as well as growth and longevity. daf-2 encodes the only homolog of the mammalian insulin/IGF receptor family in the C. elegans genome sequence (Kimura et al. 1997; Ruvkun and Hobert 1998). DAF-2 activity promotes reproductive growth, whereas lack of activity causes animals to arrest at the dauer stage, a diapause arrested state (Riddle et al. 1981; Malone and Thomas 1994; Kimura et al. 1997). Decreased daf-2 signaling during the adult stage causes accumulation of lipids (Kimura et al. 1997) and a greatly extended life span (Kenyon et al. 1993). daf-2 functions in a pathway that is congruent at many points with the mammalian insulin signaling pathway, including homologs of the catalytic subunit of PI-3 kinase (age-1; Morris et al. 1996), the serine/threonine kinases Akt/PKB (akt-1 and akt-2) and PDK1 (pdk-1; Paradis and Ruvkun 1998; Paradis et al. 1999), and the fork head transcription factors AFX, FKHR, FKHRL1 (daf-16) that are negatively regulated by the upstream kinase cascade (Lin et al. 1997; Ogg et al. 1997; Datta et al. 1999). Laser ablation of specific sensory neurons results in constitutive dauer arrest (Bargmann and Horvitz 1991) that depends on daf-16 gene activity, suggesting that these sensory neurons normally produce insulin-like signals that activate DAF-2 to prevent dauer-specific differentiation.
DAF-2 insulin-like ligands have not been identified by genetic analysis. We describe a systematic search of the essentially complete C. elegans genome sequence for genes encoding members of the insulin superfamily and characterize the action of those insulins most closely related to mammalian insulin, as well as the action of human insulin itself, in the daf-2 pathway. Our genetic analysis shows that the closest relative of mammalian insulin acts in the daf-2 pathway. Human insulin can similarly affect daf-2 pathway signaling. Our analysis suggests that the insulin superfamily in humans and other vertebrates could be much more extensive than thought previously.
Results
Identification of insulin gene superfamily members
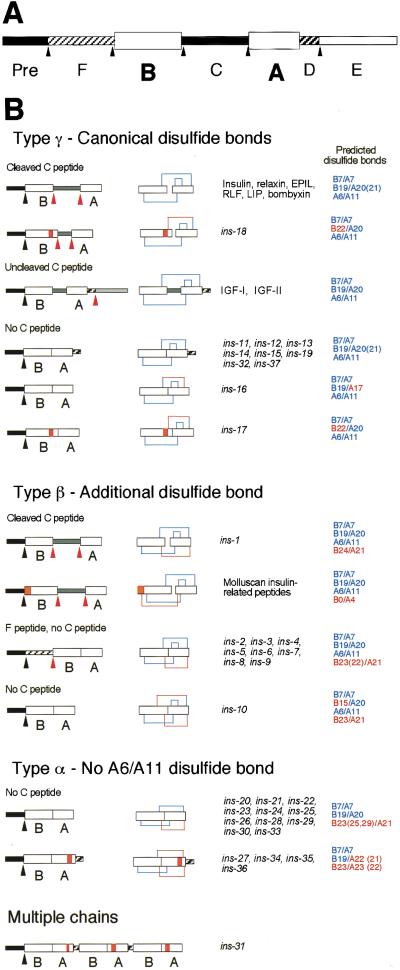
Proteins of the insulin superfamily are synthesized as precursors (preproinsulins) consisting of four domains (Pre, B, C, A; Fig. 1A). Most preproinsulins are processed to excise the Pre and C domains, forming proteins consisting of variable length B and A peptides joined by disulfide bonds (Fig. 1B). Precursors of the IGF subfamily have two additional carboxy-terminal domains (D and E). In these proteins, the C peptide is not removed; however, the carboxy-terminal E peptide is removed by proteolytic processing (Fig. 1B). Whereas the lengths of the secondary structure elements vary and the loops connecting them can take on several conformations, the basic fold is shared by all superfamily members for which structures are known; the B chain contains a single α-helix that lies across the two parallel α-helices formed by the A chain (Murray-Rust et al. 1992).
Figure 1.
Domain structure and genomic organization of the ins gene family. (A) All insulin superfamily genes encode at least a signal sequence, B and A domains. The signal sequence is removed during transit into the endoplasmic reticulum. (Pre) Signal sequence. Arrowheads indicate possible sites of proteolytic processing. (B) Domain organization of prepro- and mature insulin superfamily proteins. Canonical disulfide bonds are those contained in vertebrate insulin. Orange boxes indicate amino acid insertions into loops between the α-helices of the B and A chains. Black arrowheads indicate predicted processing sites for removal of the signal sequence, and red arrowheads indicate other predicted proteolytic processing sites. Predicted disulfide bonds and cysteine locations in blue indicate locations identical to disulfide bonds in vertebrate insulin. Disulfide bonds and cysteine locations in red indicate additional predicted disulfide bonds not contained in vertebrate insulin and cysteine locations that vary from the exact spacing in vertebrate insulin. We established the cDNA sequence of ins-1 through ins-31 and a transcript from ins-33 has been identified independently (M. Hristova and V. Ambros, pers. comm.). Because all of the tested genes are expressed, few, if any, of the predicted genes are pseudogenes.
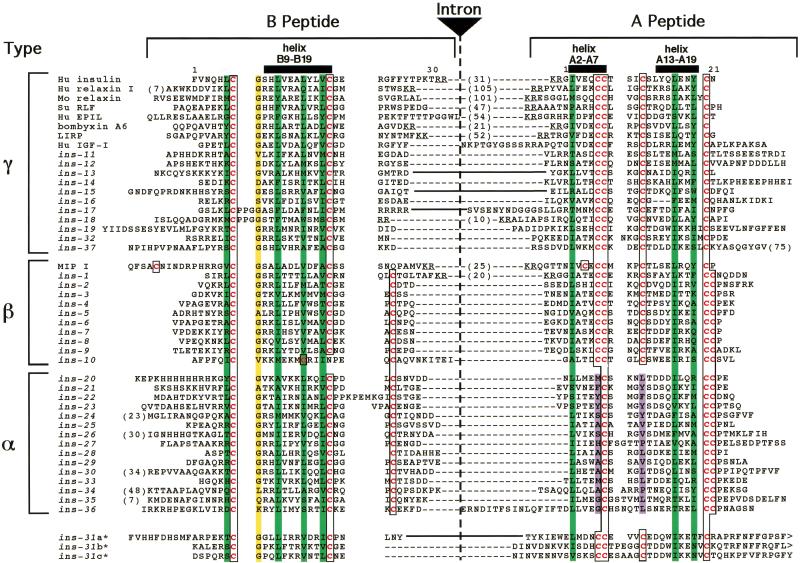
To comprehensively identify C. elegans insulin superfamily genes, we analyzed the entire C. elegans genomic sequence with multiple sequence searching tools, including BLAST (Altschul et al. 1990), FASTA (Pearson and Lipman 1988), Markoff model-based searches (Eddy et al. 1995), and Regular Expression Searches. We examined candidate ORFs encoding at least a putative signal sequence, B chain, and A chain using a stereochemical restriction model derived from structural elements common to insulin superfamily proteins (Murray-Rust et al. 1992) as follows: (1) Two helices within the A peptide joined by a loop; (2) an extended amino-terminal coil within the B peptide followed by a tight turn and a central helix; (3) a hydrophobic core that forms the interface between the A and B peptides; and (4) at least three disulfide bonds formed by conserved cysteine residues (Fig. 2).
Figure 2.
Sequence diversity of ins genes. Sequence alignment of B and A domains. Cysteine residues are in red type and boxed. Hydrophobic residues important for helix formation are overlaid in green. The conserved glycine at B8 is overlaid in yellow. The nonpolar residues that replace the A16/A11 disulfide bond are overlaid in purple. An intron is inserted at the end of the B domain except where indicated by solid lines between the domains. The sizes (in residues) of cleaved or potentially cleavable C domains are indicated by numbers in parentheses between the B and A domain sequences. Actual or predicted dibasic residue cleavage sites for prohormone convertase are underlined. The ins genes defined previously are ins-2 through ins-7, ins-11, and ins-21 through ins-23 (Duret et al. 1998), ins-1 (Gregoire et al. 1998), and ins-18 (Kawano et al. 2000).
This procedure revealed 37 candidate genes, ins-1 through ins-37 (Figs. 1B and 2). In addition to conservation of structural domains, 34 of the 37 ins genes have an intron/exon organization identical to most previously described members of the superfamily; the B and A chains are encoded by individual exons separated by an intron at the 3′ end of the B chain coding region. This search protocol successfully detected all 12 of the insulin superfamily members identified previously (Duret et al. 1998; Gregoire et al. 1998; Kawano et al. 2000).
