Abstract
MBD2 and MBD3 are closely related proteins with consensus methyl-CpG binding domains. MBD2 is a transcriptional repressor that specifically binds to methylated DNA and is a component of the MeCP1 protein complex. In contrast, MBD3 fails to bind methylated DNA in murine cells, and is a component of the Mi-2/NuRD corepressor complex. We show by gene targeting that the two proteins are not functionally redundant in mice, as Mbd3(−/−) mice die during early embryogenesis, whereas Mbd2(−/−) mice are viable and fertile. Maternal behavior of Mbd2(−/−) mice is however defective and, at the molecular level, Mbd2(−/−) mice lack a component of MeCP1. Mbd2-mutant cells fail to fully silence transcription from exogenous methylated templates, but inappropriate activation of endogenous imprinted genes or retroviral sequences was not detected. Despite their differences, Mbd3 and Mbd2 interact genetically suggesting a functional relationship. Genetic and biochemical data together favor the view that MBD3 is a key component of the Mi-2/NuRD corepressor complex, whereas MBD2 may be one of several factors that can recruit this complex to DNA.
Keywords: Methylation, chromatin, transcription, MBD proteins, behavior, embryogenesis
DNA methylation is essential for mammalian development, as mice lacking either the maintenance methyltransferase DNMT1 (Li et al. 1992) or de novo methyltransferases DNMT3α and DNMT3β (Okano et al. 1999) fail to complete development. The primary effect of DNA methylation is to repress transcription (Bird 1992), notably in situations where CpG islands become methylated, such as in genomic imprinting (Bartolomei and Tilghman 1997), X chromosome inactivation (Heard et al. 1997) and tumor-suppressor gene inactivation in cancer cells (Herman and Baylin 2000). DNA methylation also has been shown to cause repression of repetitive DNA element promoters during murine embryogenesis (Walsh et al. 1998). Recently, it has been demonstrated that DNA methylation is important for the appropriate activation of stage-specific genes in Xenopus development (Stancheva and Meehan 2000) and for silencing of tissue-specific genes and repetitive DNA elements in murine fibroblast cultures (Jackson-Grusby et al. 2001). DNA methylation-mediated transcriptional silencing is achieved at least in part through an indirect mechanism in which a methyl-CpG binding protein specifically binds to methylated DNA to bring about transcriptional repression (Boyes and Bird 1991; for reviews, see Bird and Wolffe 1999; Hendrich and Bird 2000). The first methyl-CpG binding activity to be identified was the Methyl-CpG Binding Protein 1 (MeCP1; Meehan et al. 1989). MeCP1 is a large protein complex of 400–800 kD that binds >12 methylated CpGs, irrespective of sequence context. Transcriptional repression by MeCP1 was found to correlate with methylation density (Boyes and Bird 1992). In contrast to MeCP1, MeCP2 consists of a single polypeptide that is capable of binding to a single symmetrically methylated CpG dinucleotide in vitro (Lewis et al. 1992). Two functional domains have been identified within the MeCP2 protein: a methyl-CpG binding domain that is necessary and sufficient to target the protein to methylated DNA (Nan et al. 1993, 1996), and a transcriptional repression domain (TRD) that recruits histone deacetylase (HDAC) activity (Jones et al. 1998; Nan et al. 1997, 1998) to bring about transcriptional silencing.
In order to identify potential components of MeCP1, the EST databases were searched for genes capable of encoding protein motifs similar to the methyl-CpG binding domain of MeCP2. A family of four proteins, MBD1–MBD4, was identified based upon the presence in each of a methyl-CpG binding domain (Cross et al. 1997; Hendrich and Bird 1998). Three of these proteins, MBD1–MBD3, are implicated in transcriptional repression (Bird and Wolffe 1999) whereas MBD4 is a mismatch repair protein (Hendrich et al. 1999). A short form of the MBD2 protein (MBD2b; Hendrich and Bird 1998) was reported to directly demethylate DNA through the removal of a methyl group from 5-methylcytosine (Bhattacharya et al. 1999). As methylation is associated with gene silencing, a demethylase would be expected to act as a transcriptional activator. Contrary to this expectation, MBD2 was subsequently found to repress transcription through the recruitment of histone deacetylases (Ng et al. 1999). Further, both MBD2 and histone deacetylases were shown to be components of MeCP1 in HeLa cells.
The MBD3 protein is 70% identical to MBD2b, but lacks the 152 amino-acid N-terminal extension of MBD2 and contains an acidic C-terminal tail (Hendrich and Bird 1998) (Fig. 1a). The two proteins are encoded by genes showing an identical intron/exon structure and are likely to have arisen by gene duplication from a single ancestral gene (Hendrich et al. 1999). Despite these similarities, only the mammalian MBD2 protein has been found to bind methylated DNA both in vitro and in vivo (Hendrich and Bird 1998). A Xenopus MBD3-like protein, however, has been shown to bind methylated DNA in vitro (Wade et al. 1999). MBD3 was found to be a component of the NuRD or Mi-2 histone deacetylase and nucleosome remodeling complex purified from both HeLa cells and Xenopus egg extracts (Tong et al. 1998; Wade et al. 1998, 1999; Xue et al. 1998; Zhang et al. 1998, 1999). The NuRD corepressor complex is implicated in silencing in a number of contexts and in a variety of organisms including mammals, flies, nematodes, and plants (Ahringer 2000). MBD2 was not detected in the purified NuRD complex, but could be detected in an affinity-purified HDAC1 corepressor complex (Zhang et al. 1999), which probably included MeCP1 (Ng et al. 1999). In addition, MBD2 was shown to be capable of recruiting the NuRD complex to methylated DNA in vitro (Zhang et al. 1999).
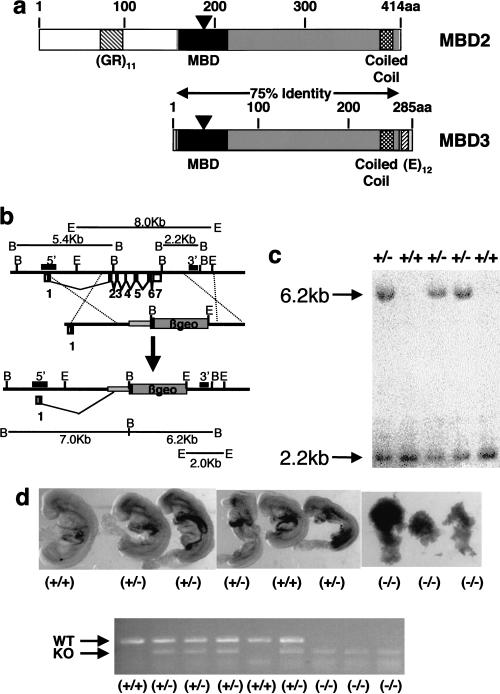
Figure 1.
Targeted deletion of Mbd3. (a) Schematic diagrams of the MBD2 and MBD3 proteins. Amino acids to the right of the black triangles are not encoded by the targeted alleles. The portion shown in dark gray is 75% identical at the amino-acid level between the two proteins. The methyl-CpG–binding domains are shown as black boxes, the putative coiled-coil domains are shown as stippled boxes, and the simple sequence repeats are shown as cross-hatched boxes. (b) The exons of the Mbd3 gene are indicated below the line with coding sequences indicated as black-filled boxes. Exons 2–7 were replaced by the βgeo cassette to generate the targeted allele. Probes used in Southern blots are indicated as hatched boxes above the line labeled either 5′ or 3′, and restriction fragments used for genotyping are indicated. (B) BglII, (E) EcoRI. (c) Southern blot of BglII digested DNA derived from five mice probed with the 3′ probe indicated in part a. Genotypes of the mice are indicated at the top. (d) 8.5 d.p.c. embryos from a single litter are shown at the top, with genotypes determined by the PCR results shown below. The order of embryos is the same as that for the PCR. The top band of the PCR is derived from the wild-type allele, whereas the lower band is derived from the targeted allele. Primers are visible at the bottom of the gel.
