Abstract
Heliothis zea nudivirus 1 (HzNV-1 or Hz-1 virus), previously regarded as a nonoccluded baculovirus, recently has been placed in the Nudivirus genus. This virus generates HzNV-1 HindIII-I 1 (hhi1) and many other transcripts during productive viral infection; during latent viral infection, however, persistency-associated gene 1 (pag1) is the only gene expressed. In this report, we used transient expression assays to show that hhi1 can trigger strong apoptosis in transfected cells, which can be blocked, at least partially, by the inhibitor of apoptosis genes Autographa californica iap2 (Ac-iap2) and H. zea iap2 (Hz-iap2). In addition to these two genes, unexpectedly, pag1, which encodes a noncoding RNA with no detectable protein product, was found to efficiently suppress hhi1-induced apoptosis. The assay of pro-Sf-caspase-1 processing by hhi1 transfection did not detect the small P12 subunit at any of the time intervals tested, suggesting that hhi1 of HzNV-1 induces apoptosis through alternative caspase pathways.
INTRODUCTION
Heliothis zea nudivirus 1 (HzNV-1 or Hz-1 virus) is an enveloped, rod-shaped, nonoccluded virus with a circular double-stranded DNA genome composed of approximately 154 open reading frames (4–8, 17, 21). This virus previously was regarded as a member of the baculovirus family, but due to its different genetic content and life cycle, it has been reclassified into a new genus of nonoccluded viruses called the Nudivirus. (47, 48). The HzNV-1 virus has a relatively broad host range and has been reported to productively and latently infect many insect cell lines (4, 14, 24, 31, 34, 35, 40). Previously, we found that more than 100 transcripts are produced during productive HzNV-1 virus infection (6); during this phase of infection, a gene located on the HzNV-1 HindIII-I fragment (hhi1) was found to generate an abundant 6.2-kb transcript within 0.5 h postinfection (hpi) (50, 51). In contrast to the expression pattern of many early genes of baculovirus, the expression of hhi1 requires the assistance of HzNV-1 viral infection (51). In another experiment, we showed that hhi1 expression also can be transactivated by Autographa californica multiple nucleopolyhedrovirus (AcMNPV). Such transactivation is due to the function of ie1 and p35 genes of this heterologous virus (50). We have reported before that hhi1 can serve as a transcriptional activator and is involved in viral reactivation from latently infected cells (50, 51). Also, during latent viral infection, only one transcript derived from persistency-associated gene 1 (pag1) was detectable. pag1 gives rise to a unique noncoding RNA, named persistency-associated transcript 1 (PAT1), which was found to be responsible for, or at least involved in, the establishment of latent viral infection (5, 6).
HzNV-1 virus infection of host Sf-21 cells results in the necrosis of most of the infected cells and produces high yields of virus progeny (28, 29), whereas the superinfection of HzNV-1 latently infected Sf-21 cells induces quick apoptosis and yields relatively lower levels of virus progeny (28, 29). Previous experiments showed that viral antigen could be detected in necrotic cells but not in apoptotic cells (29). Thus, apoptosis seems to serve as a part of the host's defense mechanism against successful virus infection in insect cells latently infected with HzNV-1 virus. However, the viral gene(s) involved and the mechanism for the induction of apoptosis in cells infected with HzNV-1 virus are not yet clear.
Caspases are a group of cysteine proteases that are the central component of apoptosis machinery. These proteases also were identified in insects. Sf-caspase-1 is the main effector caspase of Sf cells (32). To overcome the antiviral defense response of the host (i.e., apoptosis) and thus to ensure the viral infection process, baculoviruses express two types of genes known to suppress cell death: p35 and inhibitors of apoptosis (iap family) (1, 9, 49). P35 is a stoichiometric inhibitor of caspases and can function in phylogenetically disparate organisms to block apoptosis induced by a variety of signals (16, 28, 43). The IAP family is another group of apoptosis inhibitors. Some of them have been reported to be possible substitutes for P35 to block apoptosis during the infection of baculovirus AcMNPV in insect cells (44). Most IAP proteins contain a specific motif called the baculovirus IAP repeat (BIR) which is responsible for mediating apoptosis suppression. Currently, the baculovirus iap genes are thought to act as a sink for IAP antagonists and thereby inhibit apoptosis (3, 9).
In this report, we further demonstrated that the early gene hhi1 of HzNV-1 possesses the ability to directly or indirectly promote apoptosis in host cells. We also found that although p35 can prevent ie1-induced apoptosis in Sf-21 cells, it was unable to prevent hhi1-induced apoptosis. In contrast, AcMNPV iap2, which has not previously been found to function as an inhibitor of apoptosis in lepidopteran cells, was able to suppress hhi1-induced apoptosis in Sf-21 cells. In addition, we discovered that the noncoding RNA PAT1, which was found to function in the establishment of latent viral infection, could suppress hhi1-induced apoptosis. The mechanism underlying this apoptosis suppression was analyzed, and it was found that apoptosis induced by hhi1 can be blocked by two totally different mechanisms, including the functions of PAT1 and IAP.
MATERIALS AND METHODS
Cells and virus.
Spodoptera frugiperda IPLB-Sf-21 was incubated in TC-100 insect cell culture medium containing 10% fetal bovine serum (FBS) at 26°C (Gibco BRL) (38, 50, 51). Standard HzNV-l virus was derived by a serial dilution of the stock viral solution and isolated by plaque purification. The titers of the virus clones were estimated by both quantitative PCR (qPCR) (33) and 50% tissue culture infective doses (TCID50) (38).
Plasmid construction.
Plasmid pKSh was constructed by inserting the hsp 70 promoter (p-hsp) (11, 22) into plasmid pBluescript II KS(−) (Stratagene). Unless otherwise stated, His tags were added to the coding regions of egfp, hhi1, p35, Autographa californica iap1 (Ac-iap1), Ac-iap2, H. zea iap1 (Hz-iap1), Hz-iap2, Sf-caspase-1, and ie1 by PCR (Table 1), and the PCR fragments were inserted downstream of plasmid pKSh to generate the expression plasmids pKShE (egfp), pKShH1 (hhi1), pKShp35 (p35), pKShAiap1 (Ac-iap1), pKShAiap2 (Ac-iap2), pKShHiap1 (Hz-iap1), pKShHiap2 (Hz-iap2), pKShC1 (Sf-caspase-1), and pKShIE (ie1), respectively. Plasmids pKShH1ΔH, pKShHiap2ΔH, and pKShIEΔH, which express untagged HHI1, IAP2, and IE1, also were constructed. These plasmids function either to induce apoptosis (HHI1 and IE1) or to block hhi1-induced apoptosis (IAP2) in Sf-21 cells with levels undistinguishable from that induced by their counterplasmids without His tags (data not shown). The HzNV-1 viral early promoter regions of pag1 (+29 to ∼727) (51) were obtained by PCR using HzNV-1 viral DNA as a template. Promoter regions were ligated into pGL3-basic vector (Promega) to obtain plasmid pGLpL for activity assay with or without pKShp35 transfection. To assay the suppression of hhi1-induced apoptosis by pag1, we obtained the pag1 coding region by PCR (51), and the PCR product was inserted downstream of the pag1 promoter (p-pag1) (5) of plasmid pKSp to generate the expression plasmid pKSpP1. All of the insertions of newly constructed plasmids were confirmed by sequencing.
Table 1.
