Abstract
We recently reported that Ras-GTPase-activating protein-binding protein 1 (G3BP1) interacts with hepatitis C virus (HCV) nonstructural protein (NS)5B and the 5′ end of the HCV minus-strand RNA. In the current study we confirmed these observations using immunoprecipitation and RNA pulldown assays, suggesting that G3BP1 might be an HCV replication complex (RC) component. In replicon cells, transfected G3BP1 interacts with multiple HCV nonstructural proteins. Using immunostaining and confocal microscopy, we demonstrate that G3BP1 is colocalized with HCV RCs in replicon cells. Small interfering RNA (siRNA)-mediated knockdown of G3BP1 moderately reduces established HCV RNA replication in HCV replicon cells and dramatically reduces HCV replication-dependent colony formation and cell-culture-produced HCV (HCVcc) infection. In contrast, knockdown of G3BP2 has no effect on HCVcc infection. Transient replication experiments show that G3BP1 is involved in HCV genome amplification. Thus, G3BP1 is associated with HCV RCs and may be co-opted as a functional RC component for viral replication. These findings may facilitate understanding of the molecular mechanisms of HCV genome replication.
INTRODUCTION
Hepatitis C virus (HCV) is a member of the family Flaviviridae and possesses a single-stranded, 9.6-kb positive-sense RNA genome, encoding a single polyprotein which is cleaved co- and posttranslationally into at least 10 individual polypeptides with the following protein order: 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′ (25). The single open reading frame is flanked by highly conserved 5′ and 3′ untranslated regions (UTRs). The 5′ UTR contains an internal ribosome entry site (IRES) to regulate HCV translation (20), while sequences upstream of the HCV IRES are essential for positive-strand RNA replication (4, 14). The 3′ UTR is proposed to be involved in negative-strand RNA replication and is composed of three sequential elements: a nonconserved variable region (30 to 50 nucleotides), a poly(U/C) stretch (20 to 200 nucleotides), and a conserved 98-nucleotide sequence, termed the 3′X region, which contains three stem-loop structures (13, 21, 48).
Like many of the positive-sense RNA viruses (3, 22, 36, 37), HCV replicates on endoplasmic reticulum (ER) membrane-associated structures. In parallel with other positive-sense RNA viruses (1, 26, 45), HCV may require interactions with host factors to complete its replication cycle. A long list of host factors have been reported as exploited by HCV and involved in its replication (9, 15, 16, 18, 27, 34, 39, 44, 46). Interaction of HCV nonstructural proteins with the host protein hVAP-A/B may mediate the formation of HCV replication complexes (RCs) on lipid rafts (15, 16). The host protein cyclophilin A interacts with HCV NS5A and modulates RNA replication (reviewed in reference 30). F-box/LRR-repeat protein 2 (FBL2) is required for HCV replication, which requires its geranylgeranylation (44). However, the functions of many other host factors involved in HCV replication and the molecular mechanisms of HCV RNA replication are largely unknown.
We previously reported that Ras-GTPase-activating protein-binding protein 1 (G3BP1) interacts with HCV NS5B and the 5′ terminus of the HCV minus-strand RNA [5′(−)TR] and thus might be a potential HCV RC component (40, 49). G3BP1, first identified as a binding partner for Ras-GTPase-activating protein (29), participates in mRNA metabolism (2, 42), vaccinia virus transcription (19), and T cell receptor signaling (33) and modulates Sindbis virus polyprotein expression (10). G3BP1 can initiate stress granule formation (41) and modulate ubiquitin-specific protease (USP) activity (38). The Saccharomyces cerevisiae homolog of G3BP, Bre5, functions as a cofactor in the deubiquitination of the COP-mediated transport factor Sec23 (8). Here, we found that G3BP1 was associated with, and thus may be a component of, HCV RCs. Furthermore, knockdown experiments indicated that G3BP1 is involved in HCV genome amplification.
MATERIALS AND METHODS
Cell lines.
Huh7 and Huh-7.5 (6) cells were maintained in monolayer culture in complete medium consisting of Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS). Huh7 cells harboring the S2204I bsd HCV Con1 bicistronic replicon (11), a pHCVrep1b/BB7 HCV subgenomic replicon (5) derivative that expresses blasticidin S deaminase, were generated by electroporation of in vitro-transcribed HCV replicon RNA and were grown in conditioned complete medium supplemented with 5 μg/ml of blasticidin (Invitrogen). HEK293T cells were grown in DMEM supplemented with 10% FBS.
Antibodies and plasmids.
Mouse monoclonal antibodies to HCV NS3 (217-A) or NS5A (256-A) and rabbit polyclonal antibody to NS5B (266-A) were from Virogen. Mouse monoclonal anti-γ-tubulin (T9026), anti-β-actin (A5316), and anti-Flag epitope antibodies (F1804) were from Sigma. Mouse monoclonal antibody against G3BP1 was from BD Biosciences (611126). A mouse monoclonal antibody to myc (SC-40) was from Santa Cruz. Cy3-conjugated anti-mouse IgG and Alexa Fluor 488-conjugated anti-rabbit IgG antibodies were from Rockland.
For expression in mammalian cells, DNA fragments encoding NS5A, NS5B, NS3-4A, and NS4B were generated from the HCV S2204I bsd subgenomic replicon plasmid by PCR. Primer sequences are available upon request. The amplified fragments were digested with EcoRI and XhoI and cloned into similarly digested pcDNA3.1A/3×Flag, which was made by introducing sequences encoding the 3×Flag epitope tag and cloning sites into pcDNA3.1A (Invitrogen). The resultant plasmids express the HCV proteins with a 3×Flag tag on the amino terminus.
DNA fragments encoding the 3′(+)UTR from the HCV S2204I bsd construct (nucleotides [nt] 9301 to 9605, including parts of the NS5B coding sequence, totaling 305 nt), the 5′(+)UTR (nt 1 to 341), and their reverse complementary region [5′(−)TR and 3′(−)TR] were amplified from the HCV subgenomic construct and cloned into the pSP65 vector. The plasmid for the transcription of HCV JFH1 RNA was kindly provided by Takaji Wakita. The plasmid for transcription of J6/JFH(5′C19Rluc2AUbi) HCV RNA was described previously (43).