As a first step towards determining the in vivo functions and interrelationships of MBD2 and MBD3, we have employed a gene deletion strategy in mice. We find that while MBD3 is indispensable for embryonic development, MBD2 deficient mice are viable but show a maternal nurturing defect. Furthermore, we show that MBD2-deficient cells lack MeCP1 and are unable to efficiently repress methylated reporter constructs. Double mutants provide evidence for a genetic interaction between MBD2 and MBD3.
Results
Deletion of Mbd3 causes embryonic lethality
Targeted deletion of Mbd3 was achieved by replacing a genomic fragment containing exons 2–7 of the Mbd3 gene with a promoterless βgeo cassette (Skarnes et al. 1995) (Fig. 1b) in embryonic stem (ES) cells. This removed sequences encoding all but the N-terminal 36 amino acids of the protein (Fig. 1a,b). Two independently targeted ES cell clones were used to generate chimeric mice (lines 6C and 11F). Chimeras derived from both cell lines were used to generate heterozygous animals, but no homozygous null animals were recovered after intercrossing of heterozygotes. Genotyping of 497 3-week-old offspring resulting from heterozygote x heterozygote crosses revealed the presence of wild-type and heterozygote animals at close to the expected 1:2 ratio (1:1.8, Table 1), but no Mbd3-null animals were found among these pups. Mean litter sizes resulting from Mbd3 heterozygote × heterozygote crosses were 21% smaller than normal (6.37 vs. 8.07; p = 0.0427), implying that homozygous animals died prenatally or perinatally.
Table 1.
Evidence for a genetic interaction between Mbd2 and Mbd3 genes
| Mbd3 genotype:
|
(+/+)
|
(+/−)
|
(−/−)
|
|
|---|---|---|---|---|
|
Mbd2 (+/+)
|
||||
| Number | 179 | 318 | 0 | |
| Percentage | 36.0% | 64.0% | 0% | |
| Mdb2(−/−) | ||||
| Number | 86 | 115 | 0 | |
| Percentage | 42.8% | 57.2% | 0% | |
Number of animals of the three possible genotypes born to two Mdb3(+/−) parents in the presence [Mbd2(+/+)] or absence [Mbd2(−/−)] of an intact Mbd2 gene. Results are expressed as the total number of animals found with either genotype and the percentage of the total.
To determine when Mbd3(−/−) embryos were being lost, embryos were genotyped at various stages of development. Mbd3(−/−) blastocysts were detectable at 3.5 days post coitum (d.p.c.) but no normal-looking Mbd3-null embryos were recovered after implantation. At 8.5 d.p.c. Mbd3(−/−) embryos were severely retarded and in the process of resorption (Fig. 1d). We conclude that the Mbd3 gene is absolutely required for embryonic development.
Mbd2-deficient mice are viable
To generate an Mbd2-mutant allele, exon 2 of the Mbd2 gene was replaced with the promoterless βgeo cassette (Fig. 2a). Transcription initiating at the promoter of the targeted Mbd2 locus will proceed as normal through exon 1 and intron 1 but then should terminate at the transcription stop site located in the βgeo cassette, thus preventing transcription of the remainder of the Mbd2 gene. The resulting transcript can encode the N-terminal 183 amino acids of MBD2, but translation then stops in the middle of the methyl-CpG binding domain (Fig. 1a,2a). After transfection of embryonic stem cells with the targeting construct, ∼70% of neomycin-resistant clones were found to be properly targeted.
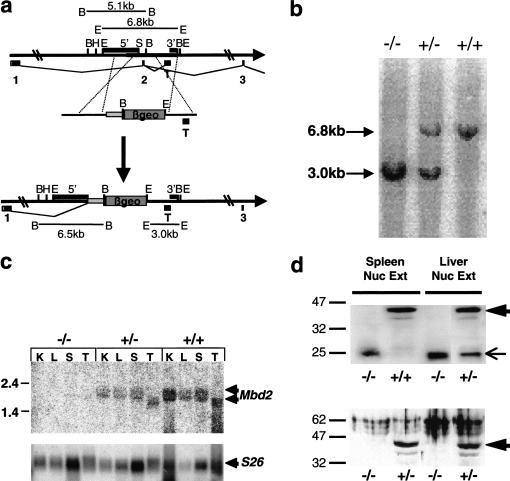
Figure 2.
Generation of Mbd2 targeted deletion. (a) Exon 2 of the Mbd2 gene was replaced by the promoterless βgeo cassette. Exons 1–3 are indicated below the line as is a testis-specific exon (T). Coding regions within the exons are shown in black. Restriction enzyme sites are indicated: (B) BglII, (E) EcoRI, (H) HindIII, (Sh) SphI. Probes used for Southern blots are shown as filled boxes above the line labeled either 5′ or 3′, and restriction fragments used for genotyping are indicated. (b) Southern blot of EcoRI-digested DNA derived from three littermates probed with the 3′ probe indicated in part a. (c) The Mbd2 transcript is visualized as a doublet on Northern blots of RNA from wild-type or heterozygous animals, but no Mbd2 transcript is detectable in Mbd2(−/−) animals. RNA was made from kidney (K), liver (L), spleen (S), and testis (T). The blot was stripped and rehybridized with a probe for the ribosomal S26 gene as a loading control. (d) Full-length MBD2 protein is absent in nuclear extracts derived from Mbd2(−/−) spleen or liver. The top panel was stained with the S923 antibody, which recognizes the full-length MBD2 protein (large arrow) as well as a smaller protein of ∼25 kD (small arrow), which is derived from the targeted allele. The bottom panel is stained with the R593 antibody, and the location of the full-length MBD2 protein is indicated with a large arrow. Protein size markers are indicated in kilodaltons.
Two independent targeted clones (5B and 10A) were used to inject blastocysts to create chimaeric mice. Intercrossing two Mbd2 heterozygous (+/−) animals produced progeny in a ratio of 13(+/+):31(+/−):12(−/−), which is not significantly different from the expected 1:2:1 ratio, leading us to conclude that, in contrast to Mbd3, a functional Mbd2 gene is not required for murine development. Mbd2(−/−) mice were viable, fertile, and of normal appearance. No differences between the two independently derived Mbd2-deleted lines were detectable in any of our assays. All results shown below were obtained with the mice derived from the 5B cell line. Northern blots showed a reduction of the Mbd2 transcript in heterozygous animals, and no normal Mbd2 transcript was detectable in (−/−) animals (Fig. 2c). RT–PCR of RNA derived from Mbd2(−/−) animals using primers located in exons 1 and 3 revealed the presence of a small amount of read-through transcript that is not detectable in wild-type animals. Neither of the detected read-through transcripts is capable of encoding more of the MBD2 protein than is included in exon 1 due to the presence of stop codons immediately after the exon 1 sequences in the targeted allele (data not shown). Western blots of spleen and liver nuclear extracts were used to verify the lack of MBD2 protein in Mbd2(−/−) animals. An antibody raised against the C terminus of MBD2 (R593; Ng et al. 1999) failed to detect any MBD2 protein in extracts derived from animals homozygous for Mbd2 disruption (Fig. 2d). A second antibody recognizing the N terminus of MBD2 (S923; Ng et al. 1999) revealed the lack of full-length protein in the Mbd2(−/−) tissues, but recognized a smaller band of ∼25 kD in heterozygous and homozygous mutant tissues. This is predicted to be encoded by exon 1 sequences, which remain unaffected in the targeted allele (Fig. 2a). This peptide may migrate slower than its predicted molecular weight (18 kD) because of the highly repetitive and basic nature of these amino acids. Any functions of MBD2 that are encoded solely by exon 1 are unlikely to involve targeted repression of methylated sites, as only the N-terminal half of the methyl-CpG binding domain is encoded in exon 1 (Hendrich et al. 1999), and the repression domain (Boeke et al. 2000) and putative coiled coil domain are encoded by downstream exons (Fig. 1a). We therefore conclude that our targeted Mbd2 allele is incapable of producing any of the known functional domains of the intact MBD2 protein, though any independent functions associated with the repetitive N terminus may be unaffected in this allele.