Primer sequences used in this study
| Primer namea | Primer sequence (5′-3′) |
|---|---|
| hhi1 F | ATTCCCGGGCTCTCCTCTACAATCATGTCTACCGTG |
| hhi1 R | ATTCCCGGGCTCAGATTCACAGTATGGTTCACG |
| p35 F | ATTGAATTCACCATTGCAAAATGTGTGTA |
| p35 R | ATTCTGCAGTTTTAACATTTATTTAATTG |
| Ac-iap1 F | ATTGAATTCTTTAACGAGCTAAAATGAACG |
| Ac-iap1 R | ATTCTTACACCACATTTATGCAGCACCAT |
| Ac-iap2 F | ATTGAATTCGGAATAAACTATAAAATGAA |
| Ac-iap2 R | ATTCTGCAGAATGTTTACTGAGGTAATGT |
| Hz-iap1 F | ATTGAATTCCAGCGAGATGAATGAAATTGA |
| Hz-iap1 R | ATTCTGCAGCACTAAATATTTAAACAACAA |
| Hz-iap2 F | ATTGAATTCTATCGTAAAATGTCGACCGC |
| Hz-iap2 R | ATTCTGCAGATAATTATATTTAAAGTAAAT |
| Sf-cas1 F | GCGGATCCATACTGGACGGAAAACAA |
| Sf-cas1 R | GCGAATTCTCTGTGGGACTGCTTCTT |
| ie1 F | GCCCATGGGTACGCAAATTAATTTTAAC |
| ie1 R | GCCTCGAGATTAAATTCGAATTTTTTATA |
F, forward; R, reverse.
DNA transfection into cells.
A total of 2 × 105 cells per well were seeded into a 24-well culture plate (Corning) and then transfected with 0.5 μg of appropriate plasmid DNAs using Cellfectin (Invitrogen) according to the manufacturer's protocol (Gibco BRL). For the apoptosis assay and hhi1 expression level assay, plasmids expressing hhi1 coding sequence were cotransfected with pKShE, pKSpP1, pKShH1, pKShC1, pKShAiap1, pKShAiap2, pKShHiap1, pKShHiap2, or pKShIE. The nucleotide sequences of the hhi1 and pag1 genes used in these experiments were submitted to GenBank, and the accession numbers assigned were AF264019 (hhi1) and NC004156 (pag1), respectively (51).
Annexin V-FITC staining and viability assay.
Cells were collected at different time points, washed with phosphate-buffered saline (PBS), and stained with fluorescein isothiocyanate (FITC)-conjugated annexin V (BD Biosciences) for 20 min at room temperature in the dark. The stained cells then were analyzed by flow cytometry (Beckman Coulter). Apoptotic cells were identified as those with annexin V-FITC staining only, and the results were expressed as the proportion of such cells among the total number of cells analyzed. Upon the transfection of Sf-21 cells with the hhi1 construct, cells were stained with calcein AM (Live/Dead Viability kit; Molecular Probes) to quantify the effect of hhi1 on cell viability. Data (means ± standard deviations) were collected from triplicate assays of three independent experiments.
Caspase activity assay of apoptotic cells.
hhi1-transfected cells were lysed at different time points posttransfection (hpt) and stained with fluorogenic substrates specific for different caspases immobilized in the wells of 96-well plates (ApoAlert caspase assay plates; Clontech). At the same time, untransfected cells were exposed to UV light (50 mJ/cm2 for 30 min) to induce apoptosis in these cells as a control.
TUNEL assay of apoptotic cells.
For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays, Sf-21 cells (4 × 105/well) were seeded in a 24-well culture plate (Corning, Acton, MA) and then left untransfected or transfected with hhi1 DNA using Cellfectin (Invitrogen, Carlsbad, CA). We then tested the apoptosis inhibitors DEVD-CHO and zVAD-fmk (BD Biosciences), using concentrations ranging from 2 to 12 μM, and found that 10 to 12 μM gave the best results for both inhibitors; therefore, 10 μM was selected for further assays. Cells were collected at different time points after treatments, fixed with 4% methanol-free formaldehyde, and stained with fluorescein-12-dUTP (Promega) for 60 min at 37°C. The stained cells then were analyzed by a fluorescence plate reader (Victor III; Perkin Elmer) using the enhanced green fluorescent protein filter set (485 nm/535 nm).
Western analyses.
Sf-21 (2 × 105) cells were transfected with 1 μg of plasmids (pKSh, pKShE, pKShH1, pKShC1, pKShAiap1, pKShAiap2, pKShHiap1, pKShHiap2, pKShH1ΔH, pKShHiap2ΔH, or pKShIEΔH) using Cellfectin (Invitrogen). At 4 hpt, the transfection reagent was replaced with TC-100 medium containing 10% FBS. Transfected cells were inoculated (multiplicity of infection [MOI], 1) with AcMNPV or HzNV-l virus and incubated at 26°C with gentle rocking. Sf-21 cells transfected with different plasmids and/or infected with different viruses (as indicated) were suspended in 0.2 M Tris-HCl buffer containing 4% SDS, 18% glycerol, 2% β-mercaptoethanol, and 0.004% bromophenol blue. Proteins were transferred to nitrocellulose filters (Schleicher & Schuell) by electroblotting for 1 h in 200 mM glycine, 2.5 mM Tris-HCl, and 20% methanol. Filters were blocked with PBS containing 5% nonfat dried milk, 0.05% Tween 20 and incubated with primary antibodies against His tag followed by alkaline phosphatase (AP)-conjugated anti-mouse antibody (Dako).
Quantitation of viral early gene expression.
RNA of transfected cells was extracted using an RNeasy Minikit (Qiagen). The RNA pellet was dissolved in 30 μl of diethyl pyrocarbonate (DEPC) water and used for cDNA synthesis. cDNA synthesis was performed using the SuperScript III first-strand synthesis system for reverse transcription-PCR (RT-PCR) by following the manufacturer's protocol (Invitrogen). HzNV-1 early genes were RT-PCR amplified using the following primers: pag1-F, 5′-ACGGGAATTCAGTGTCGAGGACTT-3′; pag1-R, 5′-CATGTCTAGAACCCTACCTACCT-3′; actin-F, 5′-CGTGATGGTGGGCATGGGTCAG-3′; and actin-R, 5′-CTAATGTCACGCACGTATTCC-3′.
Briefly, amplification was carried out by adding 1 μl of cDNA to the Taq polymerase master mix (MBI Fermentas, Vilnius, Lithuania). The resulting DNA products (Amplicon) were analyzed on an agarose gel (1.5%) after electrophoresis at 100 V for 30 min.
Luciferase activity assay.
Luciferase assays were conducted as described previously (50). Luciferase activity was measured with a luminometer (Lumat LB 9501; Berthold) by injecting 50 μl of 0.2 mM luciferin (Promega) into each well. The results were plotted as average luciferase activity against time of infection. In these experiments, luciferase activity, cellular viability, apoptosis, and caspase activity assays all were derived from triplicate assays of three independent experiments.
RESULTS
hhi1 induces apoptosis in Sf-21 cells.