The C. elegans insulin gene family members were classified into four major classes on the basis of predicted arrangement of disulfide bonds (Fig. 1B) (Duret et al. 1998). Type-γ insulin-like molecules have the arrangement of three disulfide bonds found in vertebrate insulins, IGFs, and most other previously identified members of the superfamily. The three canonical disulfides in the vertebrate genes include interchain bonds between cysteine pairs B7/A7 and B19/A20 or A21 and an intrachain bond in the A chain between A6/A11. Eleven ins genes fall into this class as follows: ins-11 through ins-19, ins-32, and ins-37. ins-17 and ins-18 also have a three-amino-acid insertion in the B chain that moves the cysteine at B19 to position B22. Type-β proteins contain the three canonical disulfide bonds as well as an additional interchain disulfide bond between B22, B23, or B24 and A21. The only previously described members of this class are the seven mollusc MIP proteins, which contain an extra disulfide between B0/A4 (Smit et al. 1996). The C. elegans genes ins-1 through ins-10 fall into this class. Eight of the type-β ins genes encode proteins containing a novel domain between the Pre sequence and the B peptide, which we have termed the F peptide (Figs. 1 and 2). The F peptide is bounded at the amino terminus by the signal peptidase cleavage site and at the carboxyl terminus in most members by the sequence RXRR. RXRR sites are cleaved in the endoplasmic reticulum by furin family proteases. The furin-related gene bli-4 has been cloned from C. elegans (Thacker et al. 1995). Type-α proteins lack the A6/A11 disulfide bond. When normally folded, the A6/A11 cysteine pair is completely buried in the core of the insulin molecule, and stabilizes a tight turn between the two A chain α-helices. In ins-21, ins-22, and ins-23, the cysteines are replaced by amino acids containing aromatic side chains (tyrosine or phenylalanine) (Figs. 1B and 2). ins-31 is a unique member of the gene superfamily; it has a signal sequence followed by three insulin repeats organized in the sequence (B/A)(B/A)(B/A) (Figs. 1B and 2). This gene may encode a single-secreted protein containing three insulin-like structural domains or a pre-proprotein that is cleaved into multiple insulin-like peptides.
One hallmark that distinguishes insulin, a regulator of mammalian metabolism, from IGF-1 and IGF-II, which do not regulate metabolism, is the presence of the C peptide that is cleaved by proteases that recognize flanking dibasic residues. Two of the thirty-seven C. elegans insulin superfamily members, INS-1 and INS-18, are predicted to have cleaved C peptides.
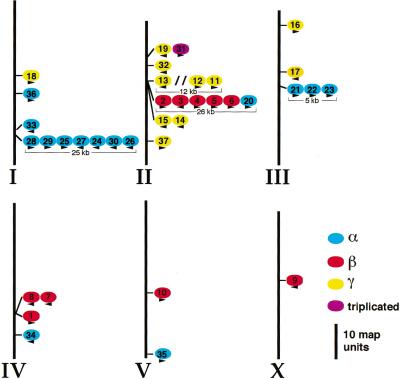
Many members of the C. elegans ins gene family are organized into clusters, usually with members of the same class, in which the ins genes are tandemly arrayed (Fig. 3). Within a cluster, the direction of transcription of the genes tends to be aligned rather than random (Fig. 3). However, the genes within a cluster are still highly divergent, showing for example only 25%–40% amino acid identity in the B and A peptide regions.
Figure 3.
Genomic organization of the ins gene family on C. elegans chromosomes (Roman numerals). Many ins genes are organized in clusters of from two to seven genes of the same class (Fig. 1B). γ-type ins genes are shown in yellow, β-type genes in red, α-type genes in blue, and the gene encoding a triplicated INS protein in purple. The sizes of clusters of three or more genes are indicated. The only intervening genes within the clusters are three genes between ins-13 and ins-12 (indicated by //). Arrowheads indicate relative direction of transcription. The left end of each chromosome is at the top. The relative distances between the genes are in map units (scale bar, 10 map units).
To determine whether the diverse C. elegans insulin superfamily sequences could assume insulin-like tertiary structures, we modeled representative INS proteins using algorithms based on homology modeling, simulated annealing, and energy minimization (data not shown). Despite wide variation in amino acid sequence from previously identified insulin-related proteins, INS-1, INS-4, and INS-21 are predicted to adopt the characteristic insulin fold. This is similar to the findings of Duret et al. (1998), who modeled INS-3, INS-22, and INS-23, using energy minimization alone, and supports their hypothesis that the aromatic amino acids replacing the A6/A11 cysteine pair can interact to stabilize the tertiary structure. The structures calculated for INS-1 were most similar to the structure of mammalian insulin.
INS-1 is the most closely related to human insulin by primary sequence comparison (Figs. 1 and 2), structural homology models, and because it has a probable cleaved C peptide. Consequently, we explored its action in the DAF-2 signaling pathway.
Overexpression of ins-1 antagonizes daf-2 signaling
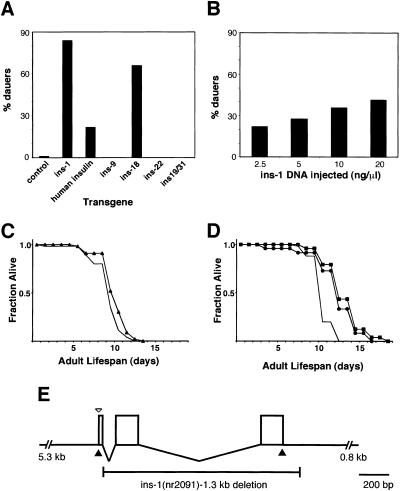
To investigate whether ins gene family members encode DAF-2 ligands and can regulate dauer arrest, transgenic animals with increased ins gene dosage were constructed. We first analyzed the closest relative to insulin, ins-1, under the control of its own promoter (ins-1 transgene) (Fig. 4E) or the heat shock hsp16/2 promoter (heat shock promoter∷ins-1). At the permissive temperature of 20°C, animals harboring either of two temperature-sensitive daf-2 mutations, daf-2(e1370) and daf-2(e1365), mapping to the kinase and ligand binding domains, respectively (Kimura et al. 1997), rarely arrest at the dauer stage (1%) (Table 1). In contrast, an ins-1 transgene providing approximately twenty times the normal gene dosage of ins-1 causes 43% and 84% of the animals, respectively, to arrest at the dauer stage (Table 1; Fig. 4A), suggesting that ins-1 acts antagonistically to DAF-2. In addition, when wild-type animals, which rarely arrest as dauers (0.3%), carry the ins-1 transgene, 5% arrest at the dauer stage at 26°C (Table 1). Similar results were obtained from two independent lines carrying integrated heat shock promoter∷ins-1 (Table 2). The heat shock promoter∷ins-1 fusion gene also dramatically enhances the L1 larval arrest phenotype of daf-2(e1370) (Table 2). This enhancement of L1 arrest is even more pronounced in animals subjected to a heat shock at the L1 stage (Table 2). The L1 arrest phenotype induced by heat shock promoter∷ins-1 in the daf-2 mutant backgrounds may be the result of even more severe antagonism of DAF-2 signaling, because it is similar to the phenotype caused by complete loss of both maternal and zygotic daf-2 signaling (Gems et al. 1998).
Figure 4.
Increased gene dosage of ins-1 and human insulin enhances daf-2(e1365) dauer formation and lengthens wild-type life span. (A) The percentage of dauers formed at 20°C by daf-2(e1365) animals carrying the indicated transgenes was scored 3 d after eggs were laid. These data are from Table 1. (B) ins-1 transgenic daf-2(e1365) animals were derived from founders injected with the indicated concentrations of ins-1 DNA. Dauer arrest was assayed as in A and the average for six independent lines at each ins-1 DNA concentration is presented. The percent dauer arrest ranges for the six lines were 0%–52% (2.5 ng/μL), 0%–80% (5 ng/μL), 6%–64% (10 ng/μL), and 5%–70% (20 ng/μL). (C) Mean adult life span at 26°C was 10.6 d for ins-1(2091);him-5 (▴, n = 46) and 9.8 d for him-5 (solid line, n = 46). (D) Mean adult life span of wild-type transgenic worms at 26°C was 13.0 d for the ins-1 (•, n = 48), 13.7 d for the Pins-1∷human insulin (▪, n = 48), and 11.1 d for no (solid line, n = 50) transgene. (E) Diagram of the ins-1 genomic region. The open arrowhead indicates the point of fusion in Pins-1∷GFP. The solid arrowheads indicate the points at which the human insulin cDNA is fused in Pins-1∷hIns. The 1.3-kb ins-1(nr2091) deletion removes the second and third exons.
Table 1.
