Abstract
Theiler's murine encephalitis viruses (TMEV) are divided into two subgroups based on their neurovirulence. Persistent strains resemble Theiler's original viruses (referred to as the TO subgroup), which largely induce a subclinical polioencephalomyelitis during the acute phase of the disease and can persist in the spinal cord of susceptible animals, inducing a chronic demyelinating disease. In contrast, members of the neurovirulent subgroup cause an acute encephalitis characterized by the rapid onset of paralysis and death within days following intracranial inoculation. We report herein the characterization of a novel neurovirulent strain of TMEV, identified using pyrosequencing technology and referred to as NIHE. Complete coverage of the NIHE viral genome was obtained, and it shares <90% nucleotide sequence identity to known TMEV strains irrespective of subgroup, with the greatest sequence variability being observed in genes encoding the leader and capsid proteins. The histopathological analysis of infected brain and spinal cord demonstrate inflammatory lesions and neuronal necrosis during acute infection with no evidence of viral persistence or chronic disease. Intriguingly, genetic analysis indicates the putative expression of the L* protein, considered a hallmark of strains within the persistent subgroup. Thus, the identification and characterization of a novel neurovirulent TMEV strain sharing features previously associated with both subgroups will lead to a deeper understanding of the evolution of TMEV strains and new insights into the determinants of neurovirulence.
INTRODUCTION
Theilioviruses have positive-sense, single-stranded RNA genomes and are members of the Picornaviridae family and the Cardiovirus genus. Several serotypes have been identified, causing disease in rodents, such as Theiler's murine encephalomyelitis virus (TMEV) (36, 37) and rat Theiler-like virus (RTV) (28), and humans, such as Vilyuisk human encephalomyelititis virus (VHEV) (6) and Saffold viruses (7, 13, 44). TMEV is an enteric pathogen that follows the fecal-oral route of infection; however, when inoculated intracranialy (IC), it can cause severe pathology in susceptible mouse strains. As a natural mouse pathogen, TMEV provides a valuable experimental model for understanding host-pathogen interactions. The TMEV serotype is divided into two subgroups based on the pathogenesis of the disease. Members of the TO subgroup, so named because they resemble Theiler's original strains (36), cause a biphasic infection. The early phase is an acute poliomyelitis characterized by viral replication in the neurons of the brain (1, 18). During the late phase of infection, the virus progresses from the gray to the white matter of the spinal cord, where, depending on the genetics of the mouse strain (4), it can persist for the life of the animal within various cell types (2, 21), causing in some instances chronic demyelination (19). In contrast, the second subgroup, consisting of the GDVII and FA strains, are neurovirulent, and their pathology is characterized by an acute and rapidly fatal encephalitis (37). These viruses replicate mainly in the neurons of the brain, and for the few animals that survive primary infection, there does not appear to be viral persistence in the central nervous system (CNS) (18). Both neurovirulent strains were isolated in 1937 by Theiler and Gard; however, full-length sequences and infectious clones have been generated only for the GDVII strain. For the FA strain, only partial genomic sequence covering the 5′-untranslated region (5′UTR) through the end of the P1 region are available (GenBank accession no. U32924 and M80883). Within these 3,262 bp, there is 99.7% identity to the GDVII strain, which has led to speculation that GDVII and FA actually are separate isolates of the same strain due to the time and place of isolation (27).
Here, we report the phenotype and genetic analysis of the complete genome of a novel neurovirulent strain of TMEV that we have designated NIHE based on its isolation at the National Institute of Hygiene and Epidemiology in Hanoi, Vietnam. We demonstrate that the kinetics of disease and the phenotype of infected animals mimic those published for GDVII and FA strains. Moreover, histopathological analyses demonstrate that tissue and cellular tropism in the CNS are consistent with those observed following the inoculation of neurovirulent strains, including the inability to persist past the acute phase in the small percentage of inoculated animals that do survive. Phylogenetic analysis reveals NIHE to be less divergent from the TO strains than GDVII. Interestingly, we present evidence for the presence of an alternate open reading frame (ORF) which is predicted to encode the full-length L* protein, facilitated by a canonical AUG start sequence and previously shown to be present only in the TO subgroup strains (15). These data illustrate the need for continued investigation to establish the genetic and host immune determinants of neurovirulence and the evolution of viral persistence in this family of viruses.
MATERIALS AND METHODS
Ethics statement.
All animal experiments described in the current study were conducted at the Institut Pasteur according to the European Union guidelines for the handling of laboratory animals The protocol was approved by the Institut Pasteur animal care and use committee. Inoculations were performed under anesthesia, and all efforts were made to minimize animal suffering.
Mice and infections.
Outbred Swiss mice were bred and maintained in the animal facility of the National Institute of Hygiene and Epidemiology in Hanoi, Vietnam. This colony originated from animals imported from France. The initial phenotype was observed in 2008 following the low-number serial passage of brain homogenate from apparently healthy donors. Samples were transferred to the Institut Pasteur in Paris for further characterization. Female 5- to 7-week-old SJL and C57BL/6 mice were purchased from Charles River Laboratories and housed in our SPF animal facility at the Institut Pasteur in Paris. The handling of mice and experimental procedures were performed in accordance with institutional guidelines for animal care. Inoculations were performed in mice via the intracranial injection of 30 to 35 μl of mouse brain homogenates. Specifically, brains from infected animals were prepared as 10% homogenates by mechanical disruption in sterile serum-free Dulbecco's modified essential medium (DMEM). Homogenates were clarified by centrifugation (10,000 × g for 20 min). Control animals received 30 to 35 μl of clarified supernatants from naïve animals at the indicated dilutions. All dilutions were prepared in sterile phosphate-buffered saline (PBS) or DMEM.
RNA isolation and preparation for 454 sequencing.
Clarified supernatants were generated from the brains of animals demonstrating a disease phenotype. Aliquots of supernatant were combined with the AVL buffer supplied in the QIAamp viral RNA Minikit (Qiagen) and processed according to the manufacturer's specifications. Aliquots were combined, and 454 sequencing was performed by Eurofins MWG Operon (Ebersberg, Germany).
BLAST analysis and bioinformatics.
FASTA files containing individual sequencing reads were obtained from the sequencing reactions. The nonredundant protein (nr) and nucleotide (nt) sequence databases were downloaded from NCBI and updated August 2010 prior to query. All sequences were queried against the nr/nt databases using BLAST+ (5). Metagenomic analysis was performed using MEGAN software (version 3.9), which resulted in the grouping of sequences according to phylogeny (11). The de novo assembly of sequences greater than 80 bp was performed using the local alignment algorithm within CodonCode Aligner software (version 3.5), and the resulting consensus sequence was checked for inserted/deleted sequences that can be an artifact of 454 sequencing homopolymeric sequences. Twenty-three sites were manually confirmed and corrected throughout the sequence. The corrected consensus sequence was used in all subsequent comparative analyses.