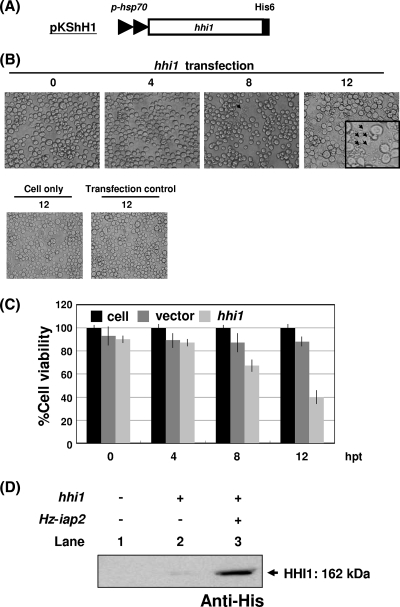
In previous experiments, we showed that hhi1 can activate viruses in latent cells and be a major transactivator stimulating the expression of many early transcripts. To further characterize the function of hhi1, we constructed plasmid pKShH1 (51), which expressed His-tagged HHI1 (Fig. 1A). Plasmid pKShH1 was transfected into Sf-21 cells, and the transfected cells were observed under light/fluorescence microscopy. Interestingly, significant apoptosis was observed in Sf-21 cells transfected with hhi1-expressing vector at 8 hpt. At 12 hpt, up to 60% of the cells were found to be apoptotic bodies (Fig. 1B). Cells were stained with calcein AM (Live/Dead viability kit; Molecular Probes) to quantify the effect of hhi1 on cell viability. In the cells transfected with hhi1, we found that cell viability was reduced to approximately 40 to 50% of that of the control cells transfected with the original vector. This result indicates that hhi1 could result in more than 50% cell death of the transfected cells by 12 hpt (Fig. 1C).
Fig. 1.
Induction of apoptosis in Sf-21 cells upon transfection with or without hhi1. (A) Schematic of the His-tagged hhi1-expression vector, pKShH1, which contains an hsp70 promoter (p-hsp) to drive hhi1 expression. (B) Monolayers of Sf-21 cells were transfected with pKShH1 by Cellfectin. Cells then were examined by light microscopy and photographed at 0, 4, 8, and 12 hpt. The arrows indicate a cluster of apoptotic bodies. Healthy and pKShE-transfected (transfection control) Sf-21 cells served as negative controls. (C) Cells were stained with calcein AM at the indicated time points, and viable cells, indicated by the exclusion of the dye, were counted under a microscope. (D) Sf-21 cells were transfected with a His-tagged hhi1 gene with or without untagged Hz-iap2 and then analyzed by Western blotting to study the expression of HHI1. Lane 1, cells only; lane 2, cells transfected with hhi1 only; lane 3, cells cotransfected with hhi1 and the antiapoptotic gene Hz-iap2.
To verify that HHI1 protein was properly expressed in the Sf-21 cells, plasmid pKShH1 was transfected into cells. Western analysis showed that the His-tagged HHI1 signal is weak in the transfected Sf-21 cells (Fig. 1D); this is likely due to a quick induction of cellular apoptosis by the transfection of hhi1 (Fig. 1B). Plasmid pKShH1 then was cotransfected into Sf-21 cells with plasmid pKShHiap2ΔH, which expresses the Hz-IAP2 protein without a His tag. In later experiments we found that this gene, with or without the His tag, can specifically block the apoptosis induced by hhi1. The cotransfection of Hz-iap2 allows us to better detect the HHI1 signal by Western blot analysis (Fig. 1D).
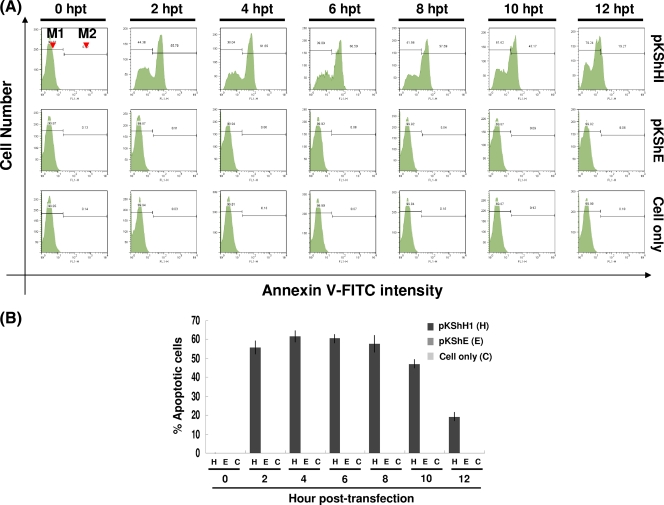
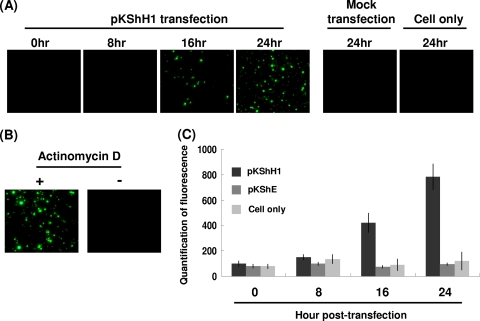
We further used dyes that stained early and late apoptotic markers to monitor the progression of apoptosis in host cells. First we used annexin V, which recognizes the early apoptotic marker phosphatidylserine (PS) on the plasma membrane, to observe the initiation of hhi1-induced apoptosis in the cells. Cells transfected with hhi1 were stained with annexin V (annexin V-FITC apoptosis detection kit I; Clontech). By flow cytometry, we found that apoptosis signals could be detected as early as 2 h after transfection, and the intensity of the signal declined after 8 hpt (Fig. 2A and B). We also studied DNA fragmentation in the late apoptotic cells. The results of TUNEL staining (DeadEnd fluorometric TUNEL system; Clontech), which stained fragmented DNA, showed that DNA starts to fragment at about 16 h after hhi1 transfection (Fig. 3A). As a control, cells also were treated with actinomycin D, and the treated cells were subjected to a TUNEL assay. DNA fragmentation was detected in cells treated with actinomycin D, whereas no DNA fragmentation was detected in cells not treated with actinomycin D (Fig. 3B). Based on these results, we concluded that hhi1 could induce apoptosis in transfected cells (Fig. 3C).
Fig. 2.
Assessment of apoptosis resulting from hhi1 transfection in Sf-21 cells by annexin V assay. (A) Representative histograms of flow cytometry analysis showing the fluorescence intensity of annexin V-labeled apoptotic cells transfected with pKShH1 or pKShE. The annexin V-positive cells can be identified in the M2 region. M1, nonapoptotic cells; M2, apoptotic cells. (B) Bar graph showing the percentage of apoptotic cells at different time points posttransfection resulting from annexin V labeling.
Fig. 3.
Assessment of apoptosis resulting from hhi1 transfection in Sf-21 cells by TUNEL assay. (A) TUNEL assay of Sf-21 cells transfected with pKShH1 at different time points. TUNEL-positive cells were observed in Sf-21 cells transfected with pKShH1 beginning from 16 hpt. (B) Sf-21 cells left untreated or treated with the apoptosis-inducing agent actinomycin D (20 μM) were TUNEL stained at 24 h posttreatment to serve as positive and negative controls, respectively. (C) Quantification of fluorescence of TUNEL-positive cells in Sf-21 cells transfected with pKShH1 or pKShE and Sf-21 cells only at different time points.
Activation of caspases in hhi1-transfected cells.