Enhancement of dauer arrest by high gene dosage of ins genes and human insulin
| Dauer constitutive mutation
|
Transgene
|
Phenotype at 20°C (%)
|
Phenotype at 26°C (%)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| L4 larvae and adult
|
Dauer
|
Otherb
|
Na
|
L4 larvae and adult
|
Dauer
|
Otherb
|
Na
|
||
| Wild type | control | 97.0 | 0 | 3.0 | 365 | 96.6 | 0.3 | 3.0 | 923 |
| Wild type | ins-1 | 97.0 | 0.2 | 2.8 | 666 | 91.1 | 5.3 | 3.6 | 924 |
| Wild type | human insulin | 96.1 | 0 | 3.9 | 916 | 78.5 | 8.9 | 12.7 | 1398 |
| daf-2(e1370) | none | 86.1 | 1.2 | 12.7 | 833 | 0 | 94.5 | 5.5 | 825 |
| daf-2(e1370) | ins-1 | 47.1 | 42.9 | 10.0 | 310 | 0 | 86.4 | 13.6 | 309 |
| daf-2(e1365) | control | 98.7 | 1.0 | 0.3 | 1856 | 0.1 | 96.1 | 3.8 | 813 |
| daf-2(e1365) | ins-1 | 16.3 | 83.7 | 0 | 1162 | 0 | 94.2 | 5.8 | 943 |
| daf-2(e1365) | human insulin | 76.2 | 21.6 | 2.2 | 1191 | 0.4 | 96.1 | 3.5 | 513 |
| daf-7(e1372) | control | 77.5 | 22.5 | 0 | 494 | 0 | 97.3 | 2.7 | 332 |
| daf-7(e1372) | ins-1 | 51.4 | 47.8 | 0.8 | 1146 | 0 | 98.9 | 1.1 | 747 |
| daf-7(e1372) | human insulin | 50.2 | 47.9 | 1.8 | 707 | 0 | 97.3 | 2.7 | 332 |
| Wild type | none | 100 | 0 | 0 | 541 | 99.5 | 0 | 0.5 | 553 |
| Wild type | Pins-1::GFP | 100 | 0 | 0 | 326 | 100 | 0 | 0 | 264 |
| daf-2(e1370) | none | 94.7 | 5.3 | 0 | 418 | 0 | 96.8 | 3.2 | 539 |
| daf-2(e1370) | Pins-1::GFP | 96.1 | 1.5 | 2.5 | 203 | 0 | 92.5 | 7.5 | 199 |
| daf-2(e1365) | none | 100 | 0 | 0 | 773 | 0 | 100 | 0 | 500 |
| daf-2(e1365) | Pins-1::GFP | 100 | 0 | 0 | 357 | 0 | 100 | 0 | 220 |
| daf-7(e1372) | none | 97.8 | 2.2 | 0 | 405 | 0 | 98.8 | 1.2 | 341 |
| daf-7(e1372) | Pins-1::GFP | 97.4 | 2.4 | 0.2 | 580 | 0 | 98.1 | 1.9 | 620 |
| Wild type | Pins-18::GFP | n.d. | n.d. | n.d. | n.d. | 77.5 | 2.5 | 20.0 | 912 |
| Wild type | control | 99.3 | 0 | 0.7 | 1589 | 96.5 | 0 | 3.5 | 1554 |
| Wild type | ins-9 | 98.6 | 0 | 1.4 | 355 | 95.1 | 0 | 4.9 | 550 |
| Wild type | ins-22 | 99.6 | 0 | 0.4 | 1895 | 99.3 | 0 | 0.7 | 1828 |
| Wild type | ins-19/31 | 90.1 | 0 | 9.9 | 1358 | 76.8 | 0 | 23.2 | 1238 |
| Wild type | ins-18 | 82.6 | 13.9 | 3.5 | 1643 | 37.3 | 58.3 | 4.4 | 1695 |
| daf-2(e1365) | control | 98.8 | 0.2 | 1.0 | 654 | 0.6 | 98.0 | 1.4 | 783 |
| daf-2(e1365) | ins-9 | 98.6 | 0 | 1.4 | 436 | 0 | 76.0 | 24.0 | 555 |
| daf-2(e1365) | ins-22 | 98.8 | 0 | 7.1 | 862 | 0 | 98.1 | 1.9 | 1052 |
| daf-2(e1365) | ins-19/31 | 85.5 | 0 | 14.5 | 553 | 0 | 81.8 | 18.2 | 511 |
| daf-2(e1365) | ins-18 | 32.8 | 65.9 | 1.2 | 1127 | 0 | 99.7 | 0.3 | 1472 |
(n.d.) Not determined.
Total number of transgenic animals scored. Numbers represent the summary of at least two separate trials from two independent or a single (control, Pins-1::GFP, and ins-9) transgenic line of each genotype.
Includes embryonic and L1 larval arrest.
Table 2.
Enhancement of dauer arrest by heat shock promoter::ins-1 transgenes
| Dauer constitutive mutation
|
Transgenea
|
Phenotype at 20°C (%)
|
Phenotype at 25°C (%) after heat shock
|
Phenotype at 25°C (%)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dauer
|
L1 arrest
|
N
|
Dauer
|
L1 arrest
|
N
|
Dauer
|
L1 arrest
|
N
|
||
| Wild type | epIs6 | n.d. | n.d. | n.d. | 1.6 | 0 | 248 | n.d | n.d. | n.d. |
| Wild type | epIs7 | n.d | n.d. | n.d. | 0 | 1.9 | 264 | n.d. | n.d. | n.d. |
| Wild type | none | n.d. | n.d. | n.d. | 0 | 0 | 291 | n.d. | n.d. | n.d. |
| daf-2(e1370) | epIs6 | 1.9 | 0 | 209 | 4.5 | 95.5 | 156 | 13.3 | 86.7 | 196 |
| daf-2(e1370) | epIs7 | 13.3 | 0 | 195 | 5.5 | 94.5 | 238 | 6.7 | 93.3 | 194 |
| daf-2(e1370) | none | 0 | 0 | 256 | 84.3 | 15.7 | 229 | 88.8 | 11.2 | 143 |
| daf-2(m41) | epIs6 | 13.1 | 0 | 268 | 100 | 0 | 127 | n.d. | n.d. | n.d. |
| daf-2(m41) | epIs7 | 100 | 0 | 402 | 100 | 0 | 107 | n.d. | n.d. | n.d. |
| daf-2(m41) | none | 1.9 | 0 | 155 | 100 | 0 | 87 | n.d. | n.d. | n.d. |
| daf-7(e1372) | epIs6 | 0 | 0.7 | 293 | 100 | 0 | 90 | n.d. | n.d. | n.d. |
| daf-7(e1372) | epIs7 | 78.6 | 0 | 248 | 100 | 0 | 71 | n.d. | n.d. | n.d. |
| daf-7(e1372) | none | 0 | 0 | 117 | 100 | 0 | 67 | n.d. | n.d. | n.d. |
(n.d.) Not determined.
epIs6 and epIs7 are independent integrated heat shock promoter::ins-1 transgenes.
The INS-1 protein is responsible for the enhancement of the daf-2 mutant phenotype. First, the ins-1 and heat shock promoter∷ins-1 transgenes cause similar induction of dauer arrest, although INS-1 expression is under the control of different promoters (Tables 1 and 2). Second, high gene dosage of a transgene in which green fluorescent protein (GFP) is expressed under the control of the ins-1 promoter (Pins-1∷GFP) (Fig. 4E) has no effect on dauer arrest in any of the genetic backgrounds tested (Table 1), showing that it is not the ins-1 promoter that causes daf-2 enhancement.
Because we expected the DAF-2 ligands, like the insulin and IGF-I ligands, to act as receptor agonists, it was surprising that overexpression of ins-1 antagonizes rather than activates DAF-2 signaling. DAF-2 antagonism caused by increased ins-1 gene dosage could reflect the normal role of INS-1 as an antagonist, or chronic overexpression of a DAF-2 agonist could lead to DAF-2 receptor down-regulation and loss of DAF-2 signaling, similar to the insulin resistance caused by long term exposure of cultured cells to insulin (Garvey et al. 1986). If the antagonism is due to DAF-2 down-regulation, a lower level of ins-1 gene dosage might be expected to have the opposite effect and suppress the daf-2 mutant phenotype at the restrictive temperature. However, a tenfold decrease in ins-1 gene dosage continues to show an antagonistic effect on daf-2(e1365) (Fig. 4B), consistent with INS-1 action as an antagonist at all gene dosages studied. In addition, at the restrictive temperature of 26°C, daf-2(e1365) is not suppressed by any of the lower gene dosage ins-1 transgenes (data not shown). These data suggest that INS-1 is a DAF-2 antagonist.
ins-1 and heat shock promoter::ins-1 transgenes also enhance dauer arrest in animals bearing a null mutation in daf-7 (see Materials and Methods), which encodes the ligand for a transforming growth factor-β (TGF-β) pathway that acts synergistically and in parallel to the DAF-2 pathway (Tables 1 and 2). Because the daf-7 mutation that is enhanced by ins-1 is a null allele, and because mutations in the daf-7 and daf-2 pathways are synergistic, the enhancement of dauer arrest by ins-1 overexpression in the daf-7(e1372) background may be mediated by enhancement of daf-2.