Histology and immunohistochemistry.
Infected or control animals were anesthetized and perfused with PBS followed by 4% paraformaldehyde in PBS. Brains and spinal cords were dissected and fixed in 4% paraformaldehyde overnight at 4°C. Tissues were embedded in paraffin by following established procedures, and 5-μm serial sections from the brain and spinal cord were stained with hematoxylin-eosin (HE). Cross-reactivity with TMEV antigens was tested by immunohistochemistry on paraffin sections incubated with a primary rabbit polyclonal antibody raised against Theiler's virus capsid proteins (kindly provided by J. F. Bureau and M. Brahic). Slides were counterstained with hematoxylin.
Phylogenetic analysis and alignment.
Nucleotide and associated protein sequences for the following viruses were obtained from GenBank (data concerning three domains were collected: protein sequences coding for the 3D polymerase [3Dpol], P1 [protein 1], and VP1 [viral protein 1]): GDVII (M20562 and NC_001366), FA (U32924 and M80883 combined), DA (M20301), Yale (EU723238), BeAn8386 (M16020), TOB15 (EU718732), VIE415HTR (EU718733), TO4 (U33045 and M80885 combined), WW (M80889 and U33046 combined), Mengo virus (DQ294633), encephalomyocarditis virus (NC_001479), porcine encephalomyocarditis (DQ517424), Saffold virus (FM207487, NC_009448, and EF165067), cardiovirus D (EU681176), human TMEV-like cardiovirus (NC_010810 and EU376394), RTV NGS910 (AB090161), RTV-1 (EU542581 and EU815052), and Vilyuisk (EU723237, M80888, and M94868 combined). Multiple-sequence alignment was performed using the Muscle algorithm implemented within SeaView (version 4.2.4). The 3Dpol data set included 20 taxa and 1,404 characters, the P1 data set had 23 taxa for 2,617 nucleotides, and the VP1 matrix involved 23 taxa for 868 characters.
Phylogenetic reconstruction.
The phylogenetic reconstructions of three protein-coding sequences (3Dpol, VP1, and V1) from three sets of nucleotides were inferred separately by a maximum-likelihood analysis on matrixes composed of 20, 20, and 23 taxa, respectively. These phylogenetic analyses were implemented by the BEAST software package (version 1.6.1). As this method required a predetermined evolution model estimated on the data set, we specified as one of the models selected by Jmodeltest (30) on each matrix (3Dpol, VP1, and P1). Using the results of Jmodeltest, the three-codon partition model of evolution was used (under the GTR substitution model with a gamma site heterogeneity distribution [GTR+G]), so that each codon position (first, second, and third) could follow its own evolution rate. The Yule speciation process (specified as tree prior) and constant-population-size coalescent models show similar estimates for each data set. The Monte Carlo Markov chain was set to 5,000,000 states for 3Dpol and VP1 data sets and to 10,000,000 for the P1 matrix to reach adequate prior effective sampling size (ESS) values greater than 300 and 2,000, respectively. Trees were annotated with TreeAnnotator (version 1.4.8) and displayed and edited using FigTree (version 1.3.1).
Genetic analysis.
Similarity plots were generated using SimPlot (version 3.5.1) with the following parameters: 200-bp window, 20-bp step, and Kimura 2-parameter distance model (22).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper was submitted to GenBank and has been assigned the accession no. HQ652539.
RESULTS
Identification of a novel neurovirulent strain of TMEV.
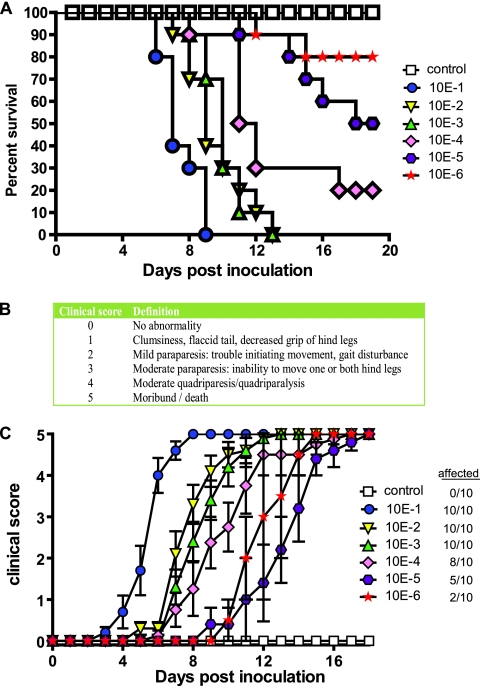
While performing animal experiments involving the serial passage of clarified lysate from mouse brain homogenates, we observed the appearance of a distinct disease phenotype. Following inoculation, several animals displayed a flaccid tail and an abnormal gait, which progressed to unilateral or bilateral limb paralysis 9 to 10 days postinoculation. Following the next passage, 100% of inoculated animals succumbed to infection; continued serial passage has demonstrated consistent lethality. To determine the kinetics of death, groups of C57BL/6 mice (n = 10) were inoculated IC with 10× dilutions (10−1 to 10−6) of infected mouse brain homogenate (IMBH), and survival was monitored (Fig. 1A). Animals receiving a 10−1 dilution had a mean survival time of 7 days, whereas animals receiving 10−2, 10−3, and 10−4 dilutions had mean survival times of 9, 10, and 11.5 days, respectively. The 50% lethal dose (LD50) was determined to be a 10−5 dilution of IMBH. Control animals received a 10−1 dilution of 10% brain homogenate derived from naïve animals with no apparent phenotype (>45 days postinoculation). To quantitatively describe the disease progression in mice, a clinical scoring method was established (Fig. 1B), and disease progression was monitored daily (Fig. 1C). To accurately describe disease progression, only symptomatic animals were included; the number of symptomatic animals per group is indicated in the figure legend.
Fig. 1.
Survival and kinetics of disease progression for animals inoculated with NIHE. (A) Five-week-old C57BL/6 mice were inoculated IC with 10× dilutions (10−1 to 10−6) of infected mouse brain homogenates (n = 10 per group); control mice (white boxes) received 10−1 brain homogenate derived from noninfected animals. Animals were monitored daily, and Kaplan-Meier curves illustrate the kinetics of survival. (B) Clinical scoring system used to evaluate disease progression in mice following IC inoculation with NIHE. (C) Groups of animals were monitored daily for the presence of disease. Clinical scores for symptomatic animals within each group were averaged. The number of infected animals per group is noted in the figure key.