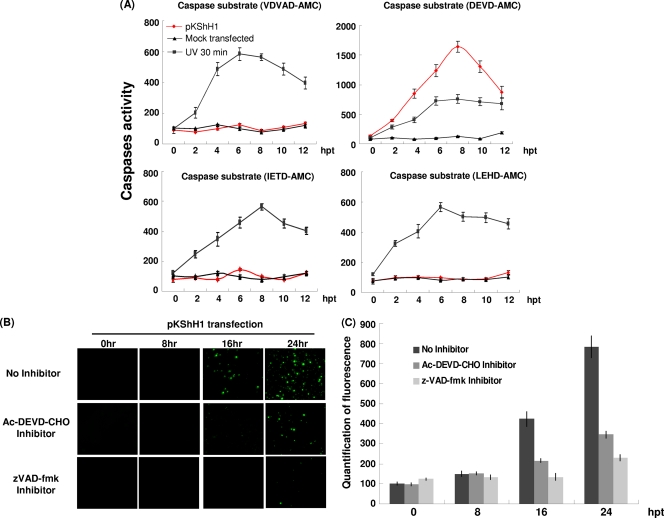
Previous reports identified a cysteine-aspartic protease (caspase) in Sf-21 cells, which has been named Sf-caspase-1 (1, 26, 44). When Sf-21 cells were infected with p35 knockout baculovirus, these cells entered apoptosis through an Sf-caspase-1 pathway. The caspase substrate DEVD-AMC (mammalian caspase-3 substrate) can be recognized and cleaved by Sf-caspase-1 in Sf-21 cells infected with p35 knockout virus (16, 20, 44). To investigate which caspase is involved in hhi1-induced apoptosis, we measured caspase activity in hhi1-transfected cells by using different substrates: VDVAD-AMC (mammalian caspase-2 substrate), DEVD-AMC (mammalian caspase-3 substrate), IETD-AMC (mammalian caspase-8 substrate), and LEHD-AMC (mammalian caspase-9 substrate) (41). These caspase substrates are cleaved predominantly by either initiator (IETD-AMC, LEHD-AMC, and VDVAD-AMC) or effector (DEVD-AMC) caspases (41). To determine whether there are differences between hhi1-induced and AcMNPV infection-induced caspase activation, we harvested cell lysates of hhi1-transfected cells at different time points posttransfection and incubated them in the wells of 96-well plates with immobilized fluorogenic substrates that can be preferentially cleaved by different caspases (ApoAlert caspase assay plates; Clontech). At the same time, untransfected cells were exposed to UV light (50 mJ/cm2 for 30 min) to induce apoptosis in these cells as a control. The result showed that hhi1 expression only resulted in the cleavage of DEVD-AMC, an effector caspase substrate (Fig. 4A). Effector caspase activity started to increase at 2 h after hhi1 transfection and reached a maximum level at 8 h after hhi1 transfection before declining (Fig. 4A). The decline probably was due to cell death as a result of apoptosis, which subsequently affected the effector caspase activity.
Fig. 4.
hhi1 induces activation of an insect caspase-3-like protein. (A) Assay of the caspase activity induced by hhi1 transfection. An ApoAlert caspase assay kit (Clontech) was used to measure caspase activities in Sf-21 cells transfected with pKShH1 or pKShE (mock) at 0, 2, 4, 6, 8, 10, and 12 hpt or after being subjected to UV irradiation. The substrates VDVAD-AMC, DEVD-AMC, IETD-AMC, and LEHD-AMC were added to the cell supernatant individually. After incubation for 1 h at 37°C, enzyme activity was measured through fluorogenic cleaved substrates. (B) TUNEL assay of the hhi1-transfected cells. Sf-21 cells were transfected with pKShH1 at different time points and then incubated with or without the caspase inhibitor Ac-DEVD-CHO (10 μM) or zVAD-fmk (10 μM) before being subjected to TUNEL assay. TUNEL-positive signals were detected at 16 and 24 hpt in the absence of caspase inhibitor, but signals were clearly detected only at 24 hpt in the presence of the caspase inhibitors. (C) Bar graph representations of the TUNEL assay showing the fluorescence intensity of TUNEL-positive cells at different time points posttransfection with or without caspase inhibitor Ac-DEVD-CHO or zVAD-fmk.
To confirm that hhi1 can activate caspase, we added the caspase inhibitors Ac-DEVD-CHO (a mammalian caspase-3 inhibitor) and zVAD-fmk (a broad caspase inhibitor) separately to cells transfected with hhi1 and then detected cell apoptosis by TUNEL assay. DNA fragmentation, the result of apoptosis, was clearly observed in hhi1-transfected cells at 16 hpt (Fig. 4B). However, with the addition of the caspase inhibitor Ac-DEVD-CHO or zVAD-fmk, DNA fragmentation was much reduced and appeared only at 24 hpt (Fig. 4B and C). The observation that both Ac-DEVD-CHO and zVAD-fmk could efficiently suppress hhi1-induced apoptosis suggests that hhi1 activates caspase(s) (Fig. 4B and C).
hhi1 does not activate the processing of pro-Sf-caspase-1.
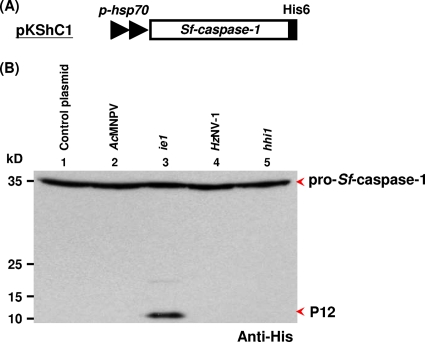
It is well known that the IE1 protein of AcMNPV can induce apoptosis in infected Sf-21 cells. To determine whether IE1 and HHI1 could cause the processing of pro-Sf-caspase-1 to its active subunits, we examined lysates of pKShC1-transfected cells (His-tagged pro-Sf-caspase-1) (Fig. 5A) with virus infection or additional plasmid transfection. The presence of the His tag allows the detection of full-length pro-Sf-caspase-1 and the smaller subunit (P12) of the active enzyme (16, 26). We found that pro-Sf-caspase-1 was not processed to its active subunit in mock-transfected and HzNV-1-infected cells (Fig. 5B, lanes 1 and 4). Seshagiri and Miller (44) have shown that both the expression level of pro-Sf-caspase-1 and the small subunit (12 kDa) of caspase 1 declined after 24 to 30 hpi upon AcMNPV infection. Similarly, the apoptosis induced by hhi1 expression may be blocked by the infection of HzNV-1 due to the expression of other viral genes, thereby rendering the possible cleavage of pro-Sf-caspase-1 to the small subunit difficult to detect.
Fig. 5.
hhi1 does not cause the processing of pro-Sf-caspase-1. (A) Schematic diagram of plasmid pKShC1, which expresses His-tagged Sf-caspase-1. (B) Assay monitoring the processing of Sf-caspase-1 by viral infections or plasmid transfections. Sf-21 cells first were transfected with pKShC1. At 4 hpt, cells were inoculated with AcMNPV (MOI, 1; lane 2) or HzNV-l virus (MOI, 1; lane 4). The same His-tagged Sf-caspase-1 also was cotransfected into Sf-21 cells with plasmids pKSh (control; lane 1), pKShIEΔH (expresses untagged IE1; lane 3), or pKShHIΔH (expresses untagged HHI1; lane 5) separately. Cells then were collected at 72 hpt, and both the uncleaved pro-Sf-caspase-1 and its processed P12 subunit were detected by Western blot analysis using anti-His-tagged antibody (44).
To study the function of hhi1 independently from the effect of other viral genes, hhi1 (Fig. 5B, lane 5) alone was transfected into cells together with ie1 alone as a positive control (Fig. 5B, lane 3). In this experiment, the expression level of pro-Sf-caspase-1 was detected at 72 hpt, a time point at which the small subunit of caspase-1, which is generated by baculovirus infection, should have been degraded (Fig. 5B, lanes 2) (44). However, without baculovirus interference, we found that the small P12 subunit processed from pro-Sf-caspase-1 still was detectable in cells transfected with IE1-expressing plasmid at 72 hpt (Fig. 5B, lane 3). Interestingly, at the same time point, the expression of hhi1 did not result in the processing of Sf-caspase-1 (Fig. 5B, lane 5), suggesting that pro-Sf-caspase-1 likely is not processed by hhi1 transfection.