To investigate the effect of decreased ins-1 function, we isolated ins-1(nr2091), which deletes the second and third exons of ins-1, encoding the B and A chains, respectively (Fig. 4E). ins-1(nr2091) does not enhance or suppress dauer arrest in wild-type, daf-2, or daf-7 genetic backgrounds (Table 3). In addition, ins-1(nr2091) has no effect on wild-type life span (Fig. 4C), which is dramatically lengthened by mutations in daf-2 and other components of the daf-2 pathway (Kenyon et al. 1993; Larsen et al. 1995). Thus, although overexpression of ins-1 is sufficient to enhance dauer arrest in these backgrounds, ins-1 function is not required for dauer arrest or wild-type life span. Given that ins-1 is 1 of 37 insulin superfamily members, redundancy is possible.
Table 3.
Deletion of ins-1 does not affect dauer arrest
| Genotype
|
Phenotype at 20°C (%)
|
Phenotype at 26°C (%)
|
||||||
|---|---|---|---|---|---|---|---|---|
| L4 larvae and adult
|
Dauer
|
Other
|
Na
|
L4 larvae and adult
|
Dauer
|
Other
|
Na
|
|
| Wild type | 100 | 0 | 0 | 1501 | 99.2 | 0 | 0.8 | 1468 |
| ins-1(nr2091) | 99.8 | 0 | 0.2 | 1237 | 100 | 0 | 0 | 1762 |
| daf-2(e1370) | 86.8 | 7.5 | 5.7 | 967 | 0 | 100 | 0 | 792 |
| daf-2(e1370); ins-1(nr2091) | 91.9 | 6.3 | 1.8 | 2127 | 0 | 100 | 0 | 1109 |
| daf-2(e1365) | 99.1 | 0 | 0.9 | 1169 | 0.1 | 99.9 | 0 | 1091 |
| daf-2(e1365); ins-1(nr2091) | 99.0 | 0 | 1.0 | 1215 | 0 | 100 | 0 | 1218 |
| daf-7(e1372) | 42.3 | 52.2 | 5.5 | 1856b | 0 | 99.8 | 0.2 | 578 |
| daf-7(e1372; ins-1(nr2091)) | 37.0 | 58.3 | 4.7 | 1545b | 0 | 93.8 | 6.2 | 533 |
Total number of animals scored. Numbers represent the summary of at least two separate experiments with each genotype, except as noted.
Numbers represent the summary of at least two separate trials from three independent outcrossed lines of this genotype. During the course of this project we found a high degree of variability in the percentage of dauers formed at 20°C by daf-7 strains from different sources. Combining data from three independent isolates of each genotype demonstrated that variability among the lines was not a result of the ins-1 mutation.
ins-18, with a C peptide, antagonizes daf-2 signaling
To determine whether other C. elegans ins gene family members also genetically interact with the daf-2 pathway, representative members of each ins gene type were studied. Transgenic animals carried the particular ins gene genomic region from the next gene upstream to the next downstream gene.
One other ins gene is predicted to include a C peptide, ins-18. Like high gene dosage of ins-1, high gene dosage of ins-18 induces dauer arrest in wild-type animals at 26°C and enhances dauer arrest in daf-2(e1365) at 20°C (Table 1; Fig. 4).
In contrast to the dauer-enhancing gene activity of high ins-1 and ins-18 gene dosage, increased gene dosage of other ins genes without predicted C peptide cleavage sites, ins-9, ins-22, ins-31, and ins-19, does not affect dauer arrest in wild-type or in daf-2(e1365) (Table 1). ins-9, although lacking predicted C peptide cleavage sites, encodes, like ins-1, a type β protein. In addition, ins-9 is only expressed in neurons implicated in regulation of dauer arrest (see below). ins-22 encodes a type α protein, making it highly diverged from ins-1. ins-31 was studied because of its unusual triplet structure and ins-19, encoding a type γ protein, was included with ins-31 because the two genes are transcribed in opposite directions with only 600 bp between them. High ins-9 gene dosage causes some embryonic and L1 larval arrest and high ins-31/19 gene dosage causes some L1 arrest. These other phenotypes may be mediated by daf-2, because some daf-2 alleles exhibit a similar early arrest phenotype. However, among the ins genes tested, only ins-1 and ins-18 cause dauer arrest.
Expression patterns of ins genes
We constructed gene fusions of the upstream promoter and enhancer regions for 15 of the ins genes, including ins-1, ins-18, ins-9, and ins-22, to the coding region of GFP. These ins genes are expressed primarily in subsets of neurons throughout most of the life cycle (Fig. 5A–D; Table 4). All ins genes tested except ins-11 are expressed in at least some amphid sensory neurons, on the basis of the positions and morphologies of the GFP-expressing processes and cell bodies (White et al. 1986). ins-1 and ins-9 are expressed in the amphid sensory neurons ASI and ASJ (Fig. 5A,C), which regulate dauer arrest (Bargmann and Horvitz 1991). ins-1 is also expressed in ASH and three other unidentified neurons in the nerve ring, as well as NSM (Fig. 5A,C). Whereas ins-9 is expressed exclusively in amphid sensory neurons, ins-1 is also expressed in other neurons, intestine, and vulval muscles. Most ins genes are also expressed in a small number of other neurons. Although we have not yet unambiguously identified the specific neurons expressing many of the reporter constructs, most ins genes are expressed in distinct but overlapping sets of neurons. Several ins genes are also expressed in non-neuronal tissues, including the hypodermis (epidermis), vulva, and pharynx (Fig. 5D; Table 4). Because the GFP reporter constructs that were tested utilize only promoter and enhancer elements located upstream of the ins gene coding regions (see Materials and Methods), and are expressed from extrachromosomal arrays, the observed patterns may not accurately display the entire expression pattern of the endogenous genes.
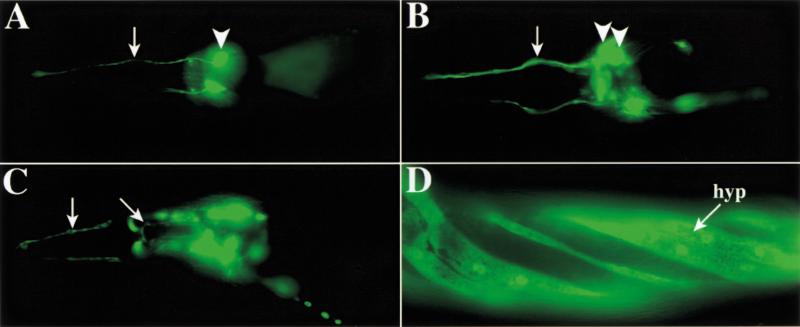
Figure 5.
Spatial expression of representative ins genes. All animals are adults oriented with anterior to the left. (A) GFP expression under control of the ins-9 promoter in a single bilaterally symmetric amphid neuron process (arrow) and cell body (arrowhead). (B) GFP expression driven by the ins-22 promoter in multiple amphid cell bodies (arrowheads mark two) and fasciculated sensory processes (arrow). (C) ins-1∷GFP expression in amphid neurons (vertical arrow marks sensory processes) and pharyngeal neurons (diagonal arrow marks cell process). (D) GFP expression under control of the ins-4 promoter in the hypodermis (hyp; or epidermis) at the surface of the animal. Regions with reduced fluorescence overlie positions of the body wall muscles, in which the hypodermal layer is much thinner. The hypodermis is helically twisted because of the transformation marker rol-6. GFP expression in neurons is not visible in this focal plane.
Table 4.
Summary of GFP expression under the control of representative ins promoters
| Geneb
|
Stagesa
|
Tissues
|
|||||
|---|---|---|---|---|---|---|---|
| Embryo
|
Larvae
|
Adult
|
Nerve ringc
|
Sensory neuronsd
|
Other neuronsd
|
Non-neuronal tissuese
|
|
| ins-1 | 4 | L1–L4 | + | ●●● | a | v, la, t, p | vm, in |
| ins-2 | b, 2, 4 | L1–L3 | − | ●●● | a, l | v, t | ph, vul |
| ins-3 | 4 | L1–L4 | + | ●● | a, l | v, la, d | |
| ins-4 | 2, 4 | L1–L4 | + | ●● | a, l | v, d, t | h |
| ins-5 | 2, 4 | L1–L4 | + | ●● | a, l | v, la, t | vul |
| ins-6 | 2, 4 | L1–L4 | + | ● | a, l | v, t | |
| ins-7 | 4 | L1–L4 | + | ●● | a, l | v, t | |
| ins-8 | 4 | L1–L4 | + | ●● | a, l | v, t | vul |
| ins-9 | L1–L4 | + | ● | a | |||
| ins-11 | 4 | L1–L4 | + | ● | l | v, t | |
| ins-18 | 4 | L1–L4 | + | ●● | a | v, t, p | in |
| ins-21 | 4 | L1–L4 | + | ● | a | v, t | |
| ins-22 | 4 | L1–L4 | + | ●●● | a, l | v, la, t | |
| ins-23 | 4 | L1–L4 | + | ● | a, l | v | |
Stages: b, bean-stage embryo; 2, twofold elongated embryo; 4, fourfold elongated embryo; L1, larval stage L1; L2, larval stage L2; L3, larval stage L3; L4, larval stage L4.