The DNA fragment encoding full-length human G3BP1 was amplified from pGEM-G3BP1 plasmid kindly provided by Derek Kennedy (Griffith University, Nathan, Queensland, Australia), digested with EcoRI and BglII, and then cloned into similarly digested pCMV-Myc (Clontech), resulting in plasmid pCMV-Myc-G3BP1 expressing G3BP1 with an amino-terminal myc epitope tag. Constructs with carboxyl-terminal deletions of the RGG or both RRM and RRG motifs were similarly generated, resulting in plasmids expressing amino acids 1 to 428 (G3BP1-ΔRGG) or 1 to 340 (G3BP1-ΔRGG/ΔRRM). Primer sequences are available upon request. For in vitro translation, G3BP1 or mutant fragments were digested from pCMV-Myc with EcoRI and BglII and ligated into similarly digested pcDNA3.1C to generate pcDNA3.1C-G3BP1 (or mutant forms).
In vitro transcription.
To generate RNA for electroporation, S2204I bsd HCV replicon DNA was digested with ScaI, while the remaining plasmids were linearized with XbaI followed by mung bean nuclease (New England BioLabs) digestion. For RNA pulldown experiments, plasmids were linearized with SmaI to produce DNA templates for SP6 runoff transcription. Plasmid pSP65 was also linearized and used as a template to transcribe a similar-length (300-nt) vector-derived RNA as a control. Prior to transcription, DNA was extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. RNAs were synthesized using the RiboMAX kit (Promega) according to the manufacturer's instructions. Transcription was terminated by the addition of 1.2 U of RNase-free DNase per μg of plasmid DNA and a 30-min incubation at 37°C. RNAs were extracted by Trizol reagent LS (Invitrogen) according to the manufacturer's instructions and quantified by optical density at 260 nm.
RNA electroporation and virus production.
For electroporation, subconfluent cells were trypsinized and detached by rinsing with DMEM containing 0.5% FBS. After washing once, cells were resuspended in Eppendorf hypoosmolar electroporation buffer at 107 cells per ml. Ten micrograms of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension. Cells were pulsed five times for 40 ms at 270 V using an Eppendorf Multiporator and a 0.2-cm-gap-width cuvette (Eppendorf). After incubating on ice for 5 min, cells were immediately transferred to 10 or 14 ml of prewarmed complete medium and then plated into 35- or 100-mm-diameter dishes. Alternatively, cells were electroporated as described previously (23) using a BTX 830 system. For virus production, Huh-7.5 cells were electroporated with in vitro-transcribed HCV genome, the medium was harvested at 3 days postelectroporation, and virus was concentrated by polyethylene glycol precipitation as described previously (23).
RNA interference.
Duplex Stealth small interfering RNAs (siRNAs) were purchased from Invitrogen. The siRNA sequences targeting human G3BP1 were as follows: siG3BP1-1, 5′-AAA CCC ACC AAA GAC CUC AUC UUG G; siG3BP1-2, 5′-UAA UUU CCC ACC ACU GUU AAU GCG C. The siRNA sequence targeting human G3BP2 was as follows: siG3BP2, 5′-UAA ACU UUC UUU CUG GUU GUC CAC U. Negative-control siRNA duplex was purchased from Invitrogen. Cells were transfected with siRNAs using RNAiMAX (Invitrogen) using forward transfection protocols provided by the manufacturer. Four hours after transfection, the medium was switched to complete medium supplemented with antibiotics. The effect of siRNA transfection on cell growth was determined by cell enumeration after siRNA transfection using the Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.).
Immunoprecipitation.
For coimmunoprecipitation, Huh7 or HEK293T cells were seeded onto a 60-mm-diameter tissue culture dish and were transfected 16 h later using Fugene 6 (Roche) according to the manufacturer's recommendations. At 48 h after transfection, the cells were washed three times with ice-cold phosphate-buffered saline (PBS) and lysed with 300 μl buffer consisting of 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 5 mM EDTA, 5 mM EGTA, 15 mM MgCl2, 60 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 0.1 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and 1× proteinase inhibitor cocktail (Roche). Detergent-insoluble materials were removed by centrifugation at 15,000 × g for 10 min at 4°C. The whole-cell soluble lysates were incubated with 1 μg monoclonal antibody or normal mouse IgG at 4°C for 2 h. Preequilibrated protein A/G-agarose beads were then added and, after 2 h of incubation, were collected by centrifugation and then gently washed three times with 500 μl lysis buffer. Bound proteins were eluted by boiling in SDS sample buffer and subjected to SDS-PAGE and Western blot analysis.
For immunoprecipitation in replicon-containing cells, cells were seeded onto a 60-mm-diameter tissue culture dish and transfected 16 h later with 4 μg of pCMV-Myc-G3BP1 plasmid. After an additional 24 h, the cells were expanded into a 100-mm-diameter dish and 24 h later were transfected again with 4 μg of plasmid. At 48 h later, the cells were lysed in 600 μl hypotonic buffer (10 mM Tris-HCl, pH 7.8, 10 mM NaCl) and the lysate was clarified by centrifugation twice at 1,000 × g for 5 min at 4°C. The supernatants were incubated for 2 h with 1 μg anti-myc monoclonal antibody. Preequilibrated protein A/G-agarose beads were then added, and after 2 h of incubation, the beads were collected by centrifugation and then gently washed three times with 600 μl PBS containing 1% bovine serum albumin (BSA). The bound proteins were eluted by boiling in SDS sample buffer and subjected to Western blot analysis after separation by 10% SDS-PAGE.
RNA affinity pulldown assay.
An RNA pulldown assay using biotinylated RNAs was performed as described previously (40). Briefly, RNA was transcribed in vitro using the MEGAscript SP6 kit (Ambion) with Bio-11-UTP (Perkin-Elmer). Purified RNA was incubated with the S10 cytoplasmic fraction of Huh7 cells or in vitro-translated G3BP1. In vitro-translated G3BP1 and mutant forms were prepared using pCDNA3.1C-G3BP1 (and mutants) with the TNT quick-coupled transcription/translation system (Promega) according to the manufacturer's recommendations.
Luciferase assays.