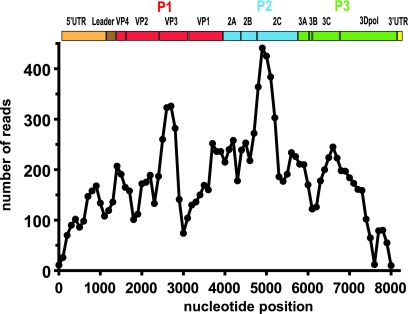
To determine the identity of the infectious agent, brains from symptomatic mice were pooled and homogenized, and viral RNA was enriched for sequencing. Mass sequencing was performed using 454 GS-FLX technology (Roche). Approximately 700,000 sequencing reads were obtained with an average read length of 330 nucleotides. BLAST analyses were performed, and mouse genomic sequences were excluded. The remaining sequences were blasted against available viral genomes in the NCBI nr/nt databases. Of the identified sequences, 6.9% (∼5,100/73,000) showed similarity to members of the Cardiovirus genus, with the greatest sequence identity to TMEV isolates. Sequences were assembled de novo into contigs using a local alignment algorithm (CodonCode Aligner, version 3.5.5). Complete and redundant genomic coverage of a virus (designated NIHE) was obtained with an average of 177 reads per 100-bp segment (Fig. 2). Based on these analyses, a consensus sequence was generated and translated to reveal a full-length polyprotein. The nucleotide and amino acid sequences of NIHE were used to compare percent identities to those of full-length neurovirulent (GDVII) and persistent (BeAn, DA, Yale, TOB15, and Vie415HTR) TMEV strains available in GenBank. Strains with partial sequences were also considered, namely, neurovirulent (FA and ASK-1) and persistent (TO4 and WW) TMEV strains (Tables 1 and 2).
Fig. 2.
Complete and redundant coverage of the NIHE genome was obtained by pyrosequencing. Individual sequencing reads (average length of 330 bp) were assembled de novo, and the number of sequencing reads was determined for each 100-bp segment along the length of the genome. An organizational map of the TMEV genome is aligned at the top of the figure. Gene regions are drawn to scale, and P1 (red), P2 (blue), and P3 (green) are highlighted.
Table 1.
Comparison of nucleotide sequence identities for TMEV strainsa
| Strain | % Identity to: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| NIHE | GDVII | FAb | DA | BeAn8386 | TOB15 | VIE415HTR | Yale | TO4/WWc | |
| GDVII | 88 | ||||||||
| FA | 87 | >99 | |||||||
| DA | 88 | 88 | 83 | ||||||
| BeAn8386 | 88 | 90 | 89 | 89 | |||||
| TOB15 | 88 | 91 | 91 | 88 | 93 | ||||
| VIE415HTR | 85 | 84 | 82 | 86 | 85 | 84 | |||
| Yale | 89 | 91 | 90 | 90 | 91 | 93 | 85 | ||
| TO4/WW | 85 | 83 | 83 | >99 | 85 | 83 | 86 | 83 | |
| ASK-1d | 87 | 87 | 88 | 81 | 85 | 87 | 81 | 87 | 87 |
Nucleotide sequences were aligned, and a percent identity was determined for each sequence pair.
Partial sequence was obtained by combining sequences U32924 and M80883 (accession numbers), which yielded a 3,265-nt fragment.
Partial sequence was obtained by combining sequences U33045 and M80885 (accession numbers), which yielded a 3,260-nt fragment.
Only a partial sequence (2,649 nt) is available for this strain.
Table 2.
Comparison of amino acid sequence identities for TMEV strainsa
| Strain | % Identity to: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| NIHE | GDVII | FAb | DA | BeAn8386 | TOB15 | VIE415HTR | Yale | TO4/WWc | |
| GDVII | 95 | ||||||||
| FA | 94 | >99 | |||||||
| DA | 95 | 94 | 91 | ||||||
| BeAn8386 | 95 | 95 | 94 | 95 | |||||
| TOB15 | 95 | 95 | 94 | 94 | 96 | ||||
| VIE415HTR | 93 | 93 | 92 | 94 | 95 | 92 | |||
| Yale | 95 | 95 | 94 | 95 | 95 | 96 | 93 | ||
| TO4/WW | 93 | 91 | 91 | >99 | 93 | 92 | 92 | 92 | |
| ASK-1d | 96 | 95 | 95 | 95 | 95 | 94 | 94 | 94 | 93 |
Amino acid sequences were aligned, and the percent identity was determined for each sequence pair. For partial sequences, amino acid numbers refer to the corresponding number of the NIHE strain.
Only a partial sequence is available; amino acids 1 to 922 were used for comparison.
Only a partial sequence is available; amino acids 1 to 920 were used for comparison.
Only a partial sequence is available; amino acids 62 to 934 were used for comparison.
Disease pathogenesis is consistent with NIHE being a neuropathogenic strain.
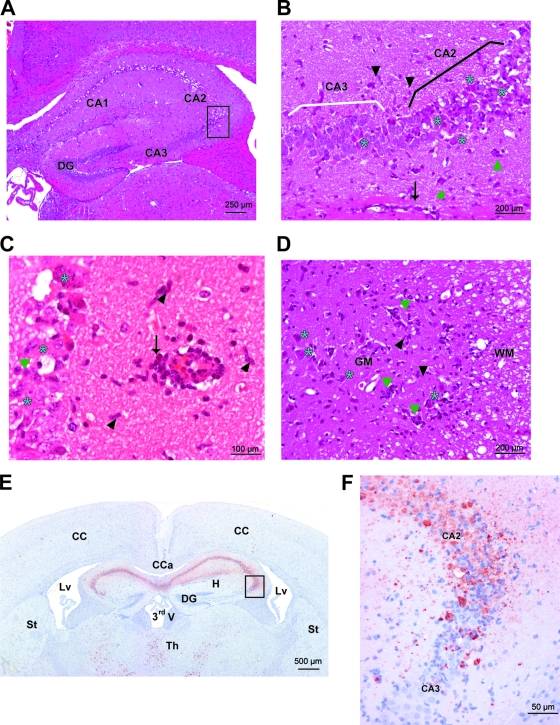
To further characterize the pathogenesis of NIHE-mediated disease, histopathological analysis was performed on brains and spinal cords isolated from SJL and C57BL/6 mice 8 days postinoculation with 10−1 to 10−7 dilutions of IMBH. In the brain, acute inflammatory lesions were observed in several regions, including the cerebral cortex, thalamus, brain stem, and cerebellum, although most of the neuronal destruction and viral antigen was observed in the hippocampus (Fig. 3). In the gray matter, perivascular cuffing was observed and was comprised mainly of mononuclear cells. Many neurons showed characteristics of necrosis and were surrounded by mononuclear cells showing evidence of neuronophagia. Examples of the histopathological findings are shown for the hippocampus (Fig. 3A to C). Infected regions of the spinal cord showed similar, albeit more severe, inflammatory lesions. Perivascular cuffs and neuronal necrosis were observed primarily in the gray matter, but perivascular infiltrates very occasionally were observed in the white matter as well (Fig. 3D). To determine if NIHE is cross-reactive to antibodies known to recognize other TMEV strains (31), immunohistochemistry was performed and representative sections are shown, demonstrating significant viral infection in the brain throughout the hippocampus and thalamus (Fig. 3E and F) as well as gray matter of the spinal cord (data not shown). Numerous foci of infected cells, particularly neurons, are evident in both tissues. Severe neuronal necrosis and TMEV staining were observed, particularly in the CA1 and CA2 pyramidal cell layers of the hippocampus, yet the CA3 and dentate gyrus areas were almost completely spared (Fig. 3A and B).