No expression was detected from the ins-20 promoter GFP fusion construct.
Indicates expression in more than eight (●●●), in 4–8 (●●), or in two (●) bilaterally symmetric pairs of neurons.
Neurons: a, amphid sensory neurons; l, labial neurons; v, ventral nerve cord neurons; la, neurons with lateral processes and/or cell bodies; d, dorsal nerve cord neurons, observed by dorsal and/or circumferential processes; t, tail neurons; p, pharyngeal neurons.
Non-neuronal tissues: vm, vulval muscle; in, intestine; ph, pharynx; vul, vulva; h, hypodermis.
Overexpression of human insulin in C. elegans antagonizes daf-2 signaling
DAF-2 is 36% identical to the human insulin receptor in the ligand binding domain (Kimura et al. 1997). Given the orthologous relationship between DAF-2 and the mammalian insulin receptor, and the sequence similarity between INS-1 and human insulin, we tested whether human insulin can genetically interact with daf-2. Human insulin was expressed in the same cells that express ins-1, using the 5′ flanking region of ins-1 to drive a human insulin cDNA (Fig. 4E). This ins-1 regulatory region drives the expression of GFP in a variety of neural and non-neural tissues. Human insulin expression causes a low level of dauer arrest in wild type at 26°C and strongly enhances dauer arrest in the daf-2(e1365) and daf-7(e1372) backgrounds at 20°C (Table 1; Fig. 4A). Thus human insulin can also engage the worm insulin signaling pathway.
Dauer arrest induced by high ins gene dosage is partially suppressed by daf-16
To establish which signaling pathways are engage by INS-1 or human insulin to cause dauer arrest, we tested the ability of mutations in the insulin and parallel TGF-β signaling pathways to suppress their dauer constitutive phenotypes at 26°C. The major output of DAF-2 signaling is the negative regulation of the DAF-16/fork head transcription factor. Mutations in daf-16 fully suppress the dauer arrest phenotype of daf-2 and other dauer constitutive mutations in the daf-2 pathway, but do not suppress mutations in the daf-7 TGF-β pathway (Gottlieb and Ruvkun 1994). Conversely, mutations in daf-3, which encodes a SMAD protein acting in the daf-7 pathway (Patterson et al. 1997), suppress the phenotypes of dauer constitutive mutations in the daf-7 pathway, but not the daf-2 pathway (Vowels and Thomas 1992). Null mutations in either daf-16 or daf-3 incompletely suppress the dauer arrest caused by high gene dosage of ins-1 or human insulin (Table 5), indicating that effects of the ins-1 and human insulin transgenes are partially dependent on the daf-2 and daf-7 pathways. Partial suppression of the dauer arrest induced by high ins-1 or human insulin by mutations in daf-16 indicates that the DAF-2/DAF-16 pathway is a target of high ins-1 and human insulin gene dosage. Another indication that these insulin transgenes affect daf-2 signaling is that they cause a small but reproducible increase in adult life span in the wild-type background (Fig. 4C). Mutations in the daf-2 pathway but not in the daf-7 pathway, cause increases in life span (Larsen et al. 1995). However, the partial suppression of high ins-1 and human insulin-induced dauer arrest by daf-16 mutations and the partial suppression by daf-3 suggests that these hormones either engage more than the DAF-2 receptor, or that there are other outputs from DAF-2 besides the major output to DAF-16. In fact, there is precedence for daf-16 independent outputs of daf-2 at high temperature (Ailion and Thomas 2000). High temperature may induce higher expression or release of INS-1, for example. The partial suppression of high ins-1 or human insulin dauer arrest by daf-3 could be due to the synergy between daf-7 and daf-2 pathway mutations; activation of daf-7 pathway signaling in a daf-3 mutant may partially compensate for the decline in daf-2 pathway signaling caused by high ins-1 or human insulin gene dosage. Animals with mutations in both daf-16 and daf-3 show similar rates of dauer arrest when carrying the ins-1 or human insulin transgene as with a control transgene (Table 5), suggesting that together the daf-2 and daf-7 pathways mediate all of the response to high gene dosage of ins-1 and human insulin.
Table 5.
Modulation of dauer arrest induced by ins-1 and human insulin by daf-2 and daf-7 pathway mutants
| Dauer defective mutation
|
Transgene
|
Phenotype at 26°C (%)a
|
|||
|---|---|---|---|---|---|
| L3 and L4 larvae and adult
|
Dauer
|
Otherb
|
Nc
|
||
| Wild type | control | 95.4 | 0 | 4.6 | 521 |
| daf-16(mgDf47) | control | 95.0 | 0 | 5.0 | 380 |
| daf-3(mgDf90) | control | 90.6 | 0 | 9.4 | 416 |
| daf-16(mgDf47); daf-3(mgDf90) | control | 79.8 | 16.5 | 3.6 | 689 |
| Wild type | ins-1d | 84.1 | 12.5 | 3.4 | 615 |
| daf-16(mgDf47) | ins-1 | 82.0 | 7.7 | 10.3 | 534 |
| daf-3(mgDf90) | ins-1 | 89.3 | 6.5 | 4.2 | 742 |
| daf-16(mgDf47); daf-3(mgDf90) | ins-1 | 84.6 | 9.1 | 6.3 | 760 |
| Wild type | human insulind | 54.3 | 35.4 | 10.3 | 477 |
| daf-16(mgDf47) | human insulin | 83.6 | 10.0 | 6.4 | 659 |
| daf-3(mgDf90) | human insulin | 85.8 | 2.7 | 11.5 | 550 |
| daf-16(mgDf47); daf-3(mgDf90) | human insulin | 80.3 | 2.9 | 16.8 | 720 |
To maximize the synchronicity of the progeny, parents were allowed to lay eggs for 2 h at 25°C.
This category includes animals that could not be classified as either dauers or L3 and older at the time of scoring, due to less synchronous growth of the progeny than in the other sets of experiments.
Total number of transgenic animals scored after 50 h at 26°C. Numbers represent the summary of at least two separate trials from a single transgenic line of each genotype.
In this set of experiments, we found that both transgenes caused a somewhat higher level of dauer formation in the wild-type background than the experiments shown in Table 2, perhaps due to fluctuations in incubator temperature between the two sets.
Discussion
The C. elegans insulin gene family is much larger and more diverse than the known human insulin superfamily, with sequence and structural deviations that go well beyond those found in previously recognized members of the insulin superfamily. Although large families of insulin-related genes have been described in Bombyx mori (Kondo et al. 1996), these genes are nearly identical to each other, lack the characteristic intron/exon structure of the superfamily, and are probably the result of a relatively recent retrotransposon-mediated gene duplication. In the C. elegans ins gene family, there is wide sequence divergence within each class with respect to the presence of C peptides and other domains, putative post-translational processing sites, and amino acid insertions or deletions in the loops connecting the predicted α-helices. However, all of these insulin-like genes bear structural features of the insulin superfamily, and most have an intron between the B and A peptide coding regions, a signature of the insulin family as defined from vertebrate superfamily studies. Therefore, this stereotypical gene structure must have been present in the common ancestors of nematodes and vertebrates, over 600 million years ago (Smit et al. 1998).
These divergent insulins could not have been detected by use of standard molecular techniques or conventional search algorithms. In addition, their detection depended on analysis of the full genome sequence rather than analysis of EST sequences. Of 37 genes in the ins family, only 15 were predicted to be genes by GeneFinder. This work reinforces the need for multiple search tools, including structural modeling, to determine the true extent of a family for which only a few members are known. For example, the insulin superfamily has traditionally been restricted to proteins with invariant number and spacing of cysteine residues within the A and B peptides. We show that the location of cysteines relative to α-helices may be a more relevant criterion, because additions and deletions to loop regions that change the spacing of cysteines in the primary structure (Fig. 2) can be accommodated without changing the insulin fold. It is clear that the traditional criteria for insulin-related molecules must be broadened to include the novel structural and sequence motifs encompassed by the C. elegans INS proteins.
Many of the C. elegans ins genes are organized into clusters of three to seven genes (Fig. 3). The clustering, tandem arrangement, and closer sequence similarity between members of a given cluster suggests that these clusters arose relatively recently by gene duplication. Linked insulin superfamily genes are also found in humans (Crawford et al. 1984; Bell et al. 1985). However, the diversity even within the clusters of C. elegans ins genes suggests that they may be rapidly evolving. DAF-2 signaling controls arrest of the dauer larva, which is specialized to survive harsh conditions. Depending on the environment, survival may be favored by antagonism of daf-2 signaling, leading to dauer arrest, or by activation of daf-2 signaling, which leads to growth and reproduction. Some INS proteins may be DAF-2 agonists, whereas others may be antagonists.