For the luciferase activity assay, cells were washed twice with PBS and lysed in passive lysis buffer (Promega). Luciferase activity was measured with the Renilla luciferase assay system (Promega) in a Lumat LB9507 luminometer (Berthold).
Quantitative RT-PCR.
RNA from cells was harvested using Trizol reagent (Invitrogen) according to the manufacturer's protocol. After treatment with DNase I, each RNA sample was reverse transcribed using random primers (dN6). cDNAs were mixed with SYBR green PCR master mix (Qiagen) and subjected to reverse transcription-PCR (RT-PCR) using an ABI PRISM 7500 (Applied Biosystems). Cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA from the same RNA sample was used as an internal control for cell number and metabolic status normalization. The primers specific for the HCV 5′ NTR were 5NTR-sen (5′-CCC TGT GAG GAA CTA CTG TCT TCA CGC-3′) and 5NTR-anti (5′-GCT CAT GGT GCA CGG TCT ACG AGA CCT-3′). The primers specific for GAPDH were 5′-GGT ATC GTG GAA GGA CTC ATG AC-3′ (sense) and 5′-ATG CCA GTG AGC TTC CCG TTC AGC-3′ (antisense). The primers specific for G3BP1 were 5′-AAG AGT GCG AGA ACA ACG AA-3′ (sense) and 5′-TGG TGA CTG TCA GGG TGT CT-3′ (antisense). The primers specific for G3BP2 were 5′-GGT CAT GGG TTT GCT GTC TA-3′ (sense) and 5′-AGC CTT TGG TGG TTC TTG TG-3′ (antisense). Forty cycles of PCR were performed with cycling conditions of 15 s at 95°C, 20 s at 55°C, and 25 s at 72°C.
Immunofluorescence staining and confocal microscopic analysis.
Cells were fixed with 3.5% paraformaldehyde and permeabilized with 50 μg/ml digitonin in PBS for 5 min. After being washed with PBS twice, samples were then incubated at 4°C overnight with antibody (NS5A, 1:200; NS5B, 1:25; G3BP1, 1:25). After being washed thrice with PBS containing 1% FBS, samples were incubated at 37°C for 1 h with Alexa 488 (1:200) or Cy3 (1:400)-conjugated secondary antibody. After being mounted on the coverslips, the cells were observed using a confocal laser scanning microscope (TCS-NT; Leica, Heidelberg, Germany) equipped with a 64× NA 1.4 oil immersion objective. Images were acquired as previously described (7) at a 1,024-by-1,024 pixel resolution. Images were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/index.html); Pearson and Spearman correlation coefficients were used to calculate colocalization as described previously (12).
Western blot analysis.
Cells were lysed directly with 2× SDS loading buffer (100 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 3% β-mercaptoethanol, 0.02% bromophenol blue) and boiled. Samples were separated by 10% SDS-PAGE, transferred to nitrocellulose membrane (Schleicher & Schuell BioScience), and blocked with 5% milk in PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, and 0.1% Tween 20, pH 7.4). Blots were probed with different primary antibodies followed by secondary antibody conjugated with horseradish peroxidase. Protein bands were visualized by ECL Plus chemiluminescence reagent (PerkinElmer).
RESULTS
G3BP1 interacts with HCV NS5B but not NS3, NS5A, or NS4B.
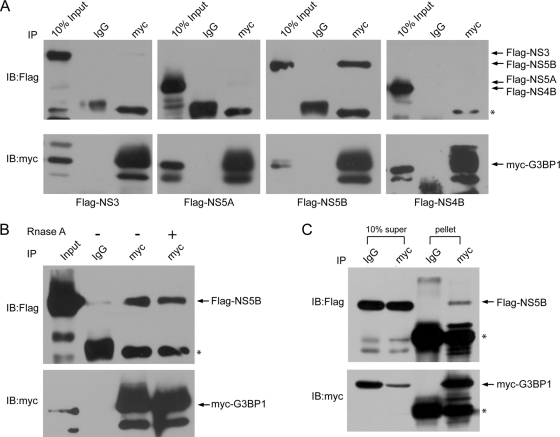
We previously reported that G3BP1 interacts with HCV NS5B, the viral RNA-dependent RNA polymerase, and the HCV 5′(−)TR RNA, suggesting that G3BP1 might be a component of the HCV replication complex (RC) (40, 49). Here, we verify the G3BP1-NS5B interaction and further investigate the interactions between G3BP1 and other HCV nonstructural proteins. Myc-tagged G3BP1 and Flag-tagged NS3-4A (which mediates self-cleavage to generate NS3), NS5A, NS5B, or NS4B were coexpressed in HEK293T cells, and their interaction was investigated by coimmunoprecipitation. G3BP1 was immunoprecipitated from cell extracts using anti-myc monoclonal antibody (or control normal mouse IgG), and NS5B was found to be associated with G3BP1 as detected by anti-Flag antibody (Fig. 1A). To rule out the possibility that G3BP1 and NS5B were coprecipitated due to an ability to bind to a common RNA, the lysates were treated with RNase prior to immunoprecipitation and Western blot analysis. As seen in Fig. 1B, RNase digestion did not abrogate the G3BP1-NS5B coprecipitation, suggesting that their interaction is mediated by a protein-protein interaction. To ensure that the interaction was not cell type dependent, the interaction was confirmed in Huh7 cells (Fig. 1C), which are permissive to HCV infection.
Fig. 1.
Interactions between G3BP1 and HCV nonstructural proteins. (A) HEK293T cells were transfected with pCMV-Myc-G3BP1 and pcDNA3.1A/3×Flag-NS3-4A, NS5A, NS5B, and NS4B, respectively. Cell lysates were immunoprecipitated (IP) with anti-myc monoclonal antibody or mouse normal IgG as a control. The immunoprecipitated fractions and 10% of the cell lysates (10% Input) were then subjected to Western blot analysis. The blot was first analyzed using anti-Flag antibody and then reprobed with anti-myc antibody after stripping. Similar results were obtained in two other independent experiments. (B) The G3BP1 interaction with HCV NS5B is insensitive to RNase treatment. Cell lysates obtained as in panel A were treated with RNase A (50 μg/ml) at room temperature for 5 min and were immunoprecipitated (IP) with anti-myc monoclonal antibody or mouse normal IgG as a control. (C). G3BP1 interacts with HCV NS5B in Huh7 cells. Huh7 cells were transfected with pCMV-Myc-G3BP1 and pcDNA3.1A/3×Flag-NS5B. Cell lysates were immunoprecipitated (IP) with anti-myc monoclonal antibody or mouse normal IgG as a control. The immunoprecipitated fractions (pellet) and 10% of the remaining supernatant fractions (10% super) were then subjected to Western blot analysis. Asterisks indicate IgG heavy-chain bands in NS3, NS5A, and NS5B blots and light-chain bands in the NS4B blot. Arrows to the right indicate the migration of the relevant proteins.