The Mbd2 and Mbd3 genes encode proteins that are over 70% identical (over the length of the MBD3 protein; Fig. 1a) and are implicated in the same protein complex (Zhang et al. 1999), but are evidently of different importance for development. In order to determine whether these two proteins interact genetically as well as physically, we crossed the Mbd3 mutation onto an Mbd2(−/−) background. Intercrossing two Mbd3(+/−) heterozygotes on an Mbd2(−/−) background produced no Mbd3-null progeny as expected, but the number of Mbd3(+/−) heterozygotes among the offspring was also significantly reduced compared to the equivalent cross on an Mbd2(+/ +) background. Table 1 shows that the ratio of Mbd3(+/−) to Mbd3(+/+) progeny was 1.3:1 in the absence of the Mbd2 gene, which is 30% less than the near-Mendelian ratio of 1.8:1 seen when the mice were wild-type for Mbd2. The results show that the viability of Mbd3(+/−) progeny is significantly reduced when Mbd2 is absent, whereas the viability of Mbd3(+/+) progeny generated in the same cross is unaffected (data not shown). The results demonstrate a genetic interaction between the Mbd2 and Mbd3 genes as the phenotype of the double mutant is stronger than that of either single mutant. It is possible that the reduced concentration of MBD3 protein in heterozygotes leads to increased failure of development when MBD2 protein is also absent. The stage at which development can interrupt is currently unknown. Other aspects of the Mbd2(−/−) phenotype (see below) are not obviously affected by heterozygosity for the Mbd3 mutation.
Normal methylation levels in MBD2(−/−) mice
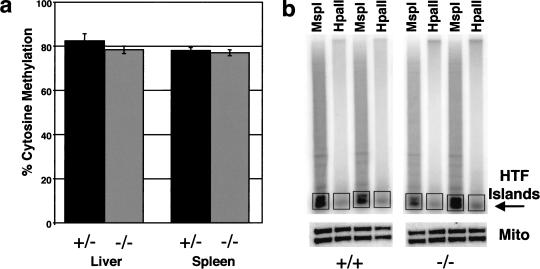
MBD2 has previously been reported to be a DNA demethylase (Bhattacharya et al. 1999), though this has not been verified by other researchers (Ng et al. 1999; Wade et al. 1999; Boeke et al. 2000). To further address this issue, we assayed genomic methylation levels in Mbd2(−/−) animals by two different methods. First we compared global levels of CpG methylation between Mbd2(+/−) and Mbd2(−/−) animals using nearest neighbor analysis. As is shown in Figure 3a, there was no significant difference in the fraction of CpG dinucleotides methylated in spleen or liver DNA derived from mice heterozygous or homozygous for the Mbd2(−/−) allele. We next looked for the presence of CpG islands, as revealed by HpaII tiny fragments (HTF Islands; Bird 1987), in DNA derived from Mbd2(−/−) mice. CpG islands are, for the most part, free of DNA methylation in normal animals. It has been suggested that these CpG-rich sequences are methylated early in development, but that methylation is subsequently removed by a demethylase (Cedar and Verdine 1999). If so, a demethylase-deficient animal should be unable to remove this methylation and should have fewer methylation-free CpG islands than normal. After complete digestion of tail DNA from four wild-type and four Mbd2(−/−) knockout animals with the restriction enzyme MspI or its methylation-sensitive isoschizomer HpaII, fragments were end-labeled with 32P. Radioactivity in the HTF fraction (see boxes in Fig. 3b) was normalized to the equivalent region of the MspI lanes. The mean ratio in the Mbd2(−/−) animals was not significantly different from than that of wild type (p = 0.14). Our findings show that the absence of the Mbd2 gene does not detectably affect global levels of genomic DNA methylation or impair the formation CpG islands.
Figure 3.
Normal levels of DNA methylation in Mbd2(−/−) animals. (a) The level of CpG methylation at MboI cut sites was determined in liver and spleen DNA derived from heterozygous (+/−) and Mbd2-deficient (−/−) animals and expressed as a percentage of total CpG (±s.e.). (b) Tail DNA from Mbd2-mutant (−/−) or wild-type (+/+) mice was digested with either MspI or HpaII, end-labeled with 32P, and fractionated on an agarose gel. The HTF island fraction is indicated, and the region of each lane used for quantitation is boxed. A portion of each digest was Southern blotted and probed with a mitochondrial DNA sequence as a digestion control.
Role of MBD2 in maternal behavior
During the maintenance of the Mbd2(−/−) mouse lines we noticed that the average litter size reared by two Mbd2(−/−) parents (4.47 ± 0.43) was approximately half that found for two Mbd2(+/−) parents (8.07 ± 0.62, p = 0.000176; Fig. 4a). Further, we found that the genotype of the father does not contribute to this effect, as the average litter size born to Mbd2-deficient mothers when bred to wild-type or heterozygote fathers is not significantly different from those born from matings to Mbd2(−/−) fathers (4.14 ± 0.96, p = 0.798). In contrast, Mbd2(−/−) fathers were capable of siring litters of normal size when mated to either wild-type or heterozygous mothers (7.33 ± 0.91, p = 0.498). Comparison of litters born to Mbd2(−/−) mothers to the sizes of those born to wild-type or heterozygous mothers, irrespective of the genotype of the father, produced a highly significant difference (4.44 ± 0.40 vs. 7.74 ± 0.52, p = 0.00000945). These data indicate MBD2 contributes to the ability of mothers to either carry or deliver viable offspring.
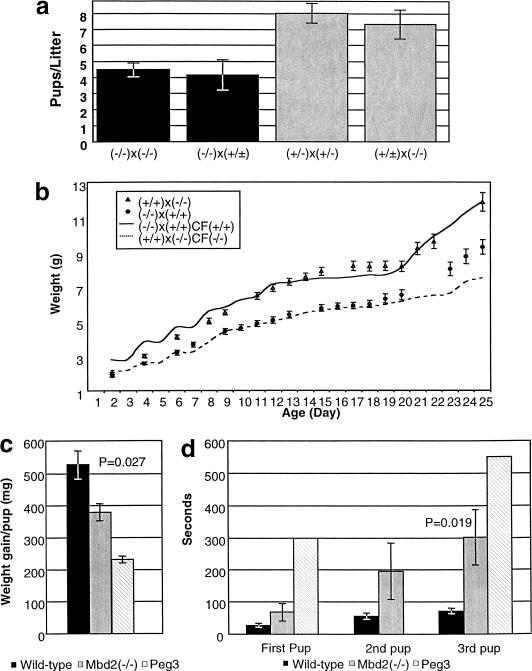
Figure 4.
Impaired nurturing behavior in Mbd2(−/−) mothers. (a) The histogram plots average litter sizes for the crosses indicated below. Genotypes refer to the Mbd2 locus. Shaded in black are litters born to Mbd2(−/−) mothers and gray for litters born to heterozygous or wild-type mothers. (b) The average weight from a litter of eight pups ±s.e. is plotted on the Y-axis, and age in days is plotted on the X-axis. Pups born to a wild-type mother crossed with a Mbd2(−/−) father are plotted as open triangles, and pups born to a Mbd2(−/−) mother and wild-type father are plotted as black circles. The average weight curve from a litter of six pups born to a Mbd2(−/−) mother and wild-type father, but fostered to a wild-type mother [(−/−) × (+/+)CF(+/+)] is plotted as a solid line, while the average weight of pups born to a wild-type mother and Mbd2(−/−) father fostered to a Mbd2(−/−) mother [(+/ +) × (-/-)CF(−/−)] are indicated as a dashed line. (c) Average 24-h weight gain per pup after a 2-h separation from the mother. Mean ±s.e. is plotted in black for pups born to wild-type mothers, gray for pups born to Mbd2(−/−) mothers, and hatched for Peg3-deficient mothers (Li et al. 1999). The P value was determined using a two-tailed t test. (d) Time, in seconds, required for mothers to retrieve pups to their nests. Plotted is the mean ±s.e. for wild-type mothers in black and Mbd2(−/−) mothers in gray. Also included as hatched boxes are the results reported for Peg3-deficient mothers (Li et al. 1999). The difference in time required to retrieve all three pups is statistically significant (two-tailed t-test).