Fig. 3.
Histopathological features and viral tropism within the brain and spinal cord of C57BL/6 mice 8 days after the intracranial inoculation of NIHE virus. (A to D) HE staining. Severe acute necrotizing polioencephalitis and poliomyelitis characterized by perivascular cuffs (arrows), neuronal necrosis (blue stars), satellitosis and neuronophagia (green triangles), and gliosis (black arrowheads). (A) Hippocampus (original magnification, ×100). (B) Hippocampus (Ammon's horn) (original magnification, ×200). (C) Hippocampus (original magnification, ×400). (D) Spinal cord (ventral horns) (original magnification, ×200). GM, gray matter of the spinal cord; WM, white matter of the spinal cord. (E and F) Immunostaining against Theiler's virus (strain DA). (E) Many viral neuronal bodies and processes are labeled in the Ammon's horn of the hippocampus and in thalamic nuclei. Original magnification, ×20. (F) In the Ammon's horn, many virus-positive neurons are seen and are necrotic in CA1 and CA2; in CA3 only a few neurons are infected and necrotic. Original magnification, ×200. CA, Ammon's horn; DG, dentate gyrus; CC, cerebral cortex; Cca, corpus callosum; Lv, lateral ventricle; St, striatum; Th, thalamus; CA1, CA2, and CA3, fields of the Ammon's horn; 3rdV, third ventricle.
A shift in the tissue tropism from the gray matter to the white matter of the spinal cord is one of the defining characteristics of the persistent TMEV subgroup. The capacity of NIHE to persist in susceptible SJL animals was examined by the histological analysis of the brain and spinal cord 45 days postinoculation, the time point generally considered the chronic phase of disease (25). Animals were inoculated IC with increasing dilutions of infected mouse brain homogenate. All animals that received a dilution of >10−4 succumbed during the acute phase of disease, 18% of the animals that received a 10−5 dilution died, and all animals that received a ≤10−6 dilution remained asymptomatic (data not shown). Histopathological analysis was performed on brain and spinal cord, and no evidence of inflammatory lesions was present in the tissues, and viral antigens were not detected following anti-TMEV staining (data not shown).
Genetic analysis of the NIHE strain in the context of known cardioviruses.
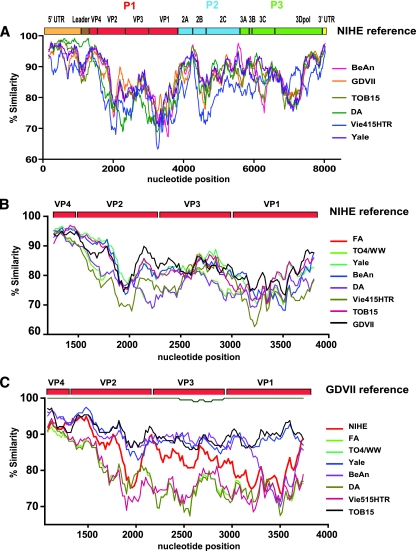
To more fully understand the basis for the neurovirulence of NIHE, as well as to determine its relationship to other TMEV group members, we performed a detailed genetic analysis. All strains with full-length sequence data were aligned and compared to the sequence of NIHE (Fig. 4A). The greatest percent identity is seen in the 5′UTR. Interestingly, the persistent strain DA maintains the highest identity to NIHE through the sequence coding for the L protein (98% identity of amino acid sequence), but it diverges significantly for other genes (Fig. 4A, green line). Regions that demonstrated the lowest percentage of nucleotide identity between NIHE and other TMEV strains were clustered in the L protein and P1 region, particularly the sequences encoding the capsid proteins VP2 and VP1 (Fig. 4A to C). A more focused comparison of the P1 region allows the inclusion of partially sequenced strains and demonstrates additional dissimilarity, specifically within the VP2 and VP1 regions, with no apparent clustering with either subgroup (Fig. 4B). Interestingly, when the same analysis was performed using the GDVII strain as the reference sequence, a separation within the TO subgroup was observed (Fig. 4C). Specifically, the Yale and TOB15 strains (blue and black lines, respectively) maintain a relatively constant degree of sequence identity throughout this region (∼90%). This is in contrast to the cluster of TO4/WW, BeAn, DA, and Vie415HTR strains, which differ from GDVII considerably (70 to 80%). NIHE (red line) appears at an intermediate position between these two groups.
Fig. 4.
Sequence similarity for the complete genome and P1 regions of TMEV strains compared to those of NIHE and GDVII. (A) Sequence identity for all five TMEV strains with complete genome sequence information was obtained by comparing 20-bp segments along their entire genome to the sequence of NIHE. (B) Sequence identity of full-length and partially sequenced TMEV strains using NIHE as a reference. (C) Sequence identity of all TMEV stains (including NIHE) compared to the GDVII strain as the reference sequence. All sliding-window graphs were generated using Simplot 3.2 software as described in Materials and Methods. The x axis for all graphs is the nucleotide position, and the y axis is the percent similarity. A genetic map of TMEV is shown at the top of each graph, illustrating the organization and relative size of the individual genomic regions.