G3BP1 interaction with the HCV minus-strand RNA 5′ terminal region [5′(−)TR] requires the RRM motif.
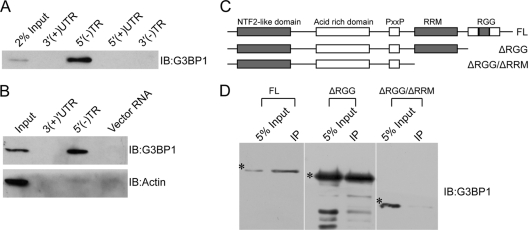
To determine whether G3BP1 directly binds to HCV 5′(−)TR RNA, we performed RNA pulldown experiments with in vitro-translated G3BP1. To confirm the RNA sequence specificity, we also included the HCV 5′(+)UTR RNA (containing the IRES) and its complementary-strand 3′(−)TR, as well as the 3′(+)UTR. In vitro-translated G3BP1 interacted exclusively with the HCV 5′(−)TR (Fig. 2A). To verify the interactions between endogenous cellular G3BP1 and the HCV 5′(−)TR RNA, biotinylated HCV 5′(−)TR RNA and its complementary-strand 3′(+)UTR, as well as an irrelevant vector-derived RNA fragment of similar length, were incubated with Huh7 S10 lysate. RNA-protein complexes were then affinity purified by immobilized streptavidin-magnetic beads and analyzed by Western blotting with anti-G3BP1 and anti-β-actin antibodies. Under these conditions, G3BP1, but not β-actin, was coisolated specifically with the HCV 5′(−)TR RNA (Fig. 2B).
Fig. 2.
Interactions between G3BP1 and HCV RNAs. (A) HCV 5′(−)TR interacts with in vitro-translated G3BP1. Biotinylated HCV 3′(+)UTR, HCV 5′(−)TR, HCV5′(+)UTR, and HCV 3′(−)TR RNAs were incubated with in vitro-translated G3BP1. RNA-protein complexes were affinity purified and detected with anti-G3BP1 antibody by Western blot analysis. (B) HCV 5′(−)TR interacts with G3BP1 from Huh7 lysates. In vitro-transcribed biotinylated HCV 3′(+)UTR RNA, HCV 5′(−)TR RNA, and a fragment of vector RNA were incubated with Huh7 S10 lysates. RNA-protein complexes were affinity purified with immobilized streptavidin-magnetic beads and detected with anti-G3BP1 and anti-β-actin antibodies by Western blot analysis. (C) Schematic of G3BP1 truncation mutants. (D) Interaction of HCV 5′(−)TR RNA with G3BP1 truncation mutants. Biotinylated HCV 5′(−)TR RNA was incubated with in vitro-translated G3BP1, G3BP1-ΔRGG, and G3BP1-ΔRGG/ΔRRM. RNA-protein complexes were affinity purified and detected with anti-G3BP1 antibody by Western blot analysis. Asterisks indicate the G3BP or mutant bands. IB, immunoblot; FL, full length. Similar results were obtained in at least one other independent experiment.
As both the RRM and RGG box of G3BP1 are suggested to mediate RNA binding (29), we tested the ability of mutant G3BP1 lacking these domains to bind to the HCV 5′(−)TR RNA. Biotinylated HCV 5′(−)TR RNA was incubated with in vitro-translated G3BP1, G3BP1-ΔRGG/ΔRRM lacking both the RRM and RGG box, and G3BP1-ΔRGG that lacks the RGG box (Fig. 2C). RNA-protein complexes were affinity purified with immobilized streptavidin-magnetic beads. G3BP1-ΔRGG could still bind to HCV 5′(−)TR RNA, although with a relative reduction in binding activity compared to that seen with full-length G3BP1 (Fig. 2D). In contrast, G3BP1-ΔRGG/ΔRRM completely lost HCV 5′(−)TR RNA binding activity. Taken together, these results indicate that the RRM motif of G3BP1 mediates its specific binding to the HCV 5′(−)TR RNA.
G3BP1 is associated with HCV RCs.
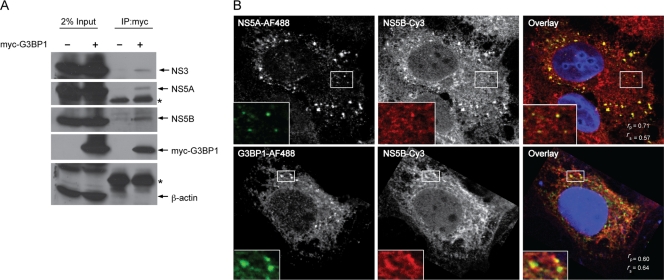
Since G3BP1 interacted with both HCV NS5B and the 5′(−)TR RNA, we wondered if G3BP1 might be a component of the HCV RC. In order to investigate this possibility, we examined the association of G3BP1 with HCV nonstructural proteins in the context of HCV replication. Myc-tagged G3BP1 was expressed in Huh7 cells harboring an HCV subgenomic replicon, and cells were broken by hypotonic buffer to maintain the integrity of the HCV RCs. Cell lysates were incubated with anti-myc monoclonal antibody to immunoprecipitate the tagged G3BP1. Under these conditions, HCV NS3, NS5A, and NS5B, but not the abundant cellular protein β-actin, coprecipitated with the myc-tagged G3BP1 (Fig. 3A). It is likely that G3BP1 interacts with HCV NS5B in a complex with NS3 and NS5A because when expressed individually, G3BP1 interacts only with HCV NS5B and not NS5A or NS3 (Fig. 1A).
Fig. 3.