In addition to the reduced litter sizes, we also noticed that the progeny born to two Mbd2(−/−) parents tended to be smaller than those born to wild-type or heterozygote parents. In order to characterize this parental effect, we compared the weights of the pups born to an Mbd2(−/−) female mated to a wild-type male to those born to the reciprocal cross of a wild-type female and Mbd2(−/−) male. As shown in Figure 4b, the pups born to the Mbd2(−/−) mother and normal father are, in general, smaller than those born to the reciprocal cross, despite all pups being of the same genotype [i.e., Mbd2(+/−)]. The most pronounced difference in weights occurs between days 13–19 after birth, though the smaller pups do regain normal weights after weaning (data not shown). In order to test whether the maternal genotype is responsible, a cross-fostering experiment was carried out in which the pups born to a Mbd2(−/−) mother were exchanged with those born to a wild-type mother and their weights monitored. As above, all pups in this experiment were heterozygous for the Mbd2-targeted allele. Fostering the pups born to Mbd2(−/−) mothers with wild-type mothers completely rescued the weight phenotype (Fig. 4b, solid line). The reciprocal experiment in which pups born to wild-type mothers were fostered with Mbd2(−/−) mothers recapitulated the low-weight phenotype (Fig. 4b, dotted line). In a second test of weight gain (Li et al. 1999), 3-day-old pups were separated from their mothers for 2 h and then returned to their nests. The total weight gained per pup in the subsequent 24 h was measured and the results are plotted in Fig. 4c. Pups born to control mothers gained an average of 526 mg after 24 h, while the weight gain of pups born to Mbd2(−/−) mothers was significantly less (379 mg, p = 0.027; Fig. 4c).
Pups resulting from both crosses apparently suckled normally and no difference in maternal milk composition could be detected by Coomassie blue staining of crude milk fractions taken from normal and Mbd2(−/−) mothers (data not shown). In general, pups born to Mbd2(−/−) mothers were not lost before weaning, despite being small. To test whether this phenotype might be a result of abnormal nurturing behavior, we assayed the ability of postpartum mothers to retrieve newborn pups to their nests (Li et al. 1999). Mbd2(−/−) mothers were marginally slower than normal to recognize and retrieve the first pup (p = 0.127), but they were significantly slower at retrieving all three pups to their nests than were normal mothers (p = 0.019, Fig. 4d). These data together indicate that the low weight gain phenotype is because of a failure of Mbd2(−/−) mothers to adequately feed their pups, possibly because of abnormal maternal behavior.
MBD2 is required for normal MeCP1 formation
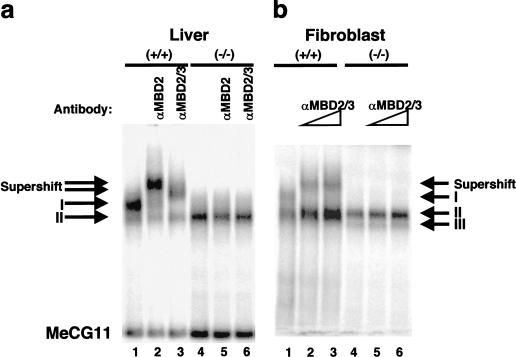
MBD2 in HeLa cells has been found to be associated with histone deacetylases (HDACs) in the MeCP1 repressor complex (Ng et al. 1999). To determine whether the abnormal maternal behavior observed in Mbd2(−/−) mice could be because of a lack of MeCP1 activity, we next asked what effect our Mbd2 deletion has upon MeCP1 formation. We found that MeCP1 in mouse liver nuclear extract consists of two bands, as originally described (Meehan et al. 1989) (Fig. 5a, lane 1, complexes I and II), whereas MeCP1 in fibroblast cell lines derived from mouse tail cells also contained a faster migrating third band (Fig. 5b, complexes I, II, and III). MeCP1 in nuclei derived from Mbd2(−/−) animals lacked complex I in both liver and fibroblast nuclear extracts, indicating that this top band is dependent upon the presence of MBD2 (Fig. 5a,b, lanes 4–6). Preincubation of the bandshift reaction with anti-MBD2 antibodies resulted in a specific supershift of complex I in both liver and fibroblast nuclear extracts (lanes 2–3,5–6), but did not affect any of the other complexes in either wild-type or Mbd2(−/−) extracts. These data indicate that the slowest migrating MeCP1 band (complex I) in murine liver and fibroblast nuclear extracts contains MBD2 and is dependent upon its presence. Thus this band is probably homologous to the single MeCP1 band of HeLa cells, which also contains MBD2. In contrast, the formation of the other one or two faster migrating complexes (complexes II or III) in murine liver or fibroblasts, respectively, is not dependent upon the presence of MBD2, nor can MBD2 be detected within them by supershifting with MBD2 antibodies.
Figure 5.
MBD2 requirement for MeCP1 formation. (a) MeCP1 from liver nuclear extracts is visualized as two different complexes (I and II) in a band shift assay using the methylated form of the probe CG11 (MeCG11). Lanes 1–3, wild-type nuclear extract; lanes 4–6, Mbd2(−/−) nuclear extract. Complex I is the major band produced in wild-type nuclei (lanes 1–3) but is absent in Mbd2(−/−) nuclei (lanes 4–6). An MBD2-specific antibody (S923) was added to lanes 2 and 5, while an antibody that recognizes both MBD2 and MBD3 (R593) was added to lanes 3 and 6. (b) Three complexes (I, II, and III) are visible in nuclear extracts derived from fibroblasts, although complex III is obscured in lanes 1–3 by the intensity of complex II. As in part a, complex I is absent in the Mbd2(−/−) extracts. The R593 antibody produces a specific shift of complex I in wild-type nuclear extract (lanes 2–3), but no shift is observed in nuclear extracts derived from Mbd2(−/−) fibroblasts (lanes 5,6).
Normal imprinting in Mbd2(−/−) animals
MeCP1 is known to be capable of repressing transcription from methylated promoters, so we next asked whether Mbd2(−/−) animals, which lack an intact MeCP1 complex, are able to repress methylated genes. DNA methylation has been shown to be necessary for repression of the paternal allele of the H19 gene, and for expression of the paternally inherited Igf2 gene (Li et al. 1993). Expression of the H19 and Igf2 genes was assayed in adult Mbd2(−/−) animals in which the maternal and paternal alleles of these genes are distinguishable by sequence polymorphism (Forne et al. 1997). No paternal H19 expression was detected in Mbd2(−/−) brain or heart RNA (Table 2). Normal paternal Igf2 expression was detected in RNA derived from heart and spleen of Mbd2(−/−) animals, while biallelic Igf2 expression was detected in brain RNA from both Mbd2(+/−) and (−/−) animals (Table 2). Expression of other imprinted genes was tested using RNAse protection and northern blot analysis of tissues from wild-type and Mbd2(−/−) mice. No differences in expression levels were detectable for the imprinted genes Igf2R, Peg1, Peg3, Zim1, Snrpn, and Znf127 in RNA derived from wild-type and Mbd2(−/−) animals (data not shown). Similarly, no expression of the Xist gene was detectable by RT–PCR of RNA derived from adult male Mbd2(−/−) animals (data not shown).
Table 2.