It is known that DAF-2 signaling and a subset of the amphid sensory neurons (Bargmann and Horvitz 1991) are required for the response to a variety of environmental stimuli that regulate dauer arrest, including food, pheromone, and temperature. Thirteen of the fifteen ins genes tested are expressed in amphid neurons. It seems likely that at least some of the INS proteins are released from amphid neurons under conditions of low pheromone and/or high food concentrations and act as ligands for the DAF-2 receptor. A diversity of C. elegans ins gene activities, perhaps in response to specific stimuli, is supported by our finding that ins-1 and ins-18 affect dauer arrest, whereas ins-9 and ins-19/31 have distinct effects on embryonic and early larval growth. In addition to dauer formation, DAF-2 signaling regulates early larval growth, life span, and metabolism. Particular INS proteins or combinations of INS proteins may regulate each of these processes independently of the others. Some of the ins genes may also mediate the other chemosensory and thermosensory functions of amphid neurons. There may also be redundancy in the C. elegans ins gene family. This may account for the failure of extensive genetic screens to identify mutations in these genes, and the lack of a detectable dauer arrest phenotype from the ins-1 knockout mutation.
INS-1 is the C. elegans INS protein most similar to human insulin at the sequence level, with 32% identity over the length of the B and A chains. INS-18 is the only other C. elegans INS protein predicted to contain a C peptide that is proteolytically removed. It may be significant that only ins-1, ins-18, and human insulin, which likely have processed C peptides, enhance dauer arrest in both wild-type and daf-2 mutant backgrounds, whereas ins-9, ins-22, and ins-19/31 do not. Cleavage of the C peptide by proteases that recognize flanking dibasic residues is associated with the loading of insulin into vesicles for regulated secretion. Vesicular insulin is released upon neural and glucose stimulation of pancreatic β cells , in a calcium-dependent manner homologous to synaptic release of neurotransmitter vesicles (Kahn and Weir 1994). ins-1 and ins-18 are expressed in many neuronal cells that may have features in common with β cells, including the ability to process out the C peptide, load-processed INS-1 into vesicles, and regulate the secretion of these hormones on the basis of dauer pheromone and other sensory inputs. Genetic analysis suggests that an unidentified DAF-2 agonist may also be secreted from neurons in a Ca2+-mediated fashion (Ailion et al. 1999).
The enhancement of the daf-2 mutant phenotype by high ins-1 gene dosage suggests that INS-1 can antagonize the function of DAF-2 when overexpressed, which may indicate an endogenous role for INS-1 as a DAF-2 antagonist. Naturally occurring antagonists for insulin/IGF receptors have not been reported. Dose response curves indicate that INS-1 has only antagonistic activity toward DAF-2 (Fig. 4). However, the lack of a dauer arrest phenotype for the ins-1 knockout mutation suggests that if INS-1 is an antagonist, it is redundant, perhaps with the other C. elegans ins genes.
Because high ins-18 gene dosage also enhances the daf-2 mutant phenotype, we predict that INS-18 may also act as a DAF-2 antagonist. In contrast, Kawano and coworkers suggest that INS-18 is a DAF-2 agonist because reducing the function of ins-18 in wild-type animals by use of RNA-mediated interference slightly increased life span (Kawano et al. 2000).
Human insulin is an agonist of the mammalian insulin receptor, but shows only antagonistic regulation of DAF-2. It is possible that human insulin engages the DAF-2 ligand-binding domain in a similar manner to its engagement of the mammalian insulin receptor, but that DAF-2 has changed in evolution to be switched off by ligand engagement rather than on in the case of the mammalian receptors. Alternatively, it is possible that the human insulin, INS-1 and INS-18 ligands, containing C peptides, antagonize DAF-2 signaling because they interfere with normal processing or secretion of the endogenous worm insulins that are agonists of DAF-2. Cell culture studies of DAF-2 signaling in the presence or absence of insulin ligand, will be needed to distinguish between these models.
Despite the diversity among the predicted INS proteins, daf-2 is the only C. elegans gene with homology to known insulin superfamily receptors. Because all of the INS proteins are predicted to adopt a similar tertiary structure, they may all bind to DAF-2. However, insulin and IGF-I (two of the most closely related members of the superfamily) bind only weakly to each other's receptor, with affinities three orders of magnitude lower than for their cognate receptor (Kjeldsen et al. 1991). Given the diversity of the C. elegans ins gene family, it is also possible that some INS proteins act through unidentified classes of receptors. There is evidence that all of the mammalian responses to insulin or IGF-I may not be mediated by the known receptors. For example, specific binding of insulin C peptide to mammalian cells is likely to be mediated by G-protein-coupled receptors (Rigler et al. 1999).
It is likely that undiscovered members of the insulin superfamily exist in vertebrates. The insulin receptor-related receptor gene encodes a close relative of the insulin receptor that is not activated by any of the known human insulin-like ligands (Zhang and Roth 1992; Watt et al. 1993), and there are other suggestions that additional insulin-like genes exist in humans (Villa-Komaroff et al. 1984; Rotwein et al. 1986). Our analysis suggests that the insulin superfamily in all metazoans may be considerably larger and more diverse than recognized previously.
Materials and methods
Strains
Strains used were as follows: wild-type N2 Bristol, daf-16(mgDf47) I, daf-7(e1372) III, daf-2(e1365, e1370, and m41) III, him-5(e1490) V, daf-3(mgDf90) X.
Identification of ins genes
The C. elegans genomic DNA database was searched with queries derived from known vertebrate and invertebrate insulin superfamily proteins. Each candidate insulin-like sequence was evaluated manually, with the aid of the GeneFinder (Eeckman and Durbin 1995) and PSORT (Horton and Nakai 1997) programs, for the ability to produce a predicted mature mRNA encoding a preprohormone with a signal sequence, B peptide, and A peptide. Specifically, candidate genes had most or all of the following features anticipated for proteins that could adopt the insulin fold: (1) spatial arrangement of cysteine residues characteristic of B and A peptides (boxed red residues in Fig. 2); (2) presence of hydrophobic residues in positions expected to contribute to the interface between the B and A peptides (shaded green in Fig. 2); and (3) glycine or other small side chain residues in the position corresponding to the tight turn immediately preceding the B peptide central helix (shaded yellow in Fig. 2).
Cloning and sequencing of ins genes
C. elegans insulin homolog cDNAs were amplified by the PCR from either an oligo(dT) primed mixed-stage cDNA library (Stratagene) or first strand cDNA made from mixed-stage C. elegans RNA, using either gene-specific primers or the spliced leader sequence SL1 (GGTTTAATTACCCAAGTTTGAG) and a gene-specific 3′ primer. For some genes it was necessary to use nested primers. PCR products were subcloned and sequenced. For cDNAs that differed from the gene prediction, clones from two independent PCR reactions were sequenced. The cDNA sequence of one of the genes identified previously (ins-5) (Duret et al. 1998), which was predicted incorrectly to be a pseudogene, revealed two additional nucleotides that change the reading frame of the B chain and show that it encodes a bona fide insulin superfamily protein.
Structural modeling of INS proteins
From sequence alignments of the A and B peptides (Fig. 2), we threaded the sequences of INS-1, INS-4, and INS-21 onto the X-ray crystal structure of porcine insulin (Baker et al. 1988) using Insight II (MSI, La Jolla, CA). By use of the amber forcefield, the starting structure was energy minimized using the steepest descent and conjugate gradient methods. In all cases, formation of the extra disulfide bond was consistent with the preliminary structures, so the additional disulfide was used in further calculations and was followed by dynamics (simulated annealing). A total of 2000 one femtosecond steps at 300°K were followed by 1000 steps at each of the following temperatures: 275, 250, 225, 200, 175, 150, 125, and 100°K and 500 steps at 90, 80, 70, 60, 50, 40, 30, 20, 10, and 0°K. The final structures were the result of further conjugate gradient minimization. Ten structures were calculated for each model. RMS deviations for the 10 calculated structures were 0.99 Å +/−0.26 Å for porcine insulin, 1.11 Å +/−0.38 Å for INS-1, 1.49 Å +/−0.38 Å for INS-4 and 1.80 Å +/−0.53 Å for INS-21. All superpositions were calculated by use of helical residues only.
Construction of ins gene, human insulin, and Pins-1∷GFP transgenic lines
For the ins transgenes, fragments spanning the ins gene region from the next upstream to the next downstream gene, as predicted by the GeneFinder program, were amplified from wild-type genomic DNA. ins-1 was not bounded by an upstream predicted gene. The sizes of the genomic regions used are as follows (upstream sequence/introns and exons/downstream sequence): ins-1, 5.7 kb/1.2 kb/1.3 kb; ins-9, 1.1 kb/0.4 kb/7.1 kb; ins-22, 2.2 kb/0.3 kb/1.6 kb; ins-18, 4.9 kb/2.1 kb/0.8 kb. The ins-19/31 region included 2.4 kb of ins-19 downstream sequence, 0.4 kb of ins-19 coding sequence, 0.6 kb of intervening sequence, 0.7 kb of ins-31 coding sequence, and 1.4 kb of ins-31 downstream sequence.