G3BP1 is associated with HCV RCs. (A) G3BP1 is associated with HCV nonstructural proteins in HCV replicon cells. Replicon cells were transfected as indicated with pCMV-Myc-G3BP1 as described in Materials and Methods. Cells were lysed in hypotonic buffer, and proteins were immunoprecipitated with anti-Myc antibody. Immunoprecipitated complexes were analyzed by Western blotting using the indicated antibodies. The asterisk indicates migration of the IgG heavy chain. Comparable results were obtained in a similar independent experiment. (B) Colocalization of G3BP1 with HCV RCs in HCV replicon cells. Replicon cells were fixed and double stained with mouse anti-HCV NS5A and rabbit anti-HCV NS5B or mouse anti-G3BP1 and rabbit anti-HCV NS5B antibodies, followed by AF488-conjugated anti-mouse IgG and Cy3-conjugated anti-rabbit IgG antibodies. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Cells were visualized by confocal microscopy and analyzed by ImageJ. Pearson's colocalization coefficient (Rp) and Spearman's colocalization coefficient (Rs) were calculated and are shown in the merged (overlay) image. An enlarged view of the boxed regions is shown in the lower left. Comparable results were obtained in two additional independent experiments.
To further verify the presence of G3BP1 within HCV RCs, we utilized confocal microscopy of Huh7 cells harboring an HCV replicon, where HCV RCs appear as small dot-like structures (47). We first verified the colocalization of HCV NS5A and NS5B in the HCV RCs. As shown in Fig. 3B, HCV NS5A and NS5B colocalized well on dot-like structures (Rp = 0.71; Rs = 0.57). We found that G3BP1 also colocalized (Fig. 3B) with HCV NS5B on dot-like structures (Rp = 0.60; Rs = 0.64). This colocalization, together with our finding of G3BP1 in a complex containing NS3, NS5A, and NS5B, suggests that G3BP1 is a component of the HCV RC.
Reduction of G3BP1 levels reduces HCV replication and infection.
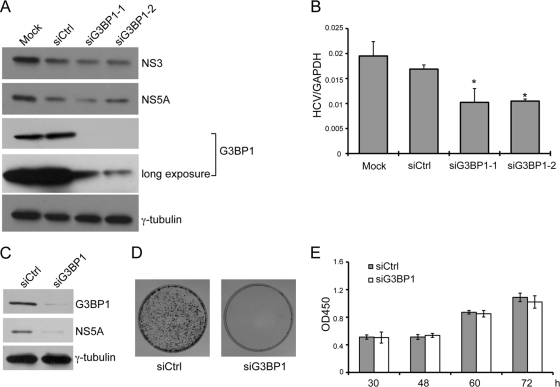
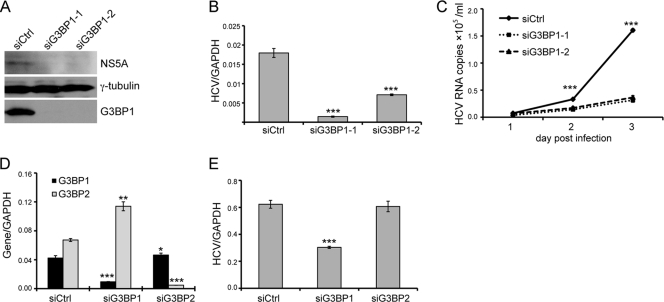
Since G3BP1 is localized to HCV RCs, we wondered whether it modulates RC formation or viral RNA replication. To address this, we examined HCV replication and infection when levels of G3BP1 were reduced by siRNA-mediated RNA silencing. First, we examined the effect of G3BP1 reduction in replicon cells in which HCV RCs have already been established. Replicon cells were transfected with duplex siRNAs targeting G3BP1 or a scrambled control duplex siRNA, and HCV RNA and protein levels were examined. Transfection of two independent G3BP1-targeting siRNAs (siG3BP1-1 and siG3BP1-2) each resulted in a slight reduction in HCV protein levels, as determined by NS3 and NS5A levels by Western blotting (Fig. 4A). HCV RNA levels in the silenced cells (Fig. 4B) were moderately, but significantly (P < 0.05), reduced compared to levels in mock-silenced cells (decreased 39% for siG3BP1-1 and 37% for siG3BP1-2). As HCV RC formation and HCV RNA replication have already been established and are relatively stable in replicon cells (47), we examined the effect of reduced levels of G3BP1 on the ability of HCV to establish and maintain replication using a colony formation assay. G3BP1-silenced or control siRNA-treated Huh7 cells were electroporated with HCV S2204I bsd replicon RNA, which after translation is capable of RC formation and genome amplification. Expression of blasticidin allows for the formation of drug-resistant colonies. G3BP1 silencing efficacy and HCV protein levels were measured by Western blot analysis 1 day after electroporation (Fig. 4C). In this situation, where the incoming viral RNA must be translated and HCV RCs formed de novo, silencing of G3BP1 resulted in a more dramatic reduction in HCV protein levels (compare Fig. 4C and A). Moreover, replication in cells with reduced G3BP1 levels failed to reach levels sufficient to establish drug-resistant colonies (Fig. 4D). To exclude the possibility that silencing of G3BP1 resulted in cell toxicity affecting HCV replication indirectly, growth of the siRNA-treated cells was determined using a cell viability assay. As shown in Fig. 4E, silencing of G3BP1 had no effect on cell growth.
Fig. 4.
Knockdown of G3BP1 reduces HCV subgenomic replicon RNA replication. (A and B) Replicon cells were left untransfected (Mock) or were transfected with an siRNA duplex targeting G3BP1 or control siRNA as indicated at a final concentration of 50 nM. After 48 h, the cells were harvested with SDS loading buffer for Western blot analysis (A) or with Trizol reagent for RNA extraction (B). (A) Western blot analysis is shown, using the antibodies indicated to the right. (B) RNA levels were measured by quantitative RT-PCR. HCV RNA levels were normalized to levels of GAPDH RNA. Mean HCV RNA values from triplicate wells are shown. Error bars indicate the standard deviation; asterisks indicate values different from the Mock value (Student's t test, *, P < 0.05). Similar results were obtained in two additional independent experiments. (C to E) Huh7 cells were transfected with siG3BP1-1 (siG3BP1) or control siRNA (siCtrl) as indicated at a final concentration of 50 nM. (C and D) Twenty-four hours later, the cells were electroporated with 10 μg of HCV replicon (S2204I bsd) RNA. (C) At 1 day postelectroporation, equal portions of the cells were analyzed by Western blotting, using the antibodies indicated to the right; γ-tubulin serves as a loading control. (D) The remaining cells were grown in conditioned media supplemented with 20 μg/ml of blasticidin for about 1 month, and colonies were visualized by crystal violet staining. (E) The effect of G3BP1 silencing on Huh7 cell growth was examined. After transfection with the indicated siRNAs, the relative cell number (OD450) was determined at the indicated time points using the Cell Counting Kit-8. For panels C to E, similar results were obtained in at least one additional independent experiment.