RNA derived from brain, heart, or spleen from Mbd2(−/−) or Mbd2(+/−) animals was subjected to RT–PCR across polymorphic sites for both the Igf2 and H19 transcripts
| Igf2
|
Paternal allele
|
Maternal allele
|
|---|---|---|
| (−/−) Brain | 5 | 12 |
| (+/−) Brain | 5 | 13 |
| (−/−) Heart | 14 | 1 |
| (+/−) Heart | 16 | 1 |
| (−/−) Spleen | 7 | 0 |
| H19 | ||
| (−/−) Heart + Brain | 0 | 69 |
PCR products were cloned and sequenced, and sequence polymorphisms were used to determine the allelic origin of the transcript from which each clone was derived. Numbers refer to numbers of independent clones sequenced. In all cases the Mus spretus allele was inherited paternally and the Mus domesticus allele was inherited maternally.
DNA methylation has been demonstrated to be required to prevent the expression of endogenous retroviral genes in somatic cell lines (Walsh and Bestor 1999; Walsh et al. 1998). To determine whether this silencing depends upon MBD2, we compared the expression of IAP retroviral elements in wild-type and Mbd2-deficient animals. No difference in expression levels was detectable between normal and Mbd2(−/−) tissues or cell lines (data not shown). Another retroviral genome that has been shown to become expressed under demethylating conditions is the moloney murine leukemia viral genome present in Mov7 and Mov10 mice (Jahner et al. 1982). These mutant viral genomes are normally transcriptionally silent and methylated, but can become transcriptionally active after treatment with the demethylating agent 5-azacytidine (Jaenisch et al. 1985). Breeding the Mov7 and Mov10 genes onto the Mbd2(−/−) background did not result in increased expression of either gene in RNA isolated from adult animals (data not shown). Thus we found no evidence that an intact Mbd2 gene is required for the DNA methylation-dependent silencing of imprinted genes, the Xist gene or of the IAP, Mov7, or Mov10 viral genomes.
MBD2 is required for transcriptional repression of a methylated promoter in cell lines
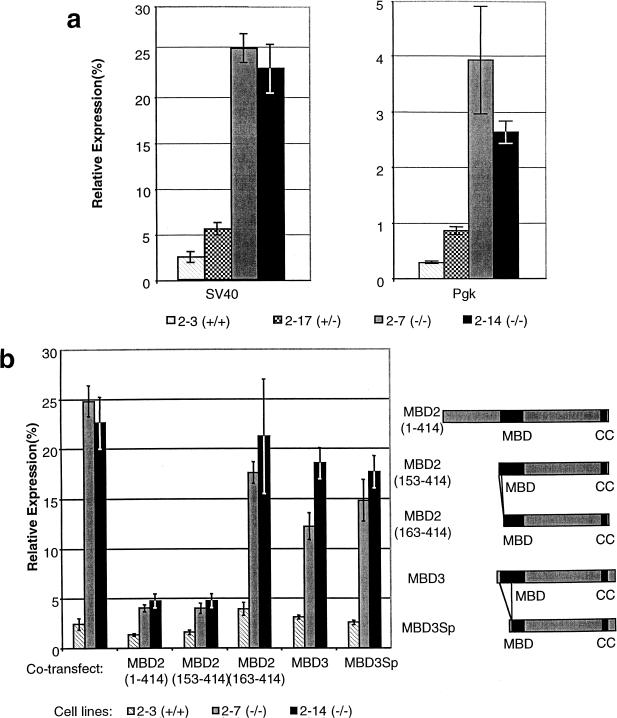
The methyl-CpG binding activity of MeCP1 present in HeLa cells and NIH3T3 cells has been implicated in repression of methylated templates (Boyes and Bird 1991). Similarly, exogenous MBD2 was shown to be capable of transcriptional repression in a transient assay (Boeke et al. 2000; Ng et al. 1999). We were, however, unable to identify an endogenous gene misexpressed in Mbd2(−/−) animals. To ask whether MBD2 is required for appropriate repression of methylated promoters in general, we assayed the ability of wild-type and MBD2-deficient cells to repress transcription of a methylated reporter construct. Cell lines derived from wild-type, heterozygous, or homozygous mutant mice were transfected with either methylated or unmethylated plasmid containing the luciferase gene under the control of the SV40 early promoter. Wild-type or heterozygous cell lines transfected with the methylated construct expressed less than 4% of the luciferase activity obtained when transfected with the same construct unmethylated (Fig. 6). In contrast, the methylated construct is expressed at levels approaching 25% of those seen for the unmethylated construct in Mbd2(−/−) cells. Significantly reduced repression also was seen for the murine phosphoglycerate kinase promoter, though to a lesser extent (Fig. 6a). We next asked whether this failure of repression could be rescued by reintroducing MBD2 into the Mbd2-deficient cells. To test this we cotransfected expression constructs encoding variants of the MBD2 protein under the control of a CMV promoter. Constructs expressing three different versions were utilized: full-length MBD2; a truncated version that lacks the N-terminal 152 amino acids of MBD2 [MBD2(153–414), previously called MBD2b; Hendrich and Bird 1998]; and MBD2(163–414), which lacks the N-terminal 9 amino acids of the methyl-CpG binding domain. As shown in Figure 6b, both full-length MBD2 and MBD2(153–414) fully restored repression of the methylated construct in Mbd2(−/−) cell lines. In contrast, MBD2(163–414), which fails to colocalize with methylated DNA in vivo (data not shown) did not restore complete repression to the Mbd2-mutant cells. Similarly, neither full-length MBD3, nor the short form encoded by an alternately spliced transcript, MBD3Sp, was capable of rescuing the silencing defect (Fig. 6b). Therefore, addition of extra MBD3 is unable to compensate for the absence of MBD2.
Figure 6.
MBD2 is required for full repression of methylated templates. The amount of luciferase activity recovered from a methylated plasmid is expressed as a percentage of that recovered from an unmethylated plasmid (Mean ±s.e.). The names and genotypes of the cell lines used are indicated below the histograms. (a) MBD2-deficient cell lines fail to fully repress the SV40 early promoter or the murine phosphoglycerate kinase (Pgk) promoter when fully methylated. (b) Repression of the methylated SV40 early promoter is restored in the Mbd2(−/−) cell lines by cotransfection of plasmids producing either full-length MBD2 or MBD2(153–414). Repression is not restored by expression of MBD2(163–414) protein, full-length MBD3 or short-form MBD3 proteins, none of which selectively bind methylated DNA in vivo. Proteins encoded by the expression constructs are diagrammed to the right of the histogram.
Discussion
MBD2 and MBD3: related proteins, different functions
Our genetic studies reveal very different phenotypes in mice lacking MBD2 versus MBD3. MBD2 is required for transcriptional silencing of methylated promoters and for proper nurturing behavior in mothers, while MBD3 is essential for successful embryogenesis. Both proteins were identified as potential methyl-CpG binding proteins, although in mammals only MBD2 specifically binds methylated DNA both in vitro and in vivo (Hendrich and Bird 1998). In other respects, however, the two proteins are closely related, sharing over 70% amino acid identity and being encoded by genes of similar genomic structure (Hendrich et al. 1999; Hendrich and Bird 1998). Both genes are activated in embryogenesis following implantation (B. Hendrich and V.A. Wilson, unpubl.), and are widely expressed in adult mice (Hendrich and Bird 1998). In addition, both proteins are associated with corepressor complexes containing histone deacetylases (Ng et al. 1999; Wade et al. 1999; Zhang et al. 1999). In vitro experiments indicate that MBD2 can interact with the NuRD complex, raising the possibility that MBD2 and MBD3 interact biochemically (Zhang et al. 1999). Recent data strengthens this possibility by detecting an in vitro interaction between MBD2 and MBD3 via their coiled-coil domains (Tatematsu et al. 2000).