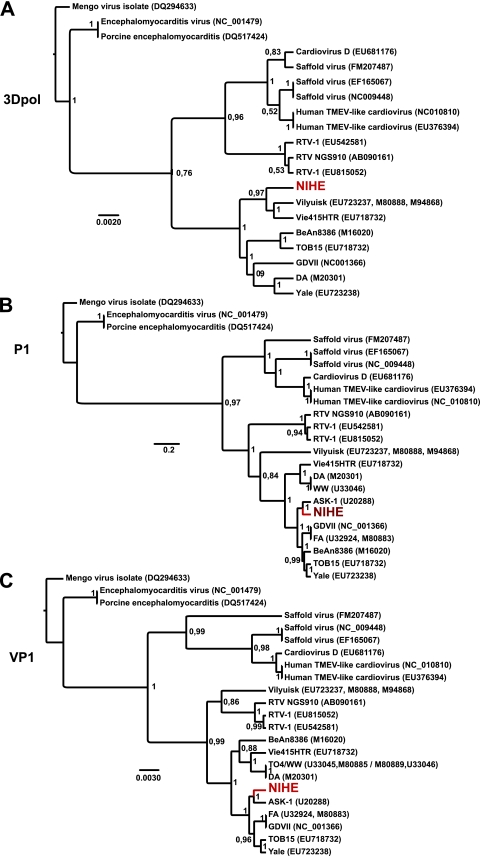
To place genetic differences within NIHE in context with other known cardioviruses, a phylogenetic analysis of three proteins or regions was performed: (i) the well-conserved 3Dpol (RNA-dependent RNA polymerase); (ii) the P1 region, which encodes the four capsid proteins; and (iii) the highly variable VP1 gene (Fig. 5). The phylogenetic analysis of the 3Dpol supports the separation of the human viruses, Thera viruses (rat), and TMEV species. NIHE clusters within the TMEV clade; however, it segregates with Vilyuisk and the mildly neurovirulent Vie415HTR strain, while GDVII groups with the TO strains. These groupings are well supported based on the high posterior probability for the nodes (Fig. 5A). Differences in the topography of the P1 region and that of 3Dpol are seen. NIHE segregates with ASK-1 and is more closely related to GDVII and FA than to the TO strains (Fig. 5B). The phylogenetic relationships established for the P1 region are echoed in the VP1 analysis, with NIHE segregating with a group comprised mostly of neurovirulent strains (Fig. 5C).
Fig. 5.
Phylogenetic relationships between NIHE and select cardioviruses based on the nucleotide sequences of 3Dpol, P1, and VP1 regions. Sequences were aligned and phylogenetic analysis was performed as outlined in Materials and Methods. Phylogenetic relationships are shown for sequences of the 3Dpol (A), P1 (B), and VP1 (C) regions of the genome. Numbers at the nodes represent posterior probabilities, and the genetic distance is indicated by the bar at the bottom of each panel. The position of the NIHE strain is marked in red.
Analysis of the L and capsid proteins.
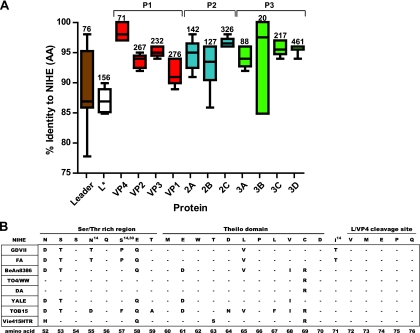
To identify possible genetic determinants of neurovirulence and persistence in the NIHE genome, we aligned and compared the amino acid sequence for all coding regions of NIHE to the other complete and partial sequences of TMEV strains. The mean percent identity (compared to NIHE) of the amino acid sequence for the indicated viral protein is shown (Fig. 6). Although significant dissimilarity is seen in the VP2 and VP1 proteins, as predicted by the similarity plots (Fig. 4), the lowest percent similarity is seen in the L (88.8%) and L* (87.3%) proteins. A comparative analysis of the amino acid sequence for the carboxy terminus of the L protein, including several distinct domains, is shown in Fig. 6B.
Fig. 6.
Percent identity for individual proteins of TMEV strains compared to NIHE proteins. Amino acid sequences for all available TMEV strains were compared to the NIHE sequence for each gene product. (A) The percent identity was determined, and whisker plots were generated showing the mean, minimum, and maximum values. Numbers above each plot identify how many amino acids are within each protein. Proteins are presented according to genome structure, and P1 (red), P2 (blue), and P3 (green) regions are colored according to the description in previous figure legends. (B) Amino acid alignments for eight TMEV strains are compared to the sequence of NIHE. The amino acid sequence for the Ser/Thr-rich region, Theilio domain, and L/VP4 cleavage sites are shown for the NIHE L protein along the top row. Dashes indicate identity to the NIHE sequence, and the single-letter codes for nonidentical amino acids are shown. Amino acid positions that have been implicated in a particular phenotype or function are annotated with a superscript number indicating the respective citation.
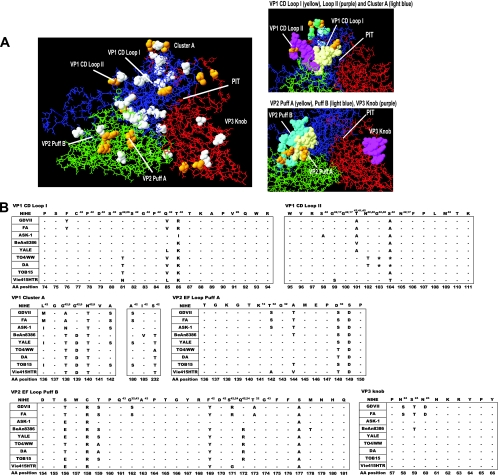
Using modeling software (Swiss-PDB viewer [10] and RasWin [33]), we compared the predicted structure of the NIHE VP1-VP3 protomer complex to the structures of BeAn and DA strains (Fig. 7). The calculated structure of NIHE VP1, VP2, and VP3 proteins (blue, green, and red, respectively) was overlaid on the BeAn structure (Fig. 7A) (23). Differences in the amino acid sequence between these two strains are indicated by white-space-filled molecules (40 amino acid changes), and sequences that are unique to NIHE (compared to all published strains) are shown in yellow (14 amino acid changes). The spatial positioning of CD loop I and CD loop II are shown (Fig. 7A, upper right), with the loops delineated by the light-yellow and purple-filled molecules, respectively. Puff A (yellow) and puff B (light-blue) regions of VP2, as well as a protrusion from VP3 known as the knob, are highlighted (Fig. 7A, lower right). To further understand the relationship between NIHE and the other TMEV strains, specifically within these critical loop structures, we compared the amino acid sequences of selected regions (Fig. 7B).
Fig. 7.