We next examined the effect of reduced levels of G3BP1 on HCV infection. Control or G3BP1-silenced Huh-7.5 cells were infected with HCV JFH1 (genotype 2a), and culture supernatants were harvested daily for determination of virion release by quantifying HCV genome in the culture media. Infected cells were harvested at 3 days postinfection to confirm G3BP1 silencing and to monitor HCV protein (Western blot analysis) and RNA (quantitative RT-PCR) levels. Silencing of G3BP1 reduced HCV NS5A protein levels (Fig. 5A) and had a marked, statistically significant (P < 0.05) effect on intracellular HCV RNA levels (Fig. 5B), with a 90% and 60% reduction in the siG3BP1-1- and siG3BP1-2-treated cells, respectively. Virion production, as measured by HCV RNA released into the culture media, was also significantly reduced in siG3BP1-treated cells (Fig. 5C).
Fig. 5.
Knockdown of G3BP1 reduces HCVcc infection. (A to C) Huh-7.5 cells were transfected with the indicated siRNAs targeting G3BP1 or control siRNA at a final concentration of 50 nM. After 24 h, the siRNA-treated Huh-7.5 cells were inoculated with JFH1 HCVcc at a multiplicity of infection of 0.1. Culture supernatants were harvested daily until 3 days postinfection; infected cells were harvested at 3 days postinfection. (A) Western blot analysis of cells harvested 3 days after JFH1 infection. The antibodies used are indicated to the right; γ-tubulin serves as a loading control. (B) Intracellular RNA levels 3 days postinfection were quantified by quantitative RT-PCR. Mean HCV RNA levels normalized to the level of GAPDH RNA from triplicate wells are shown; error bars indicate the standard deviation. (C) Levels of HCV RNA in the culture media were determined at the indicated times by quantitative RT-PCR. (D and E) Huh-7.5 cells were transfected with siG3BP1-1 (siG3BP1), siG3BP2, or control siRNA (siCtrl) as indicated at a final concentration of 50 nM. (D) Silencing efficiency was measured after 2 days by quantitative RT-PCR using primers specific for G3BP1 and G3BP2. The level of G3BP1 or G3BP2 RNA was normalized to the level of GAPDH RNA. Mean normalized values from triplicate samples are plotted; error bars indicate the standard deviation. (E) Huh-7.5 cells treated with the indicated siRNAs were inoculated 24 h later with JFH1 HCVcc. RNA was harvested 48 h after infection, and the amount of HCV RNA was determined by quantitative RT-PCR. Mean HCV RNA levels from triplicate samples, normalized for GAPDH level, are plotted; error bars indicate the standard deviation. Asterisks indicate significant differences from the control (siCtrl, Student's t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). Similar results were obtained in two additional independent experiments.
Mammalian cells have three G3BP family members (reviewed in reference 17), including G3BP1 and two isoforms of a highly related protein designated G3BP2 (31). To investigate whether G3BP2 might play a role in HCV replication, we treated cells with siRNAs targeting either G3BP1 or G3BP2. We found that the siRNAs against these two proteins were very specific, and interestingly, as has previously been reported (10), silencing G3BP1 resulted in the reciprocal induction of G3BP2 (Fig. 5D). As shown in Fig. 5E, silencing of G3BP1 reduced cell-culture-produced HCV (HCVcc) infection, as measured by HCV genome RNA levels, while silencing of G3BP2 had no effect. This suggests that G3BP2 is unlikely to be involved in HCV replication.
G3BP1 is involved in HCV genome amplification.
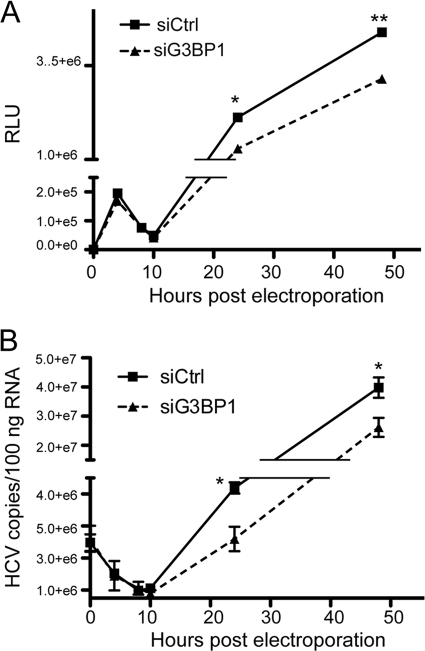
Since our findings demonstrated reduced HCV RC formation, protein production and RNA replication upon G3BP1 silencing, we wondered in which virus life cycle step G3BP1 might be involved. To address this, we took advantage of a recombinant genotype 2a HCV [J6/JFH(5′C19Rluc2AUbi), described in reference 43] expressing a Renilla luciferase reporter protein as a self-cleaving amino-terminal fusion protein, which allows very sensitive quantification of genome translation. Using this virus, translation of the genome can be monitored prior to genome RNA replication, as documented by detectable luciferase activity after electroporation of RNA containing mutations in the NS5B RNA polymerase active site (50). As shown in Fig. 6A, the initial translation of input HCV genomic RNA could be measured before 10 h postelectroporation, and levels were comparable in control cells and cells in which G3BP1 expression had been silenced. During this same early period, genome RNA levels (Fig. 6B) gradually decreased and were similar in control and G3BP1-silenced cells, suggesting similar RNA decay rates in the two cell populations. After 10 h postelectroporation, levels of HCV RNA increased (Fig. 6B), demonstrating productive RNA replication in the cells. Interestingly, despite similar early translation, silencing of G3BP1 resulted in decreased accumulation of HCV RNA at later times compared to that seen in the control cells; luciferase levels were similarly affected (Fig. 6A). That reduced G3BP1 levels did not directly affect HCV translation was verified using an HCV IRES-driven reporter system (data not shown). Taken together, the results suggest that G3BP1 functions after translation and at the genome amplification step of the virus life cycle.