MBD3 co-purifies with the human and frog NuRD histone deacetylation and nucleosome remodeling complexes (Wade et al. 1999; Zhang et al. 1999). The NuRD complex interacts with a variety of different DNA binding proteins to bring about transcriptional repression in a variety of different contexts (Ahringer 2000). Not surprisingly, then, deletion of what appears to be a core component of this roving repression complex in mice results in embryonic lethality after implantation. This is consistent with what is known about the role of a putative NuRD component in Drosophila melanogaster, an organism that also contains an MBD2/3 homolog (Tweedie et al. 1999). Flies mutant for the Drosophila Mi-2 gene are unable to develop past the second instar stage (Kehle et al. 1998), indicating that the NuRD complex also is required for embryonic development in D. melanogaster. Several likely NuRD components also have been identified in Caenorhabditis elegans (Ahringer 2000), though an MBD3 homolog is one notable exception. Extensive RNA-interference and mutant analysis of the identified genes has revealed that the C. elegans NuRD complex is involved in a wide variety of developmental processes such as patterning, vulval development, and signaling (Solari and Ahringer 2000; von Zelewsky et al. 2000). The finding of early embryonic lethality in Mbd3 mutant mice indicates that transcriptional silencing by the NuRD complex is an important part of embryonic development in many animal species extending from nematodes to mammals.
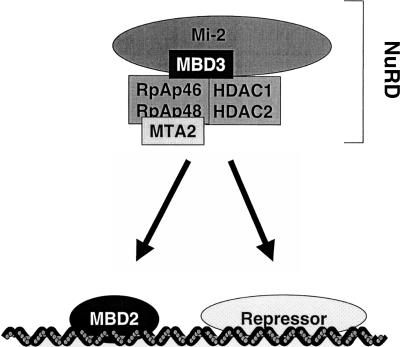
MBD2 is a component of the MeCP1 protein complex of HeLa cells, which, like NuRD, contains histone deacetylase activity (Ng et al. 1999). Indeed, MBD2 can recruit NuRD to methylated DNA in vitro (Zhang et al. 1999). Although the exact relationship between MBD2 and NuRD remains to be clarified, the data suggest that MBD2 resembles other DNA-binding repressor proteins such as MeCP2 (Jones et al. 1998; Nan et al. 1998) or Hunchback (Kehle et al. 1998), which direct histone deacetylase complexes to specific chromosomal locations (Fig. 7). In Xenopus, one form of an MBD3-like protein can specifically bind methylated DNA (Wade et al. 1999) and this also may target the NuRD complex. Thus both mammalian and amphibian NuRD complexes may be recruited to methylated DNA by MBD2/3-like proteins. The differing roles of MBD2 and MBD3 as, respectively, a DNA binding specificity domain for NuRD and a core component of the NuRD complex, may explain the very different phenotypes associated with mutations of the two genes. Deletion of the Mbd2 gene in mice would interfere with a proportion of NuRD function, leaving recruitment by other DNA binding proteins unaffected. In contrast, deletion of the Mbd3 gene probably would disrupt most or all NuRD function (Fig. 7).
Figure 7.
Proposed relationship between MBD2 and MBD3 via NuRD and MeCP1 complexes. The known components of the NuRD complex are shown, including the MBD3 protein. This complex may interact with a variety of sequence-specific DNA binding proteins to cause transcriptional repression, MBD2 being one of these. This combination of NuRD and MBD2 may be synonymous with the MeCP1 complex.
Our analysis of DNA methylation levels provides no support for the proposed role of MBD2b as a DNA demethylase (Fig. 3). Given the inability of several groups to duplicate the initial work (Ng et al. 1999; Wade et al. 1999; Boeke et al. 2000), we must conclude that the involvement of MBD2 in demethylation of 5-methylcytosine is placed in doubt.
Heterogeneity of “MeCP1”
We found that the major MeCP1 band in murine liver nuclear extracts contains MBD2 and is dependent upon the presence of MBD2 for its formation (Fig. 5a, complex I). Thus, this slow-migrating complex appears to be homologous to the single MeCP1 band found in HeLa nuclear extracts. Unlike HeLa cells, however, murine liver contains a second, faster migrating methyl-CpG binding complex that does not appear to contain MBD2, nor is it dependent upon MBD2 for its formation (Fig. 5a; complex II). Further, murine fibroblasts contain, additionally, a third complex that similarly does not require the presence of MBD2 for formation (Fig. 5b; complex III). Thus, the blanket term “MeCP1” may refer to a group of methyl-CpG binding complexes whose composition may vary between cell types. Given that the MeCP1 defined in HeLa cells appears most similar to complex I of murine nuclear extracts, we provisionally refer to this complex only as MeCP1. Complex III in fibroblasts is the Kaiso Generated Band (KGB) described by Prokhortchouk et al. (in prep.), while the composition of complex II, the middle 5-methylcytosine interacting activity (CIA), remains unknown.
Deletion of MBD2 relieved repression of two different reporter constructs that were methylated at every CpG (Fig. 6). Expression was not fully restored however, indicating that cells without MBD2 are still able to repress methylated promoters to some extent. Residual repression could be from the continued presence of one of the other methyl-CpG binding repressor proteins, as Mbd2(−/−) fibroblasts retain protein complexes that bind specifically to methylated DNA (Fig. 5b), one of which contains the known transcriptional repressor Kaiso (Prokhortchouk et al., in prep.). Alternatively, residual repression could be a result of direct interference of methylated sites with transcription factor binding (Hendrich and Bird 2000). The continued presence of methyl-CpG binding repressors MBD1, MeCP2, and Kaiso also could explain our inability to detect misexpression of endogenous methylated genes, including imprinted genes, repetitive elements, and Xist. This would imply that independent repressors cooperate to repress a methylated promoter; a possibility that can be tested by studying the consequences of combining repressor mutations in the same animal.
MBD2 and the brain
Mbd2(−/−) mothers have litters of reduced size and a behavioral defect that manifests as reduced pup weight (Fig. 5). Whether these are independent phenotypes or aspects of the same phenotype presently is unknown. It cannot be excluded that males also are behaviorally abnormal, but that this escaped detection as a result of their noninvolvement in complex nurturing behaviors. As MBD2 acts as a transcriptional repressor, the phenotype may be primarily from the inappropriate expression of some gene or genes in Mbd2(−/−) animals. The behavioral defects of Mbd2(−/−) mothers are reminiscent of those seen in mice harboring a mutation for the imprinted Peg3 gene (Li et al. 1999), though in all cases the effects are less pronounced (Fig. 5). Peg3 is one of two imprinted genes known to be important for proper maternal behavior (Lefebvre et al. 1998). It is tempting to speculate that deletion of MBD2 results in misexpression of some imprinted gene, possibly Peg3, which results in a mild maternal phenotype. We were, however, unable to detect misexpression of either Peg3, Peg1, or other imprinted genes in Mbd2 mutant animals, although it remains possible that misexpression of any of these genes at low level, or in a small region of the nervous system, would have escaped detection. Alternatively, MBD2 may be required for the expression of an as-yet unidentified imprinted gene which, like Peg1 and Peg3, plays a role in nurturing behavior. Presently, it is not known which genes are directly regulated by MBD2/MeCP1, though detailed comparison of gene expression profiles between wild-type and Mbd2(−/−) animals now can be used in an effort to identify such genes.
MBD2 and MeCP2 are the only two methyl-CpG–binding proteins for which deletions (or mutations in the case of MeCP2) have been reported, and both result in neurological phenotypes. Whereas MBD2 is required for proper nurturing behavior in mothers, mutations in MeCP2 result in Rett syndrome in humans (Amir et al. 1999). Although MeCP2 was initially thought to be essential for murine development (Tate et al. 1996), recent data (Chen et al. 2001; Guy et al. 2001) also indicate a delayed onset neurological phenotype in Mecp2-null mice. Both MBD2 and MeCP2 are expressed ubiquitously in both humans and mice, so the finding that disruption of either gene has a prominent and disproportionate effect on cerebral function is somewhat surprising. It is possible that functional redundancy of methyl-CpG–binding protein function in the brain is less than in other tissues. Alternatively, brain development may be exquisitely dependent upon DNA methylation-mediated transcriptional silencing.