Ac-iap2 and Hz-iap2 inhibit hhi1-induced apoptosis.
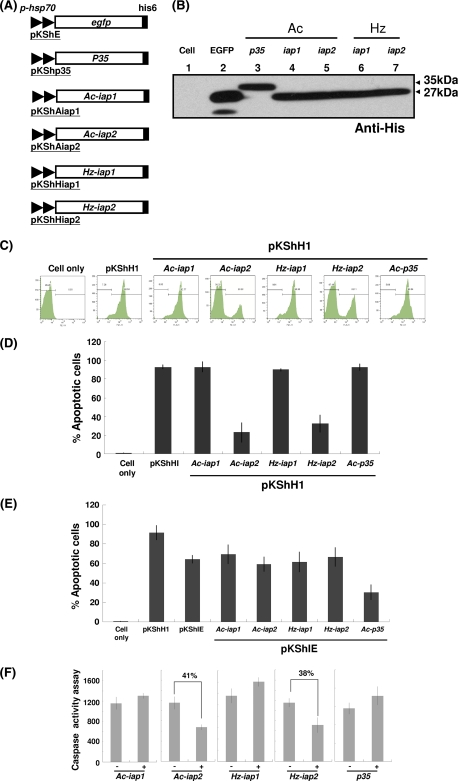
So far, two groups of baculoviral genes, p35 and iap, are known to suppress cell death (9); however, the genes involved in the antiapoptosis of cells infected with HzNV-1 have not been explored previously. Amino acid sequence analysis showed that two HzNV-1 genes, orf135 (Hz-iap1) and orf138 (Hz-iap2), possess BIR motifs, indicating that they are able to inhibit apoptosis (8). To study whether hhi1-induced cell death can be suppressed by the putative antiapoptosis genes mentioned above, we constructed several expression vectors that carry baculovirus p35, Ac-iap1, Ac-iap2, Hz-iap1, and Hz-iap2 gene sequences under the control of the Drosophila heat shock 70 promoter (Fig. 6A). The products of all of these genes contain a His tag for the detection of expressed proteins (Fig. 6B). These expression vectors were transfected into Sf-21 cells along with an hhi1 expression vector. We found that there were fewer cells showing signs of cell death in cells transfected with Ac-iap2 or Hz-iap2 under light microscopy (data not shown). Cells then were stained with annexin V, and flow cytometry analysis showed that about 80 to 90% of cells transfected with p35, Ac-iap1, or Hz-iap1 became apoptotic (Fig. 6C). In contrast, only 20 to 30% of cells transfected with Ac-iap2 or Hz-iap2 showed signs of apoptosis. These results clearly demonstrate that Ac-iap2 and Hz-iap2 are capable of suppressing hhi1-induced apoptosis (Fig. 6C and D). By using ie1-induced apoptosis as a control, we found that p35 could suppress ie1-induced apoptosis (Fig. 6E), which correlated well with previous reports (39, 43). Our results also showed that IAPs of AcMNPV and HzNV-1 had no inhibitory effects on ie1-induced apoptosis (Fig. 6E). These results indicate that hhi1 and ie1 induce apoptosis in host cells through different pathways, thereby enabling p35 to suppress ie1-induced apoptosis but not hhi1-induced apoptosis. In addition, different antiapoptotic genes may act differently on different apoptosis inducers and suppress apoptosis through different mechanisms.
Fig. 6.
Effects of antiapoptosis genes on hhi1-induced apoptosis in Sf-21 cells. (A) Schematics of plasmids containing antiapoptosis genes. (B) Western blot analysis showing proper transient expression of antiapoptotic proteins from the plasmids shown in panel A. (C) Annexin V assay of apoptosis induction in Sf-21 cells by flow cytometry. Sf-21 cells were transfected with either hhi1 (pKShH1) alone or with anti-apoptosis gene Ac-iap1, Ac-iap2, Hz-iap1, Hz-iap2, or p35 (0.5 μg of each plasmid). Transfected cells were stained by annexin V at 2 hpt and then analyzed by flow cytometry. Apoptotic cells that stained positive for annexin V were identified in the M2 region. (D) Bar graph representation of the percentages of apoptotic cells using the data shown in panel C. (E) Apoptosis induction assay in Sf-21 cells transfected with hhi1, ie1, or hhi1 and antiapoptosis genes. (F) Caspase-specific activity assay in hhi1-transfected Sf-21 cells, which were cotransfected with or without anti-apoptosis genes, using DEVD-AMC as a substrate. Percentages of apoptosis suppression by Ac-iap2 and Hz-iap2 are indicated.
We next performed caspase activity assays and showed that p35, Ac-iap1, and Hz-iap1 could not suppress caspase activity induced by hhi1. However, Ac-iap2 and Hz-iap2 could suppress the activity of caspase by approximately 40% (Fig. 6F). These results showed that Ac-iap2 and Hz-iap2 could suppress hhi1-induced apoptosis by the suppression of caspase activity. Interestingly, p35, the suppressor of apoptosis in insect cells encoded by baculovirus, did not seem to suppress hhi1-induced apoptosis.
PAT1, a noncoding RNA, inhibits hhi1-induced apoptosis.
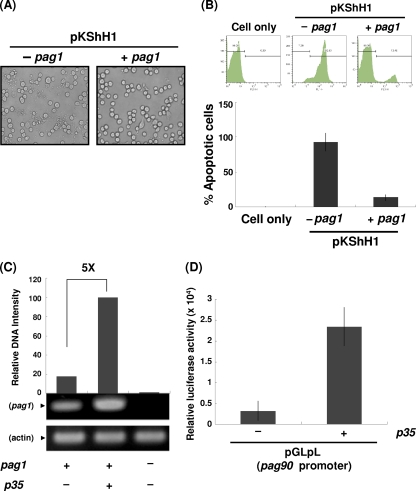
Previously, we identified the pag1 gene of HzNV-1, which expresses a noncoding RNA, PAT1, and is essential for the establishment of latent infection (5, 6). To investigate the possible interaction between hhi1 and pag1, we constructed an expression vector in which pag1 expression was controlled by its own promoter (51). We transfected hhi1 into Sf-21 cells with or without pag1 and observed the transfected cells under a microscope. At 12 hpt, we found prevalent apoptosis in cells transfected with hhi1 alone, while rather strikingly, significantly fewer apoptotic bodies were observed in cells cotransfected with hhi1 and pag1 (Fig. 7A). To estimate the level of apoptosis, transfected cells were stained with annexin V and were analyzed by flow cytometry at 4 hpt. The results showed that the apoptotic cells were reduced from 90% in the hhi1-transfected cells to 15% in the cells cotransfected with hhi1 and pag1 (Fig. 7B). Previous reports have identified P35 and the proteins belonging to the IAP family as the major antiapoptotic proteins. Our experimental data further identified a noncoding RNA that can serve a similar function in suppressing apoptosis. We currently are trying to elucidate the interaction between hhi1 and pag1, which results in the suppression of hhi1-induced apoptosis by pag1.
Fig. 7.