Fig. 6.
G3BP1 is involved in the genome amplification step of the HCV replication cycle. Huh-7.5 cells were transfected with control siRNA (siCtrl) or siG3BP1-1 (siG3BP1) as indicated and were electroporated 48 h later with 10 μg of J6/JFH(5′C19Rluc2AUbi) HCV RNA. Cells were harvested at the indicated time points with Trizol reagent for RNA analysis or passive lysis buffer for the Renilla luciferase assay. (A) Luciferase values are shown; data points show mean values from triplicate wells. RLU, relative light units. (B) HCV RNA copy number was determined by quantitative RT-PCR. Mean values from triplicate samples are plotted. For both A and B, error bars indicate the standard deviation and are sometimes obscured by the symbol. Asterisks indicate statistically different values for a given time point (Student's t test; *, P < 0.05; **, P < 0.01). Comparable results demonstrating an effect on genome amplification were obtained in similar experiments utilizing polymerase defective JFH1 RNA and replicon RNA (not shown).
DISCUSSION
In previous studies we found that G3BP1 interacts with HCV NS5B and the HCV 5′(−)TR RNA (40, 49), which raised the possibility that G3BP1 is a potential HCV RC component. In this study, we verified that G3BP1 is associated with the HCV RC. First, we further explored the interactions of G3BP1 with HCV NS5B and other nonstructural proteins (Fig. 1) and the HCV 5′(−)TR RNA (Fig. 2). Immunoisolation of G3BP1 resulted in coimmunoprecipitation of multiple HCV nonstructural proteins in replicon cells (Fig. 3A), suggesting that G3BP1 is associated with HCV RCs. Furthermore, we demonstrated that G3BP1 colocalized with the HCV RC in HCV replicon cells (Fig. 3B), as determined by colocalization with NS5B.
To demonstrate if G3BP1 is involved in HCV replication, we used siRNA-mediated silencing to reduce levels of endogenous G3BP1 and then monitored HCV RNA replication. Silencing of G3BP1 in replicon cells was very efficient, as verified by Western blot analysis (Fig. 4A), and resulted in a moderate reduction of HCV RNA replication (Fig. 4A and B). In contrast, silencing of G3BP1 reduced HCV replication to a greater extent during HCV infection (Fig. 5A, B, and C). This difference might be explained by G3BP1 additionally affecting virus entry or assembly, life cycle steps not necessary for replicon replication. Alternatively, G3BP1 already recruited to HCV RCs in the replicon cells might be refractory to siRNA-mediated silencing. To investigate the possibility that G3BP1 may be involved in virus entry or assembly, steps not relevant for HCV replicon replication, we used a colony-forming assay after electroporation of HCV replicon RNAs. Analysis by Western blot and colony formation assay demonstrated that reduced G3BP1 levels dramatically reduced viral protein expression and totally inhibited replication competent colony formation (Fig. 4C and D), which depends on viral RNA replication and genome translation. While we cannot exclude additional effects on entry or assembly, taken together the findings suggest that G3BP1 is involved at a minimum in a postentry, assembly-independent step.
It is likely that the differential effect of G3BP1 silencing of replicon cells, where RCs are already established, and of G3BP1 silencing prior to establishment of RCs by replicon RNA electroporation may be due to the timing of the silencing relative to RC establishment. These data may indicate that G3BP1 is involved in HCV replication prior to or at HCV replication establishment. We further showed that G3BP1 is involved in HCV genome amplification while not affecting translation (Fig. 6). Since G3BP1 interacts with HCV NS5B and the minus-strand copy of the genome (Fig. 1 and 2), these data raised the possibility that G3BP1 is incorporated into the HCV RC and functions as an RC component. One characteristic of HCV RCs is that fewer than 5% of the viral nonstructural proteins are required for viral RNA replication and cells contain a large abundance of excess viral proteins relative to the amount directly involved in HCV replication (24, 32). Accordingly, it is possible that host factors involved in HCV RCs are also present in excess of that needed for HCV replication. As HCV RCs were stable for up to 18 h without significant turnover (47) it is likely that in HCV replicon cells, the HCV RCs are already established and remain stable, and host factors involved in the HCV RC, including G3BP1, likely have already been recruited and remain stable. Thus, in replicon cells, silencing of G3BP1 could only moderately affect the incorporation of G3BP1 into HCV RCs. In contrast, in the colony formation assay and during HCV infection, silencing of G3BP1 prior to RC formation was able to dramatically reduce HCV replication.
G3BP1 is recruited by another positive-sense RNA virus, Sindbis virus, in which, similarly to the case with HCV, it was found to interact with the viral RNA-dependent RNA polymerase (10). For Sindbis virus, G3BP1 appears to play a role in genome translation regulation (10). We demonstrate here that while G3BP1 interacts with HCV NS5B as well as the HCV negative-strand RNA, it is able to interact directly with NS5B in an RNA-independent manner (Fig. 1B). Thus, it is likely that for both Sindbis virus and HCV, G3BP1 is recruited to replication complexes by protein-protein interactions. In contrast to the situation with Sindbis virus, where the RNA polymerase-G3BP1 interaction may function to regulate translation of newly generated positive-sense RNA, the HCV NS5B-G3BP1 interaction may be a means by which this host factor is usurped by HCV to the replication complex in order to facilitate RNA replication. Since G3BP1 binds to the HCV 5′(−)TR RNA, G3BP1 might play a role in negative-strand RNA elongation or positive-strand RNA synthesis.