Materials and methods
Gene targeting and ES cell culture
The Mbd3 targeting vector was designed to replace exons 2–7 with the promoterless βgeo cassette (Skarnes et al. 1995). The βgeo cassette was flanked by a 4-kb MboI/PmlI upstream fragment (bases 1–3943 of Genbank accession no. AF120995) and a 2.2-kb SmaI/EcoRI downstream fragment (bases 8137–10324, AF120995) in the pBluescript II KS-cloning vector (Stratagene). The 3′ EcoRI site is destroyed in the cloning, which allows for the use of EcoRI digestion to test for proper integration at this locus (see below). The resulting construct replaces a 4191 basepair (bp) PmlI/SmaI fragment containing exons 2–7 with the 7-kb βgeo cassette, leaving only the N-terminal 36 codons of the Mbd3 gene.
The Mbd2 targeting vector was designed to replace exon 2 with the βgeo cassette. The targeting vector was constructed by flanking the βgeo cassette with an upstream 4.1-kb HindIII/SphI fragment (bases 12969–17140, Genbank accession no. AF120983) and a 2.8-kb BglII downstream fragment (bases 17599–20337, AF120983) in the pBluescript II KS-cloning vector (Stratagene). The resulting construct replaces a 459 bp SphI/BglII fragment containing exon 2 with the 7-kb βgeo cassette.
Gene targeting was performed in the ES cell line E14 TG2a (Andrew Smith, Edinburgh University), which is derived from the mouse substrain 129/Ola (Thompson et al. 1989). ES cells were grown in gelatinized dishes in Glasgow MEM (Life Technologies) supplemented with 10% fetal bovine serum (Globepharm), 1× MEM nonessential amino acids, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol (Life Technologies) and LIF. LIF was provided by the addition of growth medium taken from a culture of Cos-7 cells transiently transfected with a human LIF expression construct (Austin Smith, Edinburgh University). ES cells were transfected with the linearized targeting vector by electroporation. Correctly targeted clones were identified by Southern blotting. Mbd3: For targeting at the 5′ end ES cell DNA was digested with BglII and the blots probed with a 1.3-kb MboI/NheI fragment (Fig. 1a). The wild-type gene gave a 5.4-kb band and correctly targeted clones were identified by the appearance of a 7.0-kb band from the targeted allele. For targeting at the 3′ end, DNA was digested with EcoRI and probed with a 0.3-kb SphI fragment. The wild-type allele gave an 8-kb band, the targeted allele produced a 2.0-kb band. Mbd2: For targeting at the 5′ end, ES cell DNA was digested with BglII and the blots probed with a 3.4-kb EcoRI/SphI fragment (Fig. 2a). The wild-type gene gave a 5.1-kb band and correctly targeted clones were identified by the appearance of a 6.5-kb band from the targeted allele. For targeting at the 3′ end, DNA was digested with EcoRI and probed with a 1.1-kb SphI–BglII fragment. The wild-type allele gave a 6.8-kb band, the targeted allele produced a 3.0-kb band.
Generation and breeding of mice
Correctly targeted ES cells were injected into C57BL/6 blastocysts as described (Hogan et al. 1994). Chimeric pups were identified by their agouti coat color and on maturity were mated with C57BL/6 mice. Heterozygous animals were identified by Southern blotting and probing of tail DNA. Mbd2 heterozygous animals were intercrossed to generate homozygotes, and the line subsequently was maintained in the homozygous state. The Mbd3 line was maintained by intercrossing heterozygotes and identifying subsequent heterozygous animals by Southern blotting. Thus the genomes of both the Mbd2 and Mbd3 targeted lines are, on average, 50% derived from the 129/Ola genome of the ES cells, and 50% derived from C57BL/6 animals.
Mbd3(+/−) mice used to produce embryos for genotyping, however, were the products of four generations of backcrossing to pure C57BL/6 animals. Embryos were incubated in 1 × PCR buffer, 0.45% NP40, 0.45% Tween-20, and 200 μg/mL Proteinase K at 55°C for 2 h, and 95°C for 15 min. 1–5 μL of these lysates then was used for PCR using primers Mbd3P26 (5′-TG TAGCCACCTAGCTCAAGG-3′) and Mbd3P27 (5′-CGCTG GCGACTCTTATTC-3′), which amplify the wild-type locus, and primers Mbd3P26 (as above) and EnP1 (5′-TCCG CAAACTCCTATTTCTG-3′), which amplify the targeted locus. Cycling conditions were as follows: 94°C for 10 sec, 56°C for 10 sec, and 72°C for 40 sec, for a total of 30–35 cycles.
Expression of the imprinted genes H19, Igf2, Igf2R, Peg1, Peg3, Zim1, Snrpn, and Znf127 were assayed by RNase protection using the RPA III kit (Ambion) according to manufacturer's instructions. Probes for the above genes were generated using primers derived from published sequence. Primer sequences are available upon request. RNA derived from adult mice containing the Mus spretus alleles of the H19 and Igf2 genes (derived from SD7 mice; Forne et al. 1997; Wolf Reik, The Babraham Institute) was reverse-transcribed and PCR amplified with primers H19P1 (5′-AAGAGCTAACACTTCTCTGC-3′) and H19P2 (5′-CAGGTAGTGTAGTGGTTCTG-3′) or Igf2P2 (5′-AGTG GAGCAGAGAGATCTTAGT-3′) and Igf2P3 (5′-AGTGGG GAAGTCGATGTTGG-3′). PCR products derived from three different tissues in independent reactions were cloned and sequenced. Expression of the Igf2 gene also was monitored through the use of a polymorphic BsaAI site flanked by the Igf2P2/Igf2P3 primers. Expression of IAP elements was monitored by PCR using primers IAPGagP1 (5′-GGTTCCCG TAAAGCAGACTG-3′) and IAPGagP2 (5′-GGCTTCCT TAAGTGATTAGC-3′); or by RNase protection using a probe generated from these primers. Expression of the Xist gene was assayed by RT–PCR using primers XistP4 (5′-CACATTGCTT GATCACGCTG-3′) and XistP5 (5′-TTGCTGCTTTGCAGT GCTGG-3′) or XistP6 (5′-TTGGCAGCAAGTGCCTTTAC-3′) and XistP7 (5′- CAGCAAGCCCACAATTCTGG-3′) under the conditions described above with 30, 35, or 40 cycles of amplification. Reactions were electrophoresed on 2% agarose gels, blotted onto Hybond N + membranes (Amersham), and probed with the XistP4/5 or XistP6/7 PCR products. Expression levels were quantitated using a Storm PhosphorImager (Molecular Dynamics). Mov7 and Mov10 mice (Jahner et al. 1982) (Rudolf Jaenisch, MIT) were bred with Mbd2(−/−) animals. The presence of Mov7 and Mov10 genes in mice was monitored by Southern blot. Genomic DNA was digested with BglII and probed with a 487 bp PCR product amplified by the MovP1 (5′-TCT GAGAATATGGGCCAGAC-3′) and MovP2 (5′-CTGTAAG TAGGTCGATGAGC-3′) primers. The Mov7 allele gives a band of 2.5 kb, while the Mov10 allele gives a band of 5 kb. Expression of the Mov7 and Mov10 genes was monitored by probing northern blots with the MovP1/P2 PCR product, and by RT–PCR using these same primers.