Analysis of the NIHE capsid structure formed by VP1, VP2, and VP3 proteins. Capsid proteins (VP1, VP2, and VP3) were aligned and overlaid onto the three-dimensional structure of the BeAn8386 strain using DeepView/Swiss-pdbViewer (version 4.0.1). (A) Quaternary structure with the individual proteins VP1 (blue), VP2 (green), and VP3 (red) indicated. White space-filled molecules indicate amino acid positions that differ between the sequence of BeAn8386 and NIHE. Amino acid positions that are unique to NIHE (compared to all TMEV strains) are highlighted in yellow. Highly variable regions, loops, puffs, and a canyon or “pit” are annotated. Also shown, to the right, are a close-up of VP1 CD loops I and II and the cluster A region (upper), as well as VP2 puff A and B and the VP3 knob (lower). (B) Amino acid alignments for the indicated region of all TMEV strains compared to the NIHE sequence. Dashes indicate identity to the NIHE sequence, and the single-letter codes for nonidentical amino acids are shown. Amino acid positions and annotations are the same as those described in the legend to Fig. 6.
Genetic evidence for L*.
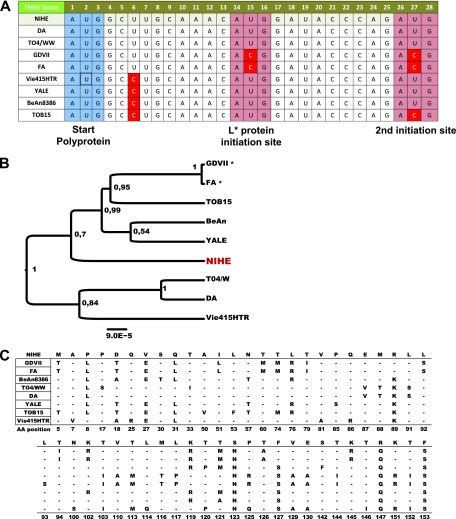
The region that was observed to have the greatest intrastrain variation coded for the L* protein (Fig. 6A). The generation of L* is the result of an additional open reading frame initiated 13 nucleotides downstream of the AUG start site that is responsible for initiating the viral polyprotein (15). This protein is not efficiently produced by the GDVII strain (and, presumably, FA) due to a polymorphism (ACG) in the sequence that initiates translation. Figure 8A depicts the sequence of this region, including the polyprotein start site and both alternative initiation sites located immediately downstream. The putative sequences coding for the L* protein of available strains were aligned, and a midrooted tree was generated (Fig. 8B). An intrastrain alignment was performed, and the difference between putative coding sequences of NIHE and other strains are shown (Fig. 8C).
Fig. 8.
Alternative AUG initiation sequence present for the L* gene within the NIHE genome. Nucleotide sequences were aligned for TMEV strains and compared to the sequence of NIHE. (A) The canonical start sequence (AUG) is outlined in blue, and both downstream initiating codon sequences are outlined in red. Sequences not identical to that of NIHE are highlighted in red. (B) Nucleotide sequences potentially coding for L* were aligned, and a midrooted tree was constructed using the maximum-likelihood method. There were a total of 156 positions in the final data set. Numbers at the nodes represent posterior probabilities, and genetic distance is indicated by the bar at the bottom of the panel. GDVII and FA strains were included in the analysis, although they do not efficiently produce L* (indicated by an asterisk). (C) Nucleotide sequences were aligned for all TMEV strains with available sequence information and compared to the sequence of NIHE. Only amino acid positions with discrepancies between strains are shown. The NIHE sequence is presented along the top row. Dashes indicate identity to the NIHE sequence, and the single-letter codes for nonidentical amino acids are shown.
DISCUSSION
A novel highly neurovirulent TMEV strain.
Our study reports the identification of a novel neurovirulent TMEV strain, the first since Theiler's characterization of the GDVII and FA strains in 1938. In fact, based on the time and location at which these two strains were discovered, as well as the current understanding of quasispecies diversity, it has been proposed that FA is a derivative of GDVII, thus making NIHE only the second fully annotated neurovirulent TMEV. A strain isolated in Japan, referred to as ASK-1 (6), has been proposed to be a member of the neurovirulent subgroup (3, 20); however, no characterization of this strain has been published, and only a partial sequence has been submitted to GenBank (24).
Several characteristics of the NIHE strain led us to classify it as a member of the neurovirulent subgroup: (i) the rapid onset of disease following IC inoculation (Fig. 1); (ii) marked neuronal destruction in the brain and spinal cord, far greater than that observed for persistent strains (Fig. 3); and (iii) a lack of evidence (pathology or viral antigen) that NIHE persists in the CNS of mice surviving low-dose inoculation (data not shown). We do, however, realize the inherent paradox in studying highly virulent TMEV strains, in that it is difficult to distinguish between the inability of the virus to persist and the failure of a productive infection due to low viral inoculation.
The use of next-generation sequencing (NGS) techniques allowed for the complete, redundant sequencing as well as the de novo assembly of the entire NIHE genome (Fig. 2), suggesting the identification of a novel TMEV strain. The degree of identity shared by NIHE and known TMEV strains is <90% at the nucleotide level and <95% in its primary amino acid sequence (Tables 1 and 2). This is in contrast to the >99% identity for quasispecies variation seen for the TO4, WW, and DA strains (27) or the laboratory derivatives of S2 BeAn (14) or GDVII substrains (16, 29).
The discovery of NIHE provides new information about the determinants of neurovirulence versus persistence and offers a novel basis of comparison on which to examine the relevance of amino acid differences previously identified between the GDVII and TO strains. Intriguingly, our analysis suggests that although NIHE has a decidedly neurovirulent phenotype, it shares high sequence identity with the TO strains in many important regions, including L, L*, and capsid proteins.
Features of NIHE L, L*, and capsid proteins segregate with TO stains.
The analysis of the sequence comparison between NIHE and the other TMEV sequences demonstrates clusters of differences in the amino acid sequence (9, 12, 17, 26, 32, 40, 41, 45). Although we present clear evidence that NIHE is neurovirulent, we were surprised to observe the number of instances in which the NIHE sequence segregated with the persistent strains. Here, we restrict our discussion to instances in which NIHE segregates with TO strains within regions important for modulating the host antiviral response, viral persistence, or interaction with the coreceptor sialic acid.
The L protein is produced by all members of the Cardiovirus genus and has been implicated in in vivo persistence because it modulates the host antiviral response (8, 34, 39). Although analysis revealed that the L protein is the second most divergent region across TMEV strains (Fig. 6), there were no unique amino acids predicted by the NIHE sequence. The majority of the sequence variation between NIHE and the other strains was clustered within the Ser/Thr-rich domain (62.5%) and the Theilo domain (79.5%). Interestingly, two positions in the Ser/Thr-rich region (L-55 and L-57) and one in the L-VP4 cleavage site (L-70) have been identified previously as distinguishing between the GDVII and TO strains (27, 35). In all three instances, the NIHE sequence segregates with the TO strains.