The human insulin transgene was constructed by use of a PCR fusion strategy. First, three independent PCR reactions were performed. The ins-1 promoter was amplified from genomic DNA by use of a 3′ primer with complementarity to the 5′ end of the human insulin cDNA (provided by H. Goodman, Massachusetts General Hospital). The human insulin cDNA (0.4 kb) was amplified by use of a 5′ primer with complementarity to the ins-1 promoter and a 3′ primer with complementarity to the ins-1 3′ UTR. The ins-1 3′ UTR was amplified with a 5′ primer with complementarity to the human insulin cDNA. The three PCR products were then fused by amplifying with a set of nested primers. The Pins-1∷GFP and Pins-18∷GFP transgenes used in the dauer assays were constructed by use of an analogous two-part fusion approach to amplify and fuse the ins-18 and ins-1 promoters to GFP sequences from pPD95.75 (provided by A. Fire, Carnegie Institute of Washington).
The final PCR products were purified and injected at 20–35 ng/μL with 25 or 50 ng/μL of Pmec-7∷GFP as an injection marker, generating transgenic lines in which the gene of interest and the injection marker are carried on an extrachromosomal array (Mello and Fire 1995). Control transgenic arrays were generated by injecting 20 ng/μL pBluescript-SK+ with Pmec-7∷GFP injection marker. With the exception of the ins-1 and control transgenic arrays in daf-2(e1365), transgenic arrays were generated in one strain background and crossed into the other genetic backgrounds tested. DNA for the ins-1 dose curve was prepared separately and injected into daf-2(e1365) at the indicated approximate concentrations with Pmec-7∷GFP.Pmec-7∷GFP, from which GFP is expressed under the control of the mec-7 promoter, was constructed by replacing the KpnI/SpeI GFP fragment of KP9 with the KpnI/SpeI GFP fragment of pPD95.75 (J.Y. Sze, pers. comm.). KP9 is a GFP derivative of pPD52.102 (J. Kaplan, pers. comm.).
Construction of heat shock promoter∷ins-1 transgenic lines
Plasmid hsp∷INS-1, which drives expression of INS-1 from the heat shock promoter hsp16-2, was constructed by cloning ins-1 genomic DNA (from start to stop codon) into vector pPD99.44 (provided by A. Fire). hsp∷INS-1 was injected into wild-type C. elegans (Mello and Fire 1995) at a concentration of 100 μg/mL plasmid plus 100 μg/mL pRF4 rol-6(d) transformation marker. The extrachromosomal array carrying hsp∷INS-1 and rol-6(d), epEx88, was integrated into the chromosomes by use of X-irradiation to produce two independent insertions, epIs6 and epIs7 (Mello and Fire 1995). Both integrant lines were outcrossed twice, and the insertions were crossed into daf-2 (e1370), daf-2 (m41), and daf-7 (e1372) by selecting the Rol phenotype of the insertion and the Daf-c phenotype of the mutant. For each mutant, two independent lines were established and shown to behave identically at 20 and 25°C.
Assays for dauer arrest
Adults were allowed to lay eggs on plates for several hours at room temperature or the assay temperature, and progeny were scored after 2 d at 26°C or 3 d at 20 and 25°C. Dauers were distinguished by a radially constricted body, darkly pigmented intestine, dauer alae, and a constricted pharynx; partial dauers, which were included in the dauer category for scoring, lacked at least one of these characteristics. For heat shocks, embryos laid over a period of several hours were allowed to hatch overnight at 25°C, shifted as L1 larvae to 33°C for 3 h, and then returned to 25°C and scored on the third day after the egg lay.
Identification of the mutation in daf-7(e1372)
daf-7(e1372) was reported previously as a CCA to CCG substitution, which would not change an amino acid position (Ren et al. 1996). We did not detect such a mutation in multiple independent isolates of daf-7(e1372), but instead found a CAGG, TAT to CAGATAT substitution at a conserved intron splicing donor position in intron 2. This mutation is located in the daf-7 pro-domain coding region upstream of the ligand coding region; the failure to properly splice the daf-7 mRNA within the pro-domain would be expected to cause a null phenotype.
Life span assays
Life span was assayed essentially as described (Paradis et al. 1999). Animals were raised at 25°C, transferred to assay plates at the L4 or young adult stage, and life span was assayed at 26°C. Animals that crawled off the plates during the assay were excluded from calculations.
ins-1(nr2091) isolation
The ins-1(nr2091) deletion mutant was isolated by screening an arrayed library of mutagenized nematodes as described (Liu et al. 1999). The mutant was backcrossed and crossed into daf-2(e1365), daf-2(e1370), and daf-7(e1372) backgrounds. The genotype was confirmed by use of a triplex PCR with three primers, two of which flank the deletion (yielding a PCR product of 1.6 kb on the deleted chromosome), and one located within the deletion (yielding a PCR product of 1.2 kb on the wild-type chromosome). The exact deletion point was confirmed by DNA sequencing. Phenotypic analysis was performed on strains that had been backcrossed at least three times. daf-7(e1372);ins-1(nr2091) strains were generated by outcrossing a twice backcrossed daf-7(e1372);ins-1(nr2091) strain to wild type, homozygosing daf-7(e1372) using the Daf-c phenotype, and isolating three homozygous ins-1(nr2091) lines and three lines that were wild type for ins-1.
ins gene expression analysis
For each of 15 ins genes, putative promoter/enhancer regions were defined in genomic sequence as regions extending from the predicted start codon of each ins gene to the next gene upstream, identified by use of the GeneFinder program. The sizes of the enhancers bounded by upstream genes were 0.6–5.7 kb. For ins-1, ins-2, and ins-21, the putative promoter/enhancer region was more than 6 kb, or not bounded by a clearly defined upstream gene in available sequence, and a 2- to 6-kb segment of upstream region was chosen for amplification. Promoter/enhancer regions were amplified by PCR from either N2 wild-type genomic DNA or cosmid DNA (C. elegans Genome Project) and ligated into pPD117.01 GFP fusion vector (provided by A. Fire). Each construct was injected into wild-type C. elegans (Mello and Fire 1995) at a concentration of 100 μg/mL plasmid plus 100 μg/mL pRF4 rol-6(d) transformation marker. Stably transformed strains were established and examined for fluorescence using an Axioplan microscope (Zeiss). Duplicate constructs were made for each fusion, and one or more transgenic lines that showed high transmission were analyzed further. Duplicate constructs were derived from independent PCR reactions for all genes except ins-4, ins-5, ins-8, ins-9, ins-20, and ins-21. The patterns of GFP expression were found to be identical for all duplicates. Embryos, larvae (L1, L2, L3, L4), and adults were examined for GFP expression, and all expressing cells or tissues were identified by their position and morphology.
Acknowledgments
Research in the Ruvkun laboratory was supported by grants R01AG16636 and R01AG14161 from the NIH and by a fellowship to S.B.P. from the Cancer Research Fund of the Walter Winchell-Damon Runyon Foundation, DRG-1500. We thank G. Duyk, D. Jacobus, S. Quinn, B. Reddy, C. Seidel-Dugan, Y. Liu, P. Delerme, C. Bargmann, A. Hart, C. Wolkow, and members of the Ruvkun laboratory for assistance and discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ruvkun@frodo.mgh.harvard.edu; FAX (617) 726-6893.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.867301.