Southern, Northern, Western, and chromatographic analysis
Tail tips were incubated overnight at 55°C in tail mix (50 mM Tris-Cl at pH 8.0, 100 mM EDTA, 100 mM NaCl, 1% SDS, 100 μg/mL Proteinase K). DNA was precipitated by the addition of isopropanol, washed in 70% ethanol and dissolved in 100 μL dH2O. Aliquots (24 μL) were used in subsequent genomic digestions. Gels were blotted onto Hybond N + membranes (Amersham). Hybridizations were performed overnight at 65°C in PEG Hyb (0.25 M NaCl, 7% SDS, 1 mM EDTA, 10% PEG 6000, 50 μg/mL sonicated salmon sperm DNA, and 125 mM sodium phosphate buffer at pH 7.2). Membranes were washed twice in 2× SSC, 2% SDS for 15 min at 65°C, followed by two washes in 0.2× SSC, 0.1% SDS. Radioactive signals were detected using a Storm PhosphorImager (Molecular Dynamics). HTF islands were detected by end-labeling HpaII or MspI-digested DNA derived from tail DNA from two male and two female wild-type or Mbd2(−/−) animals with 32P as described (Cooper et al. 1983). The amount of radioactivity in the HTF fraction was quantitated using a PhosphorImager and normalized to the equivalent-sized region from the MspI digested DNA. The P value was determined using a two-tailed t-test assuming equal variance. Of the 2 μg of DNA digested for each sample, 100 ng was used for end-labeling and the remaining 1.9 μg was electrophoresed on a 2% agarose gel, Southern blotted, and probed with a mitochondrial DNA probe to verify complete restriction enzyme digestion. The probe was the product of PCR amplification using primers 5′-CGAATGATTATAACCTAGAC-3′ and 5′-TAAAGAACACTATTAGGGAG-3′ and contains a single HpaII site. Northern blots (Hendrich and Bird 1998) and Western blots (Ng et al. 1999) were performed as described. For nearest neighbor analysis, DNA samples were digested in triplicate with MboI, labeled with [α-32P]dGTP, and the labeled nucleotide 3′ monophosphates separated by two-dimensional, thin-layer chromatography (TLC) as described (Ramsahoye et al. 2000).
Nuclear extracts and electrophoretic mobility shift assays
Nuclei were isolated according to the technique of Antequera et al. (1989) and nuclear extracts were prepared as described (Meehan et al. 1989). Electrophoretic mobility shift assay procedure was as described (Meehan et al. 1989), except that each 30 μL reaction contained 2 μg of sonicated Escherichia coli DNA as nonspecific competitor.
Cell lines, transfections, and luciferase assays
Tail biopsies from mice of known genotype were dissociated on an untreated tissue culture dish and incubated in AmnioMax complete media (Life Technologies). After the first passage, cells were transformed with SV40 virus. Tail cell lines were incubated with growth media from the SV40-expressing Ψ2-SV40 cell line (Rudolf Jaenisch, MIT) supplemented with 8 μg/mL polybrene (Sigma) and filtered through a 0.45-μm filter. Cells were infected by incubation with SV40 media under normal growth conditions for 4 h, then media was replaced with normal AmnioMax media for 1 h, and followed by another 4-h incubation in SV40 media. Established cell lines were maintained in α-MEM supplemented with penicillin/streptomycin (Life Technologies) and 10% bovine calf serum (Sigma). The genotype of resultant cell lines was verified by Southern, Northern, and Western blotting. Cell lines 2–3 (+/+) and 2–7 (−/−) were derived from male F2 littermates and the 2–17 (+/−) and 2–14 (−/−) lines were derived from male F5 littermates.
Cells of ∼50% confluence were transfected using Lipofectamine according to manufacturer's instructions (Life Technologies). Each well of a 6-well plate was transfected with 2 μg of methylated or unmethylated test plasmid [containing either the SV40 early promoter (pGL2-Promoter, Promega) or murine phosphoglycerate kinase promoter (pGLPgk) driving the firefly luciferase protein] and 50 ng of a control plasmid encoding the Renilla reniformis luciferase protein (pRL-SV40; Promega). A CMV–MBD2 expression construct (50 ng) also was added to some wells. The pGLPgk plasmid was made by cloning the murine phosphoglycerate kinase promoter from the pHA59 plasmid (Austin Smith, Edinburgh University) into the pGL2-Basic vector (Promega). Luciferase levels were measured after ∼40 h using the Dual Luciferase Assay Kit according to manufacturer's instructions (Promega). Corrected values were obtained according to the following formula: (luciferase sample-luciferase control)/(Renilla sample-Renilla control) where control values are obtained from untransfected cells. Relative luciferase values are defined as the corrected value obtained using a methylated test plasmid divided by the corrected value obtained using the unmethylated test plasmid. Plasmids were methylated to completion using M.SssI (New England Biolabs), and methylation reactions were verified by digestion with MspI, HpaII, and HhaI (New England Biolabs).
Behavioral analyses
Pups were cross-fostered on the day of birth after normalization of litter sizes. Pup retrieval and weight gain experiments were carried out essentially as described (Li et al. 1999) 5–6 h after lights were switched off.
Acknowledgments
We thank Eric B. Kaverne for advice and discussions, Andrew Smith for the E14 TG2a cell line and advice, Rudolf Jaenisch for Mov mice and Ψ2-SV40 cells, Wolf Reik for SD7 mice, Austin Smith for the LIF expression construct and pHA58 plasmid, and Donald Macleod for the mitochondrial DNA probe. We also thank David Melton for advice, members of the Bird Lab for discussions and critical reading of the manuscript, and Helen Barr, Victoria Clark, Aileen Grieg, and Joan Davidson for technical support. This work was supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Brian.Hendrich@ed.ac.uk; FAX 44-131-650-5379.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.194101.
References
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van Den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Antequera F, Macleod D, Bird AP. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Bird AP. HTF islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Boeke J, Ammerpohl O, Kegel S, Moehren U, Renkawitz R. The minimal repression domain of MBD2b overlaps with the methyl-CpG–binding domain and binds directly to Sin3A. J Biol Chem. 2000;275:34963–34967. doi: 10.1074/jbc.M005929200. [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- ————— Repression of genes by DNA methylation depends upon CpG density and promoter strength: Evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Verdine GL. The amazing demethylase. Nature. 1999;397:568–569. doi: 10.1038/17492. [DOI] [PubMed] [Google Scholar]
- Chen R, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Taggart MH, Bird AP. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S, Meehan R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;13:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- Forne T, Oswald J, Dean W, Saam JR, Bailleul B, Dandolo L, Tilghman SM, Walter J, Reik W. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc Natl Acad Sci. 1997;94:10243–10248. doi: 10.1073/pnas.94.19.10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse MeCP2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Abbott C, McQueen H, Chambers D, Cross S, Bird A. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome. 1999;10:906–912. doi: 10.1007/s003359901112. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Mammalian methyltransferases and methyl-CpG-binding domains: Proteins involved in DNA methylation. Curr Top Microbiol Immunol. 2000;249:55–74. doi: 10.1007/978-3-642-59696-4_4. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Promoter-region hypermethylation and gene silencing in human cancer. Curr Top Microbiol Immunol. 2000;249:35–54. doi: 10.1007/978-3-642-59696-4_3. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Schnieke A, Harbers K. Treatment of mice with 5-azacytidine efficiently activates silent retroviral genomes in different tissues. Proc Natl Acad Sci. 1985;82:1451–1455. doi: 10.1073/pnas.82.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJC, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M, Müller J. dMi-2, a hunchback-interacting protein that functions in Polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Nan X, Meehan RR, Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy F, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenmann RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Moss JE, Hurtley SM, Beddington RS. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc Natl Acad Sci. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solari F, Ahringer J. NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol. 2000;10:223–226. doi: 10.1016/s0960-9822(00)00343-2. [DOI] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- Tatematsu KI, Yamazaki T, Ishikawa F. MBD2–MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells. 2000;5:677–688. doi: 10.1046/j.1365-2443.2000.00359.x. [DOI] [PubMed] [Google Scholar]
- Thompson S, Clarke AR, Pow AM, Hooper ML, Melton DW. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989;56:313–321. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ng HH, Barlow AL, Turner BM, Hendrich B, Bird A. Vestiges of a DNA methylation system in Drosophila melanogaster? Nat Genet. 1999;23:389–390. doi: 10.1038/70490. [DOI] [PubMed] [Google Scholar]
- von Zelewsky T, Palladino F, Brunschwig K, Tobler H, Hajnal A, Müller F. The C. elegans Mi-2 chromatin-remodelling proteins function in vulval cell fate determination. Development. 2000;127:5277–5284. doi: 10.1242/dev.127.24.5277. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes & Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig H-P, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activites. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]