A slightly different result emerged when analyzing the L* reading frame. Several nucleotide changes are unique to the NIHE strain, resulting in distinct amino acid sequences at positions L*-123 (Ser), L*-147 (Arg), and L*-153 (Phe) compared to sequences of any of the known TMEV strains (Fig. 8C). Again, the strains encoding L* sequences most closely related to NIHE are those produced by the Yale, BeAn, and TOB15 strains. Perhaps the most notable example of this divergence is the observation that NIHE, like the TO strains, retains the genetic capacity to efficiently express L* by encoding an AUG sequence at the two initiation positions (Fig. 8). It is important to note that some L* transcripts can be produced via the ACG initiation sites, albeit less efficiently (38). What evolutionary advantage this protein provides to a neurovirulent strain is unclear, particularly given that it is not required to infect neurons during the acute phase of disease. Advantages, however, can be imagined if moderate levels of L* are required by the virus during the natural route of transmission or for the crossing of the blood-brain barrier.
Many intraspecies sequence differences mapped to exposed areas of the viral capsid (Fig. 7). Cluster A, a highly variable region within VP1, initially was believed to segregate between GDVII and persistent strains; however, sequence information for several viruses was not then available. The inclusion of recently published strains and our contribution of the NIHE genome further clarifies these observations. Although GDVII and FA segregate with ASK-1 and NIHE at two positions (VP1-139 and VP1-232), NIHE segregates with a subset of TO strains at positions VP1-136, VP1-142, and VP1-180. In addition to these differences, NIHE also encodes two unique amino acids within this domain (VP1-138 and VP1-140), as shown in Fig. 7.
Perhaps the most functionally relevant example of NIHE sequences similar to those of persistent TO strains is observed in VP2 puff B (within the EF loop). Positions VP2-161, VP2-162, VP2-163, and VP2-174 were demonstrated to be the binding site for sialic acid in the persistent strains (43). Although a cellular receptor for TMEV strains currently is unknown, this region has been considered relevant to persistence due to the observation that Ser at position VP2-162 within the GDVII/FA strains prevents VP1-161 (Glu) from forming a hydrogen bond with sialic acid. Alignment with NIHE and ASK-1 indicate that these nonpersistent strains are identical to the TO strains at these positions. VP2-171, VP2-172, and VP2-173 also influence binding to sialic acid by establishing a structural framework for Gly at VP2-174 to bond with sialic acid (24, 42, 43). NIHE is unique in this area, encoding Ser-Gln-Thr, whereas Arg-Gln-Ala is encoded in the GDVII/FA strains and Ser-Arg-Thr is in the majority of the TO strains (Fig. 7B). Interestingly, NIHE is identical to ASK-1, but the consequence on the orientation of Gly at VP2-174 remains unknown. Within this domain, there also exist three unique amino acids within NIHE (VP2-156, VP2-158, and VP2-177). None of these residues has been implicated previously in TMEV pathogenicity, and they may reflect either genetic drift in a nonessential position or compensatory mutations that promote viral entry or replication.
Evolutionary comparisons within TMEV.
To ascertain information about the relationship of NIHE to other TMEV strains and gain insight into its evolution, we performed the phylogenetic analysis of highly conserved 3Dpol as well as the divergent regions P1 and VP1. The analysis of the 3Dpol gene clearly positions NIHE within the TMEV cluster and demonstrates that NIHE is more closely related to the human Vilyuisk virus and VIE415HTR than the other murine neurovirulent strains (Fig. 5A). The position of NIHE within the topology of the P1 and VP1 phylogenies is well supported and further supports our conclusion of neurovirulence. Furthermore, the addition of the NIHE sequence clarifies the groupings by defining a neurovirulent clade with GDVII, FA, and ASK-1, particularly within the VP1 analysis (Fig. 5C). Evidence of parallel evolutionary paths are predicted for NIHE, ASK-1, and GDVII/FA strains, all of which have obtained neurovirulence, albeit via potentially distinct mutations (Fig. 5B and C). Although it would be difficult to decipher an evolutionary history with such few numbers of sequenced strains, it has been hypothesized that neurovirulent strains have evolved from persistent strains (27).
In summary, our study reports the discovery and characterization of a novel, highly neurovirulent TMEV, the first report of a new strain in this subgroup in more than 70 years. Critically, we characterized the pathogenesis of disease and obtained complete sequencing information without subjecting NIHE to conditions that promote genomic changes (e.g., in vitro expansion, which is known to introduce artifactual compensatory mutations). Perhaps the most significant advance of this work is that the discovery of the NIHE strain helps to illuminate the relevance of sequences putatively implicated in the evolution of neurovirulence versus persistence, data that previously had been based solely on comparison with the GDVII strain. NIHE also provides an opportunity to evaluate new positions as potential determinants for neurovirulence, as NIHE contains unique amino acids at several positions, particularly within the capsid proteins. Further investigation will be required to assess to what extent these positions contribute to the extreme phenotype of neurovirulence observed in the NIHE strain.
ACKNOWLEDGMENTS
This work was supported by grants from the Agence National de la Recherche (ANR) and an EURYI Award from the European Science Foundation (ESF).
We are grateful to Michel Brahic (Stanford University, Stanford, CA) for the anti-TMEV antibody and to Thomas Michels (Université Catholique de Louvain, Brussels, Belgium) for the anti-L* antibody. We also thank Patric Avé (Histotechnologie et Pathologie, Institut Pasteur, Paris, France) for his expert technical assistance in the preparation of the histology slides and Herbert Virgin IV (Washington University School of Medicine, St. Louis, MO) for advice on deep sequencing and data mining. We thank Olivier Schwartz and Felix Rey for their critical reviews of the manuscript.