Analysis of antiapoptotic activities of pag1 in Sf-21 cells. (A) Inhibition of hhi1-induced apoptosis by pag1. Sf-21 cells (2 × 105 cells per 24-well plate) were transfected with pKShH1 in the presence or absence of pKSpP1 (carrying pag1). At 12 hpt, cells were examined by light microscopy and photographed to examine the inhibition of apoptosis. (B) Flow cytometry analysis of apoptosis suppression. In this assay, apoptotic cells that stained positive by annexin V were identified in the M2 region. The bar graph below shows the percentages of apoptotic cells derived from the flow analysis. (C) PAT1 level analysis by RT-PCR. Sf-21 cells were transfected with plasmids pKSpP1 (pag1) alone or cotransfected with pKShP35 (carrying p35), and the expression of the pag1 gene product, PAT1, was analyzed by RT-PCR. The expression of actin served as a control. (D) Luciferase assay showing that p35 can activate the pag1 promoter. Plasmid pGLpL (luciferase coding region driven by the pag1 promoter) (51) was cotransfected with or without pKShP35 into Sf-21 cells. Luciferase activities from each group were measured at 48 hpt.
HzNV-1-induced apoptosis can be suppressed by p35 through activation of pag1.
Our previous experiments showed that p35 could inhibit HzNV-1 superinfection-induced apoptosis in latently infected cells (28). However, our current results show that p35 could not suppress hhi1-induced apoptosis in transient expression assays. We previously noticed that p35 expression levels varied in different stable cell lines, although this is quite normal in the stable transfection of a given gene (28). However, what we did not understand was that the levels of PAT1 in the p35 stably transfected latent cells lines were stimulated to much higher levels than those of parental latent cells (28). Since we have demonstrated that pag1 can block hhi1-induced apoptosis (Fig. 7A and B) and P35 is a well-known transactivator (13, 50), we speculated that the apoptosis inhibition by p35 reported in Lee and Chao is achieved indirectly through the stimulation of pag1 expression by p35 (28). To verify this possibility, we introduced p35- and pag1-expressing plasmids into cells and detected the expression level of pag1 in transfected cells by RT-PCR. The result showed that pag1 expression was stimulated significantly by the expression of p35, suggesting that P35 functions as an efficient transactivator for the expression of pag1 (Fig. 7C). We further confirmed this hypothesis by cotransfecting a p35 expression plasmid with another plasmid that contains a luciferase reporter gene driven by the pag1 promoter. The result showed that luciferase activity was increased in the presence of p35, indicating that p35 indeed stimulates the activity of the pag1 promoter for downstream gene expression (Fig. 7D). Therefore, the blocking of HzNV-1 superinfection-induced apoptosis by p35 is more likely an indirect result of pag1 activation, which in turn suppresses hhi1-induced apoptosis.
DISCUSSION
Many viral early genes are strong transcriptional activators, and previous experiments have shown that some of them can be strong inducers of apoptosis in infected cells (27, 39, 42). Baculovirus ie1 was found previously to be capable of inducing apoptosis in Sf-21 cells (25, 39). icp0 of herpes simplex virus (HSV) and ns1 of human influenza A virus are two other examples that have been demonstrated to induce apoptosis in host cells (18, 23, 27). Previously, we found that hhi1, an early gene and strong transactivator of HzNV-1, can activate viruses in the latently infected cells (5, 6, 51). pag1, another early gene, which, interestingly, carries only a noncoding RNA, was shown to function in the establishment and/or enhancement of latent HzNV-1 virus infection in host cells (5). In this study, we found that hhi1 not only transactivates gene expression but also plays a direct or indirect role in promoting HzNV-1-induced apoptosis. In contrast to apoptosis induced by ie1 of AcMNPV, a transient expression assay revealed that hhi1 cannot activate the processing of pro-Sf-caspase-1. Therefore, it is likely that HzNV-1 induces apoptosis in host cells through a different caspase pathway. This notion is further supported by the finding that hhi1-induced apoptosis can be blocked by Ac-iap2 and Hz-iap2 but not p35.
Previous reports have shown that apoptosis can be regarded as a cellular defense mechanism against viral infection (9, 28). Viruses, however, also have evolved to produce a group of genes that can suppress or delay cell-induced apoptosis. p35 and iap genes were first discovered in baculovirus AcMNPV (12). Although p35 has been shown to block the apoptosis triggered by ie1 during baculovirus infection, since their discovery the exact functions of the Ac-iap genes have not been determined (10). Since viruses should retain only genes that they need, it is interesting to speculate on the possible roles of these iap genes (9). In this paper, we found that iap2 of both HzNV-1 (8) and AcMNPV (3, 15) can effectively suppress apoptosis induced by hhi1.
P35 originally was identified as a general caspase inhibitor due to its ability to inhibit virus-induced caspase activity (44). Many reports have demonstrated that p35 can suppress apoptosis induced by various genes in Sf-21 cells (9, 16, 26); however, our result shows that p35 cannot suppress hhi1-induced apoptosis in this cell line. Thus, our data suggest that hhi1 induces apoptosis by activating a caspase other than Sf-caspase-1. It has been demonstrated that p35 fails to rescue cell death induced by a caspase called DRONC, in vivo and in vitro, in Drosophila cells (19, 36). In addition, a novel caspase distinct from Sf-caspase-1 that is not sensitive to p35 also was proposed to exist in Sf-9 cells (1). All of these are indications that caspases other than Sf-caspase-1 exist in insect cells, and one of them may be responsible for the hhi1 induction of apoptosis.
The main effector caspase in Sf-21 cells is Sf-caspase-1 (32). Sf-caspase-1 is generated from pro-Sf-caspase-1 (P37), which can be cleaved into short prodomains P6, P12, and P19 by a sequential proteolytic process upon apoptotic signaling (1, 16, 26, 30). In our experiment, we showed that ie1, but not hhi1, can activate Sf-caspase-1 cleavage (43). Out of four mammalian caspase substrates, we found that hhi1 induces the cleavage of DEVD-AMC only (caspase 3 substrate). However, previous reports showed that mammalian caspases 6, 7, 8, and 10, as well as Sf-caspase-1, all can act on substrate DEVD-AMC (16, 26, 37). This raises the possibility that multiple caspases are present in Sf-21 cells, and one of them may be activated by hhi1 to cleave the substrate DEVD-AMC. Further studies are required to see whether this unique caspase(s) can be found in Sf-21 cells. Although it seems unlikely, hhi1 may also act on genes downstream of Sf-caspase-1 and simply bypass Sf-caspase-1 in the caspase-induced apoptosis pathway.
Besides Sf-caspase-1, the effector caspase Sf-caspase-2 was identified previously (52). During baculovirus infection, the proteolytic processing of effector caspases Sf-caspase-1 and Sf-caspase-2 can be prevented by the caspase inhibitor P49. Although P49 is a homolog of pancaspase inhibitor P35, the latter prevents the proteolytic processing of Sf-caspase-1 but not Sf-caspase-2 (16, 26). Therefore, we cannot rule out the possibility that hhi1 induces apoptosis by activating Sf-caspase-2.
In Fig. 5B, although we used ie1 as a control, the small subunit P12 of pro-Sf-caspase-1 still was clearly detectable at 72 hpt in the transfected cells (Fig. 5B, lane 3); we could not see the same subunit resulting from hhi1 transfection. Previously, Seshagiri and Miller (44) showed that P12 could result from the cleavage of pro-Sf-caspase-1 (driven by a heat shock promoter). The P12 fragment was detected at around 12 hpi and degraded afterwards. In our experiments shown in Fig. 5, although heat shock promoters were used to drive ie1 (lane 3) and hhi1 (lane 5), cells were not heat shock treated or virus infected as in Seshagiri and Miller (44). Under these conditions, we could clearly see the existence of P12 until 72 h after ie1 transfection, suggesting that this cleaved subunit can stay in the cell for long periods without virus infection. To further validate these results, we performed a Western blot experiment to measure caspase-1 expression levels at different time points (6 to 72 hpt) and found that the level of pro-Sf-caspase-1 was highest at 72 hpt (see Fig. S1 in the supplemental material). Further experiments showed that P12 cleaved from the cotransfection of pro-Sf-caspase-1 and ie1 began to be detectable at 48 hpt, and the expression level increased up to 72 hpt (see Fig. S2, lanes 3, in the supplemental material). This is an indication that P12 is not further degraded without the infection of the virus. Nevertheless, under the same conditions, we still could not detect P12 as a result of hhi1 transfection (see Fig. S2).