G3BP1 has also been shown to facilitate vaccinia virus intermediate-stage transcription, possibly through binding to transcript 5′-poly(A) leader sequences of variable length (19). G3BP1 also selectively binds the 3′ UTR of cellular mRNAs (28, 42). Prior studies utilizing an in vitro iterative genetic selection approach identified ACCCA(U/C)(A/C)(C/G)G(C/A)AG as a consensus sequence for G3BP1 binding (42). Within the HCV 5′(−)TR are two regions with partial homology to this G3BP1 binding consensus sequence (data not shown) as well as a poly(A) tract. Determining the role of these RNA sequences in G3BP1 binding and the role of the G3BP1-RNA interaction in HCV replication requires further investigation.
In conclusion, we demonstrate that HCV co-opts G3BP1 for its replication and that reduction of this host factor inhibits the formation of new HCV RCs. Understanding the host factors that contribute to HCV RNA replication not only facilitates our understanding of the molecular mechanism of HCV replication but also may aid the development of new antiviral therapeutic approaches.
Acknowledgements
This work was supported by the Chinese State Basic Research Foundation Grant (grant no. 2009CB522504), the National Mega Project for Infectious Diseases (2008ZX10002-002), the Shanghai Program for Outstanding Medical Academic Leaders, the National Institutes of Health (grant CA057973, C.M.R.), the Greenberg Medical Research Institute, and the Starr Foundation. Zhigang Yi is the recipient of a Chinese National Natural Science Grant (30700738).
We thank T. Wakita for HCV strain JFH1 and D. Kennedy for the G3BP1 plasmids.
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Ahlquist P., Noueiry A. O., Lee W. M., Kushner D. B., Dye B. T. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atlas R., Behar L., Elliott E., Ginzburg I. 2004. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 89:613–626 [DOI] [PubMed] [Google Scholar]
- 3. Bienz K., Egger D., Pfister T., Troxler M. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binder M., et al. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 81:5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blight K. J., Kolykhalov A. A., Rice C. M. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 6. Blight K. J., McKeating J. A., Rice C. M. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bolte S., Cordelieres F. P. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 [DOI] [PubMed] [Google Scholar]
- 8. Cohen M., Stutz F., Dargemont C. 2003. Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J. Biol. Chem. 278:51989–51992 [DOI] [PubMed] [Google Scholar]
- 9. Costa-Mattioli M., Svitkin Y., Sonenberg N. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cristea I. M., et al. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 84:6720–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans M. J., Rice C. M., Goff S. P. 2004. Genetic interactions between hepatitis C virus replicons. J. Virol. 78:12085–12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. French A. P., Mills S., Swarup R., Bennett M. J., Pridmore T. P. 2008. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3:619–628 [DOI] [PubMed] [Google Scholar]
- 13. Friebe P., Bartenschlager R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friebe P., Lohmann V., Krieger N., Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047–12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao L., Aizaki H., He J. W., Lai M. M. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamamoto I., et al. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473–13482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irvine K., Stirling R., Hume D., Kennedy D. 2004. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 48:1065–1077 [DOI] [PubMed] [Google Scholar]
- 18. Isken O., et al. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 13:1675–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katsafanas G. C., Moss B. 2004. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 279:52210–52217 [DOI] [PubMed] [Google Scholar]
- 20. Kieft J. S., Grech A., Adams P., Doudna J. A. 2001. Mechanisms of internal ribosome entry in translation initiation. Cold Spring Harbor Symp. Quant. Biol. 66:277–283 [DOI] [PubMed] [Google Scholar]
- 21. Kolykhalov A. A., Mihalik K., Feinstone S. M., Rice C. M. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kopek B. G., Perkins G., Miller D. J., Ellisman M. H., Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 24. Miyanari Y., et al. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301–50308 [DOI] [PubMed] [Google Scholar]
- 25. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 26. Noueiry A. O., Ahlquist P. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77–98 [DOI] [PubMed] [Google Scholar]
- 27. Okamoto T., et al. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ortega A. D., Willers I. M., Sala S., Cuezva J. M. 2010. Human G3BP1 interacts with beta-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 123:2685–2696 [DOI] [PubMed] [Google Scholar]
- 29. Parker F., et al. 1996. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol. Cell. Biol. 16:2561–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poenisch M., Bartenschlager R. 2010. New insights into structure and replication of the hepatitis C virus and clinical implications. Semin. Liver Dis. 30:333–347 [DOI] [PubMed] [Google Scholar]
- 31. Prigent M., Barlat I., Langen H., Dargemont C. 2000. IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J. Biol. Chem. 275:36441–36449 [DOI] [PubMed] [Google Scholar]
- 32. Quinkert D., Bartenschlager R., Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79:13594–13605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rahmouni S., et al. 2005. Removal of C-terminal SRC kinase from the immune synapse by a new binding protein. Mol. Cell. Biol. 25:2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Randall G., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36. Schaad M. C., Jensen P. E., Carrington J. C. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwartz M., et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 38. Soncini C., Berdo I., Draetta G. 2001. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene 20:3869–3879 [DOI] [PubMed] [Google Scholar]
- 39. Tellinghuisen T. L., Rice C. M. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419–427 [DOI] [PubMed] [Google Scholar]
- 40. Tingting P., Caiyun F., Zhigang Y., Pengyuan Y., Zhenghong Y. 2006. Subproteomic analysis of the cellular proteins associated with the 3′ untranslated region of the hepatitis C virus genome in human liver cells. Biochem. Biophys. Res. Commun. 347:683–691 [DOI] [PubMed] [Google Scholar]
- 41. Tourriere H., et al. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Tourriere H., et al. 2001. RasGAP-associated endoribonuclease G3BP: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 21:7747–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tscherne D. M., et al. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang C., et al. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425–434 [DOI] [PubMed] [Google Scholar]
- 45. Wang R. Y., Nagy P. D. 2008. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3:178–187 [DOI] [PubMed] [Google Scholar]
- 46. Watashi K., et al. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111–122 [DOI] [PubMed] [Google Scholar]
- 47. Wölk B., Büchele B., Moradpour D., Rice C. M. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yi M., Lemon S. M. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yi Z., et al. 2006. Subproteomic study of hepatitis C virus replicon reveals Ras-GTPase-activating protein binding protein 1 as potential HCV RC component. Biochem. Biophys. Res. Commun. 350:174–178 [DOI] [PubMed] [Google Scholar]
- 50. You S., Rice C. M. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J. Virol. 82:184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]