Footnotes
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Aubert C., Brahic M. 1995. Early infection of the central nervous system by the GDVII and DA strains of Theiler's virus. J. Virol. 69:3197–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aubert C., Chamorro M., Brahic M. 1987. Identification of Theiler's virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb. Pathog. 3:319–326 [DOI] [PubMed] [Google Scholar]
- 3. Bandyopadhyay P. K., Pritchard A., Jensen K., Lipton H. L. 1993. A three-nucleotide insertion in the H stem-loop of the 5′ untranslated region of Theiler's virus attenuates neurovirulence. J. Virol. 67:3691–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bureau J. F., et al. 1992. The interaction of two groups of murine genes determines the persistence of Theiler's virus in the central nervous system. J. Virol. 66:4698–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camacho C., et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casals J. 1963. Immunological characterization of Vilyuisk human encephalomyelitis virus. Nature 200:339–341 [DOI] [PubMed] [Google Scholar]
- 7. Chiu C. Y., et al. 2008. Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc. Natl. Acad. Sci. U. S. A. 105:14124–14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delhaye S., van Pesch V., Michiels T. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78:4357–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu J. L., et al. 1990. Neurovirulence determinants of genetically engineered Theiler viruses. Proc. Natl. Acad. Sci. U. S. A. 87:4125–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guex N., Peitsch M. C. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 11. Huson D. H., Auch A. F., Qi J., Schuster S. C. 2007. MEGAN analysis of metagenomic data. Genome Res. 17:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jarousse N., et al. 1994. A single amino acid change determines persistence of a chimeric Theiler's virus. J. Virol. 68:3364–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones M. S., Lukashov V. V., Ganac R. D., Schnurr D. P. 2007. Discovery of a novel human picornavirus in a stool sample from a pediatric patient presenting with fever of unknown origin. J. Clin. Microbiol. 45:2144–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim B. S., et al. 1998. A spontaneous low-pathogenic variant of Theiler's virus contains an amino acid substitution within the predominant VP1(233-250) T-cell epitope. J. Virol. 72:1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kong W. P., Roos R. P. 1991. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J. Virol. 65:3395–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Law K. M., Brown T. D. 1990. The complete nucleotide sequence of the GDVII strain of Theiler's murine encephalomyelitis virus (TMEV). Nucleic Acids Res. 18:6707–6708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin X., et al. 1998. Molecular characterization of a nondemyelinating variant of Daniel's strain of Theiler's virus isolated from a persistently infected glioma cell line. J. Virol. 72:1262–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lipton H. L. 1980. Persistent Theiler's murine encephalomyelitis virus infection in mice depends on plaque size. J. Gen. Virol. 46:169–177 [DOI] [PubMed] [Google Scholar]
- 19. Lipton H. L. 1975. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 11:1147–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lipton H. L., Kumar A. S., Hertzler S., Reddi H. V. 2006. Differential usage of carbohydrate co-receptors influences cellular tropism of Theiler's murine encephalomyelitis virus infection of the central nervous system. Glycoconj. J. 23:39–49 [DOI] [PubMed] [Google Scholar]
- 21. Lipton H. L., Twaddle G., Jelachich M. L. 1995. The predominant virus antigen burden is present in macrophages in Theiler's murine encephalomyelitis virus-induced demyelinating disease. J. Virol. 69:2525–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lole K. S., et al. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo M., He C., Toth K. S., Zhang C. X., Lipton H. L. 1992. Three-dimensional structure of Theiler murine encephalomyelitis virus (BeAn strain). Proc. Natl. Acad. Sci. U. S. A. 89:2409–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo M., Toth K. S., Zhou L., Pritchard A., Lipton H. L. 1996. The structure of a highly virulent Theiler's murine encephalomyelitis virus (GDVII) and implications for determinants of viral persistence. Virology 220:246–250 [DOI] [PubMed] [Google Scholar]
- 25. McAllister A., Tangy F., Aubert C., Brahic M. 1990. Genetic mapping of the ability of Theiler's virus to persist and demyelinate. J. Virol. 64:4252–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCright I. J., Tsunoda I., Whitby F. G., Fujinami R. S. 1999. Theiler's viruses with mutations in loop I of VP1 lead to altered tropism and pathogenesis. J. Virol. 73:2814–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michiels T., Jarousse N., Brahic M. 1995. Analysis of the leader and capsid coding regions of persistent and neurovirulent strains of Theiler's virus. Virology 214:550–558 [DOI] [PubMed] [Google Scholar]
- 28. Ohsawa K., Watanabe Y., Miyata H., Sato H. 2003. Genetic analysis of a Theiler-like virus isolated from rats. Comp. Med. 53:191–196 [PubMed] [Google Scholar]
- 29. Pevear D. C., et al. 1988. Insights into Theiler's virus neurovirulence based on a genomic comparison of the neurovirulent GDVII and less virulent BeAn strains. Virology 165:1–12 [DOI] [PubMed] [Google Scholar]
- 30. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 31. Roussarie J. P., Ruffie C., Brahic M. 2007. The role of myelin in Theiler's virus persistence in the central nervous system. PLoS Pathog. 3:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato S., et al. 1996. A neutralization site of DA strain of Theiler's murine encephalomyelitis virus important for disease phenotype. Virology 226:327–337 [DOI] [PubMed] [Google Scholar]
- 33. Sayle R. A., Milner-White E. J. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 34. Stavrou S., Feng Z., Lemon S. M., Roos R. P. 2010. Different strains of Theiler's murine encephalomyelitis virus antagonize different sites in the type I interferon pathway. J. Virol. 84:9181–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takano-Maruyama M., Ohara Y., Asakura K., Okuwa T. 2006. Theiler's murine encephalomyelitis virus leader protein amino acid residue 57 regulates subgroup-specific virus growth on BHK-21 cells. J. Virol. 80:12025–12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Theiler M. 1937. Spontaneous encephalomyelitis of mice, a new virus disease. J. Exp. Med. 65:705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Theiler M., Gard S. 1940. Encephalomyelitis of mice. I. Characteristics and pathogenesis of the virus. J. Exp. Med. 72:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Eyll O., Michiels T. 2002. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J. Virol. 76:10665–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Pesch V., van Eyll O., Michiels T. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811–7817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wada Y., McCright I. J., Whitby F. G., Tsunoda I., Fujinami R. S. 1998. Replacement of loop II of VP1 of the DA strain with loop II of the GDVII strain of Theiler's murine encephalomyelitis virus alters neurovirulence, viral persistence, and demyelination. J. Virol. 72:7557–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wada Y., Pierce M. L., Fujinami R. S. 1994. Importance of amino acid 101 within capsid protein VP1 for modulation of Theiler's virus-induced disease. J. Virol. 68:1219–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou L., Lin X., Green T. J., Lipton H. L., Luo M. 1997. Role of sialyloligosaccharide binding in Theiler's virus persistence. J. Virol. 71:9701–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou L., Luo Y., Wu Y., Tsao J., Luo M. 2000. Sialylation of the host receptor may modulate entry of demyelinating persistent Theiler's virus. J. Virol. 74:1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zoll J., et al. 2009. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 5:e1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zurbriggen A., Thomas C., Yamada M., Roos R. P., Fujinami R. S. 1991. Direct evidence of a role for amino acid 101 of VP-1 in central nervous system disease in Theiler's murine encephalomyelitis virus infection. J. Virol. 65:1929–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]