There are several points worth mentioning here. First, with AcMNPV coinfection, P12 was not detected at all (Fig. 5, lanes 2; also see Fig. S2). It is likely that the cleaved P12 was quickly degraded, as shown in Seshagiri and Miller (44). It also is likely that as the MOI we used was 1, rather than 20 as used by Seshagiri and Miller (44), the resulting subunit was present at levels too low to be detected in these experiments. However, P12 was detectable by transfection with ie1, suggesting that transfection was successful in our studies. Second, although P12 was not detected after hhi1 transfection (Fig. 5, lanes 5; also see Fig. S2), hhi1 transfection clearly resulted in strong apoptosis (date not shown), suggesting that the hhi1 plasmid used in this experiment was functional.
In the current study, we found that p35 fails to suppress hhi1-induced apoptosis; however, our previous report showed that p35 can suppress apoptosis induced by HzNV-1 superinfection in latently infected cells (28). This apparent contradiction can be explained by viewing p35 in a different role. Previously, we have shown that p35 can activate early HzNV-1 genes by functioning as a transactivator (50). Here, we further showed that p35 can function as a strong transactivator of the pag1 promoter (Fig. 7C and D), and our new discovery found that pag1 is a strong inhibitor of hhi1-induced apoptosis (Fig. 7A and B). Since the pag1 product PAT1 is a noncoding RNA (5), the mechanism by which pag1 inhibits apoptosis may be different from those exploited by the known antiapoptosis proteins P35 and the IAPs.
One possible mechanism by which PAT1, the product of pag1, could inhibit apoptosis is by the production of microRNAs (miRNAs) (46). Similarly to PAT1, HSV also expresses a noncoding RNA, the latency-associated transcript (LAT), which has been shown to inhibit virus-induced apoptosis (23, 45). These studies showed that miRNAs are produced from the LAT to block the translation of the early genes icp0 and icp4 (46). Although further experiments are needed, it is reasonable to assume that virus will go into latency if the expression of major early transcripts is blocked. We are currently investigating how pag1 suppresses apoptotic cell death, and the results should help us to understand the roles played by noncoding RNAs in insects.
In this study, we have employed a transient assay to study the utilization of Hz-iap2 and pag1 by HzNV-1 virus to suppress apoptosis. HzNV-1 virus expresses both pag1 and Hz-iap2 during productive viral infection but only expresses pag1 during viral latency (5). Hence, the roles of Hz-iap2 and pag1 may overlap but functionally differ. It is likely that HzNV1 virus mainly uses Hz-iap2 to suppress apoptosis triggered by hhi1 expression during initial viral invasion into healthy cells. In other words, Hz-iap2 only functions and is used during productive viral infection. pag1, on the other hand, may be used during productive infection to control the excess hhi1 transcript and may serve as the major force to suppress possible viral reactivation initiated by hhi1 expression (51) during latent viral infection. pag1 also may be responsible for blocking possible apoptosis induced by reactivated hhi1 during viral latency.
In conclusion, we have shown that the early gene hhi1 of HzNV-1 may promote apoptosis directly or indirectly in transiently transfected Sf-21 cells. It has been reported that some virus-encoded transcription factors, such as baculovirus IE1 and human immunodeficiency virus Tat, are proapoptotic, whereas some DNA viruses trigger apoptosis via the expression of transcription factors involved in viral DNA replication or perturbing the normal cell cycle (2, 43). Thus, hhi1 might induce apoptosis by either activating the expression of prodeath genes or altering the cell cycle of the host. The apoptosis pathway triggered by HHI1 does not seem to result in the activation of Sf-caspase-1 (Fig. 5B) but does result in the cleavage of mammalian caspase 3 substrate. We also identified a functional antiapoptosis gene, Hz-iap2, which can suppress hhi1-induced apoptosis. Interestingly, although Ac-iap2 was speculated to be a nonfunctional iap (9), we found that it is able to block apoptosis induced by hhi1. Furthermore, although p35 cannot directly suppress hhi1-induced cell death, we have shown an indirect mechanism whereby p35 can stimulate the expression of the pag1 gene to inhibit apoptosis. We conclude that hhi1 provides a unique opportunity for the further detailed investigation of apoptosis induction and suppression through both caspase-dependent and -independent pathways.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Chun-Hong Chen, Jeffrey J. Y. Yen, Tzong-Yueh Chen, and Rollie J. Clem for valuable discussions and suggestions. We also thank Heiko Kuhn and Miranda Loney for revisions.
This research was funded by grants 98-2313-B-001-004-MY3 and 98-2321-B-001-031-MY3 from the National Science Council and by grant 098S0050001-AA from Academia Sinica, Taiwan.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 4 May 2011.
REFERENCES
- 1. Ahmad M., et al. 1997. Spodoptera frugiperda caspase-1, a novel insect death protease that cleaves the nuclear immunophilin FKBP46, is the target of the baculovirus antiapoptotic protein p35. J. Biol. Chem. 272:1421–1424 [DOI] [PubMed] [Google Scholar]
- 2. Alimonti J. B., Ball T. B., Fowke K. R. 2003. Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J. Gen. Virol. 84:1649–1661 [DOI] [PubMed] [Google Scholar]
- 3. Ayres M. D., Howard S. C., Kuzio J., Lopez-Ferber M., Possee R. D. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605 [DOI] [PubMed] [Google Scholar]
- 4. Burand J. P., Stiles B., Wood H. A. 1983. Structural and intracellular proteins of the nonoccluded baculovirus HZ-1. J. Virol. 46:137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chao Y. C., et al. 1998. A 2.9-kilobase noncoding nuclear RNA functions in the establishment of persistent Hz-1 viral infection. J. Virol. 72:2233–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chao Y. C., et al. 1992. Differential expression of Hz-1 baculovirus genes during productive and persistent viral infections. J. Virol. 66:1442–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chao Y. C., Young S. Y., Kimb K. S., Scott H. A. 1985. A newly isolated densonucleosis virus from Pseudoplusia includens (Lepidoptera: Noctuidae). J. Invertebrate Pathol. 46:70–82 [Google Scholar]
- 8. Cheng C. H., et al. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clem R. J. 2007. Baculoviruses and apoptosis: a diversity of genes and responses. Curr. Drug Targets 8:1069–1074 [DOI] [PubMed] [Google Scholar]
- 10. Crook N. E., Clem R. J., Miller L. K. 1993. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 67:2168–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crouch E. A., Passarelli A. L. 2005. Effects of baculovirus transactivators IE-1 and IE-2 on the Drosophila heat shock 70 promoter in two insect cell lines. Arch. Virol. 150:1563–1578 [DOI] [PubMed] [Google Scholar]
- 12. Dubrez-Daloz L., Dupoux A., Cartier J. 2008. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle 7:1036–1046 [DOI] [PubMed] [Google Scholar]
- 13. Gong M., Guarino L. A. 1994. Expression of the 39k promoter of Autographa californica nuclear polyhedrosis virus is increased by the apoptotic suppressor P35. Virology 204:38–44 [DOI] [PubMed] [Google Scholar]