References
- Ailion M, Thomas JH. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics. 2000;156:1047–1067. doi: 10.1093/genetics/156.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion M, Inoue T, Weaver CI, Holdcraft RW, Thomas JH. Neurosecretory control of aging in Caenorhabditis elegans. Proc Natl Acad Sci. 1999;96:7394–7397. doi: 10.1073/pnas.96.13.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD, et al. The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos Trans R Soc Lond B Biol Sci. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Bedarkar S, Turnell WG, Blundell TL, Schwabe C. Relaxin has conformational homology with insulin. Nature. 1977;270:449–451. doi: 10.1038/270449a0. [DOI] [PubMed] [Google Scholar]
- Bell GI, Gerhard DS, Fong NM, Sanchez-Pescador R, Rall LB. Isolation of the human insulin-like growth factor genes: Insulin-like growth factor II and insulin genes are contiguous. Proc Natl Acad Sci. 1985;82:6450–6454. doi: 10.1073/pnas.82.19.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell TL, Humbel RE. Hormone families: Pancreatic hormones and homologous growth factors. Nature. 1980;287:781–787. doi: 10.1038/287781a0. [DOI] [PubMed] [Google Scholar]
- Brown H, Sanger F, Kitai R. The structure of sheep and pig insulins. Biochem J. 1955;60:556–565. doi: 10.1042/bj0600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullesbach EE, Schwabe C. A novel Leydig cell cDNA-derived protein is a relaxin-like factor. J Biol Chem. 1995;270:16011–16015. doi: 10.1074/jbc.270.27.16011. [DOI] [PubMed] [Google Scholar]
- Chassin D, Laurent A, Janneau JL, Berger R, Bellet D. Cloning of a new member of the insulin gene superfamily (INSL4) expressed in human placenta. Genomics. 1995;29:465–470. doi: 10.1006/geno.1995.9980. [DOI] [PubMed] [Google Scholar]
- Crawford RJ, Hudson P, Shine J, Niall HD, Eddy RL, Shows TB. Two human relaxin genes are on chromosome 9. EMBO J. 1984;3:2341–2345. doi: 10.1002/j.1460-2075.1984.tb02136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Genes & Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- Duret L, Guex N, Peitsch MC, Bairoch A. New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res. 1998;8:348–353. doi: 10.1101/gr.8.4.348. [DOI] [PubMed] [Google Scholar]
- Eddy SR, Mitchison G, Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J Comput Biol. 1995;2:9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- Eeckman FH, Durbin R. ACeDB and macace. In: Epstein HF, Shakes DC, editors. Methods in cell biology. Caenorhabditis elegans: Modern biological analysis of an organism. San Diego, CA: Academic Press; 1995. pp. 583–605. [PubMed] [Google Scholar]
- Fullbright G, Lacy ER, Bullesbach EE. The prothoracicotropic hormone bombyxin has specific receptors on insect ovarian cells. Eur J Biochem. 1997;245:774–780. doi: 10.1111/j.1432-1033.1997.t01-1-00774.x. [DOI] [PubMed] [Google Scholar]
- Garvey WT, Olefsky JM, Marshall S. Insulin induces progressive insulin resistance in cultured rat adipocytes. Sequential effects at receptor and multiple postreceptor sites. Diabetes. 1986;35:258–267. doi: 10.2337/diab.35.3.258. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: Genetically interacting genes controlling dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire FM, Chomiki N, Kachinskas D, Warden CH. Cloning and developmental regulation of a novel member of the insulin-like gene family in Caenorhabditis elegans. Biochem Biophys Res Commun. 1998;249:385–390. doi: 10.1006/bbrc.1998.9164. [DOI] [PubMed] [Google Scholar]
- Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Intell Systems Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Kahn CR, Weir GC. Joslin's diabetes mellitus. Philadelphia, PA: Lea & Febiger; 1994. [Google Scholar]
- Kawano T, Ito Y, Ishiguro M, Takuwa K, Nakajima T, Kimura Y. Molecular cloning and characterization of a new insulin/IGF-like peptide of the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;273:431–436. doi: 10.1006/bbrc.2000.2971. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:404–405. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kjeldsen T, Andersen AS, Wiberg FC, Rasmussen JS, Schaffer L, Balschmidt P, Moller KB, Moller NP. The ligand specificities of the insulin receptor and the insulin-like growth factor I receptor reside in different regions of a common binding site. Proc Natl Acad Sci. 1991;88:4404–4408. doi: 10.1073/pnas.88.10.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koman A, Cazaubon S, Couraud PO, Ullrich A, Strosberg AD. Molecular characterization and in vitro biological activity of placentin, a new member of the insulin gene family. J Biol Chem. 1996;271:20238–20241. doi: 10.1074/jbc.271.34.20238. [DOI] [PubMed] [Google Scholar]
- Kondo H, Ino M, Suzuki A, Ishizaki H, Iwami M. Multiple gene copies for bombyxin, an insulin-related peptide of the silkmoth Bombyx mori: Structural signs for gene rearrangement and duplication responsible for generation of multiple molecular forms of bombyxin. J Mol Biol. 1996;259:926–937. doi: 10.1006/jmbi.1996.0370. [DOI] [PubMed] [Google Scholar]
- Lagueux M, Lwoff L, Meister M, Gotzene F, Hoffmann JA. cDNAs from neurosecretory cells of brains of Locusta migratoria (Insecta, Orthoptera) encoding a novel member of the superfamily of insulins. Eur J Biochem. 1990;187:249–254. doi: 10.1111/j.1432-1033.1990.tb15302.x. [DOI] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9:859–867. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone EA, Thomas JH. A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics. 1994;136:879–886. doi: 10.1093/genetics/136.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald N, Murray-Rust J, Blundell T. Structure-function relationships of growth factors and their receptors. Br Med Bull. 1989;45:554–569. doi: 10.1093/oxfordjournals.bmb.a072342. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Murray-Rust J, McLeod AN, Blundell TL, Wood SP. Structure and evolution of insulins: Implications for receptor binding. BioEssays. 1992;14:325–331. doi: 10.1002/bies.950140507. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum H, Ruvkun G. The Fork head transcription factor DAF-16 tranduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Storch MJ, Anderson RG, Vassalli JD, Perrelet A. Proteolytic maturation of insulin is a post-Golgi event which occurs in acidifying clathrin-coated secretory vesicles. Cell. 1987;49:865–868. doi: 10.1016/0092-8674(87)90624-6. [DOI] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes & Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes & Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GI, Koweek A, Wong A, Liu Y, Ruvkun G. The DAF-3 Smad protein antagonizes TGF-β-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes & Dev. 1997;11:2679–2690. doi: 10.1101/gad.11.20.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren PF, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Rigler R, Pramanik A, Jonasson P, Kratz G, Jansson OT, Nygren P, Stahl S, Ekberg K, Johansson B, Uhlen S, et al. Specific binding of proinsulin C-peptide to human cell membranes. Proc Natl Acad Sci. 1999;96:13318–13323. doi: 10.1073/pnas.96.23.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderknecht E, Humbel RE. The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J Biol Chem. 1978a;253:2769–2776. [PubMed] [Google Scholar]
- ————— Primary structure of human insulin-like growth factor II. FEBS Lett. 1978b;89:283–286. doi: 10.1016/0014-5793(78)80237-3. [DOI] [PubMed] [Google Scholar]
- Rotwein P, Naylor SL, Chirgwin JM. Human insulin-related DNA sequences map to chromosomes 2 and 11. Somat Cell Mol Genet. 1986;12:625–631. doi: 10.1007/BF01671947. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science. 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Satake S, Masumura M, Ishizaki H, Nagata K, Kataoka H, Suzuki A, Mizoguchi A. Bombyxin, an insulin-related peptide of insects, reduces the major storage carbohydrates in the silkworm Bombyx mori. Comp Biochem Physiol B Biochem Mol Biol. 1997;118:349–357. doi: 10.1016/s0305-0491(97)00166-1. [DOI] [PubMed] [Google Scholar]
- Schwabe C, McDonald JK. Relaxin: A disulfide homolog of insulin. Science. 1977;197:914–915. doi: 10.1126/science.887933. [DOI] [PubMed] [Google Scholar]
- Smit AB, Vreugdenhil E, Ebberink RHM, Geraerts WPM, Klootwijk J, Joosse J. Growth-controlling molluscan neurons produce the precursor of an insulin-related peptide. Nature. 1988;331:535–538. doi: 10.1038/331535a0. [DOI] [PubMed] [Google Scholar]
- Smit AB, Spijker S, Van Minnen J, Burke JF, De Winter F, Van Elk R, Geraerts WP. Expression and characterization of molluscan insulin-related peptide VII from the mollusc Lymnaea stagnalis. Neuroscience. 1996;70:589–596. doi: 10.1016/0306-4522(95)00378-9. [DOI] [PubMed] [Google Scholar]
- Smit AB, van Kesteren RE, Li KW, Van Minnen J, Spijker S, Van Heerikhuizen H, Geraerts WP. Towards understanding the role of insulin in the brain: Lessons from insulin-related signaling systems in the invertebrate brain. Prog Neurobiol. 1998;54:35–54. doi: 10.1016/s0301-0082(97)00063-4. [DOI] [PubMed] [Google Scholar]
- Thacker C, Peters K, Srayko M, Rose AM. The bli-4 locus of Caenorhabditis elegans encodes structurally distinct kex2/subtilisin-like endoproteases essential for early development and adult morphology. Genes & Dev. 1995;9:956–971. doi: 10.1101/gad.9.8.956. [DOI] [PubMed] [Google Scholar]
- Villa-Komaroff L, Gonzalez A, Song HY, Wentworth B, Dobner P. Novel insulin-related sequences in fetal brain. Adv Exp Med Biol. 1984;181:65–86. doi: 10.1007/978-1-4684-4868-9_7. [DOI] [PubMed] [Google Scholar]
- Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;139:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt VM, Shier P, Chan J, Petrisor BA, Mathi SK. IRR: A novel member of the insulin receptor family. Adv Exp Med Biol. 1993;343:125–132. doi: 10.1007/978-1-4615-2988-0_13. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans Roy Soc (London) 1986;B314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Zhang B, Roth RA. The insulin receptor-related receptor. Tissue expression, ligand binding specificity, and signaling capabilities. J Biol Chem. 1992;267:18320–18328. [PubMed] [Google Scholar]