- 14. Granados R. R. 1978. Early events in the infection of Hiliothis zea midgut cells by a baculovirus. Virology 90:170–174 [DOI] [PubMed] [Google Scholar]
- 15. Griffiths C. M., et al. 1999. In vitro host range of Autographa californica nucleopolyhedrovirus recombinants lacking functional p35, iap1 or iap2. J. Gen. Virol. 80:1055–1066 [DOI] [PubMed] [Google Scholar]
- 16. Guy M. P., Friesen P. D. 2008. Reactive-site cleavage residues confer target specificity to baculovirus P49, a dimeric member of the P35 family of caspase inhibitors. J. Virol. 82:7504–7514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hacketta K. J., Booreb A., Demingb C., Buckleya E. C. M., Shapiroa M. 2000. Helicoverpa armigera granulovirus interference with progression of H. zea hucleopolyhedrovirus disease in H. zea larvae. J. Invertebrate Pathol. 75:99–106 [DOI] [PubMed] [Google Scholar]
- 18. Hagglund R., Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hawkins C. J., et al. 2000. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 275:27084–27093 [DOI] [PubMed] [Google Scholar]
- 20. Hozak R. R., Manji G. A., Friesen P. D. 2000. The BIR motifs mediate dominant interference and oligomerization of inhibitor of apoptosis Op-IAP. Mol. Cell. Biol. 20:1877–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang Y. S., Hedberg M., Kawanishi C. Y. 1982. Characterization of the DNA of a nonoccluded baculovirus, Hz-1V. J. Virol. 43:174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ingolia T. D., Craig E. A., McCarthy B. J. 1980. Sequence of three copies of the gene for the major Drosophila heat shock induced protein and their flanking regions. Cell 21:669–679 [DOI] [PubMed] [Google Scholar]
- 23. Kather A., et al. 2010. Herpes simplex virus type 1 (HSV-1)-induced apoptosis in human dendritic cells as a result of downregulation of cellular FLICE-inhibitory protein and reduced expression of HSV-1 antiapoptotic latency-associated transcript sequences. J. Virol. 84:1034–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelly D. C., et al. 1981. Induction of a nonoccluded baculovirus persistently infecting Heliothis zea cells by Heliothis armigera and Trichoplusia ni nuclear polyhedrosis viruses. Virology 112:174–189 [DOI] [PubMed] [Google Scholar]
- 25. LaCount D. J., Friesen P. D. 1997. Role of early and late replication events in induction of apoptosis by baculoviruses. J. Virol. 71:1530–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LaCount D. J., Hanson S. F., Schneider C. L., Friesen P. D. 2000. Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J. Biol. Chem. 275:15657–15664 [DOI] [PubMed] [Google Scholar]
- 27. Lam W. Y., et al. 2008. Avian influenza virus A/HK/483/97(H5N1) NS1 protein induces apoptosis in human airway epithelial cells. J. Virol. 82:2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J. C., Chao Y. C. 1998. Apoptosis resulting from superinfection of Heliothis zea virus 1 is inhibited by p35 and is not required for virus interference. J. Gen. Virol. 79:2293–2300 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. C., Chen H. H., Wei H. L., Chao Y. C. 1993. Superinfection-induced apoptosis and its correlation with the reduction of viral progeny in cells persistently infected with Hz-1 baculovirus. J. Virol. 67:6989–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin C. C., et al. 2007. Sf-caspase-1-repressed stable cells: resistance to apoptosis and augmentation of recombinant protein production. Biotechnol. Appl. Biochem. 48:11–19 [DOI] [PubMed] [Google Scholar]
- 31. Lin C. L., et al. 1999. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J. Virol. 73:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Q., Chejanovsky N. 2006. Activation pathways and signal-mediated upregulation of the insect Spodoptera frugiperda caspase-1. Apoptosis 11:487–496 [DOI] [PubMed] [Google Scholar]
- 33. Lo H. R., Chao Y. C. 2004. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 20:354–360 [DOI] [PubMed] [Google Scholar]
- 34. McIntosh A. H., Grasela J. J., Ignoffo C. M. 2007. In vitro host range of the Hz-1 nonoccluded virus in insect cell lines. In Vitro Cell Dev. Biol. Anim. 43:196–201 [DOI] [PubMed] [Google Scholar]
- 35. McIntosh A. H., Ignoffo C. M. 1981. Establishment of a persistent baculovirus infection in a lepidopteran cell line. J. Invertebr. Pathol. 8:395–403 [Google Scholar]
- 36. Meier P., Silke J., Leevers S. J., Evan G. I. 2000. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 19:598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nicholson D. W. 1999. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6:1028–1042 [DOI] [PubMed] [Google Scholar]
- 38. O'Reilly D. R., Miller L. K., Luckow V. A. 1994. Baculovirus expression vectors: a laboratory manual.Oxford University Press, New York, NY [Google Scholar]
- 39. Prikhod'ko E. A., Miller L. K. 1996. Induction of apoptosis by baculovirus transactivator IE1. J. Virol. 70:7116–7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ralston A. L., Huang Y. S., Kawanishi C. Y. 1981. Cell culture studies with the IMC-Hz-1 nonoccluded virus. Virology 115:33–44 [DOI] [PubMed] [Google Scholar]
- 41. Salvesen G. S. 2002. Caspases: opening the boxes and interpreting the arrows. Cell Death Differ. 9:3–5 [DOI] [PubMed] [Google Scholar]
- 42. Sanfilippo C. M., Blaho J. A. 2006. ICP0 gene expression is a herpes simplex virus type 1 apoptotic trigger. J. Virol. 80:6810–6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schultz K. L., Wetter J. A., Fiore D. C., Friesen P. D. 2009. Transactivator IE1 is required for baculovirus early replication events that trigger apoptosis in permissive and nonpermissive cells. J. Virol. 83:262–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seshagiri S., Miller L. K. 1997. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc. Natl. Acad. Sci. U. S. A. 94:13606–13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shen W., et al. 2009. Two small RNAs encoded within the first 1.5 kilobases of the herpes simplex virus type 1 latency-associated transcript can inhibit productive infection and cooperate to inhibit apoptosis. J. Virol. 83:9131–9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Umbach J. L., et al. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Kleespies R. G., Huger A. M., Jehle J. A. 2007. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 81:5395–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Y., van Oers M. M., Crawford A. M., Vlak J. M., Jehle J. A. 2007. Genomic analysis of Oryctes rhinoceros virus reveals genetic relatedness to Heliothis zea virus 1. Arch. Virol. 152:519–531 [DOI] [PubMed] [Google Scholar]
- 49. Wei Y., Fan T., Yu M. 2008. Inhibitor of apoptosis proteins and apoptosis. Acta Biochim. Biophys. Sin. (Shanghai) 40:278–288 [DOI] [PubMed] [Google Scholar]
- 50. Wu Y. L., et al. 2008. Cooperation of ie1 and p35 genes in the activation of baculovirus AcMNPV and HzNV-1 promoters. Virus Res. 135:247–254 [DOI] [PubMed] [Google Scholar]
- 51. Wu Y. L., et al. 2010. The early gene hhi1 reactivates Heliothis zea nudivirus 1 in latently infected cells. J. Virol. 84:1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zoog S. J., Schiller J. J., Wetter J. A., Chejanovsky N., Friesen P. D. 2002. Baculovirus apoptotic suppressor P49 is a substrate inhibitor of initiator caspases resistant to P35 in vivo. EMBO J. 21:5130–5140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.