Abstract
Alternate forms of the general transcription machinery have been described in several tissues or cell types. However, the role of tissue-specific TBP-associated factors (TAFIIs) and other tissue-specific transcription components in regulating differential gene expression during development was not clear. Here we show that the cannonball gene of Drosophila encodes a cell type-specific homolog of a more ubiquitously expressed component of the general transcription factor TFIID. cannonball is required in vivo for high level transcription of a set of stage- and tissue-specific target genes during male gametogenesis. Regulation of transcription by cannonball is absolutely required for spermatogenesis, as null mutations block meiotic cell cycle progression and result in a complete failure of spermatid differentiation. Our results demonstrate that cell type-specific TAFIIs play an important role in developmental regulation of gene expression.
Keywords: Spermatogenesis, TAF, TFIID, Drosophila, cannonball, dTAFII80
For many genes, recruitment of the general transcription factor TFIID to the promoter is a crucial step in the initiation of transcription by RNA polymerase II (Pol II) (Orphanides et al. 1996; Roeder 1996). The TFIID complex consists of the TATA-box-binding protein (TBP) and 10–14 TBP-associated factors (TAFIIs) (Dynlacht et al. 1991; Tanese et al. 1991; Kokubo et al. 1993; Chen et al. 1994; Poon et al. 1995; Kuras et al. 2000; Li et al. 2000; Sanders and Weil 2000). Although the TAFIIs constitute the bulk of the mass of TFIID, their role in transcription is under considerable debate. Certain TAFIIs physically interact with transcriptional activators in vitro, suggesting they may mediate transcriptional activation at certain promoters (for review, see Goodrich et al. 1996). Other TAFIIs interact with DNA sequences near the start site of transcription and may affect binding of TFIID to promoter region DNA (Verrijzer et al. 1995; Burke and Kadonaga 1996; Shen and Green 1997; Wang et al. 1997; Chalkley and Verrijzer 1999). Several TAFIIs are also components of histone acetyltransferase (HAT) complexes isolated from yeast (SAGA) or human cells (PCAF), suggesting that they may affect transcription by locally altering chromatin structure (Grant et al. 1998; Ogryzko et al. 1998). Furthermore, although TAFIIs appear to be dispensable for transcription initiation at some Pol II-dependent promoters, they are required for and specifically recruited to other promoters (Holstege et al. 1998; Kuras et al. 2000; Li et al. 2000).
Homologs of TBP and tissue-specific TAFIIs have been described in several organisms including humans and Drosophila. However, the possible contributions of these proteins to differential gene expression during development were not understood (for review, see Berk 2000). The recent completion of the Drosophila genome sequence revealed that several TAFIIs identified biochemically as components of TFIID have a recognizable sequence homolog encoded by a different gene. Here we show that one of these genes encodes a tissue-specific TAFII homolog expressed only in testes, where it regulates a critical gene expression program required for male gametogenesis. Our results suggest that expression of tissue-specific components of the so-called general transcription machinery may play an important role in coordinate control of gene expression during development.
Results
The cannonball gene of Drosophila encodes a homolog of dTAFII80
Male gametogenesis is characterized by an extremely active tissue-specific transcription program (Fuller 1993). Genetic analysis in Drosophila has identified several genes required for tissue- and stage-specific gene expression in primary spermatocytes (Lin et al. 1996; White-Cooper et al. 1998). Wild-type function of the Drosophila genes cannonball (can), meiosis I arrest (mia), spermatocyte arrest (sa), and always early (aly) are required for normal accumulation of transcripts from several spermatid differentiation genes (White-Cooper et al. 1998). Loss of function of can, mia, sa, or aly results in male sterility by preventing the initiation of spermatid differentiation. In addition, cell cycle progression is blocked at the G2/M transition of meiosis I in mutant males (Lin et al. 1996).
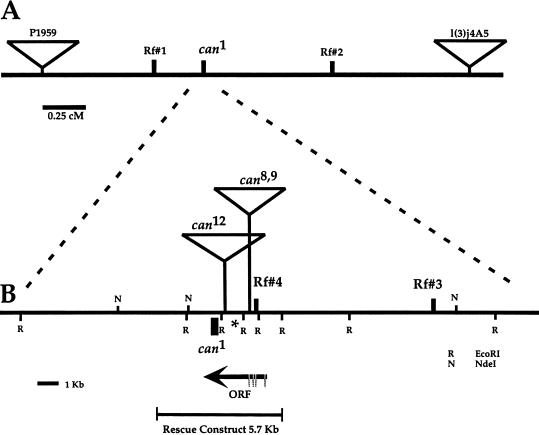
To explore the molecular mechanisms that regulate the tissue- and stage-specific transcrption program for spermatid differentiation, we positionally cloned the can gene (see Materials and Methods). We first mapped the can1 mutation between flanking marked P-element inserts by recombination and further localized can1 with respect to restriction fragment polymorphism (RFLP) markers (Fig. 1). Southern blot analysis of transposon-induced can alleles revealed that the can1 allele was associated with an ∼200-bp deletion. The can12 mutation had RFLPs consistent with an ∼7.5-kb insertion, and the can8 and can9 mutations had rearrangements consistent with ∼4.5-kb insertions (Fig. 1B). A 5.7-kb genomic DNA fragment spanning the region of the can1, can8, can9, and can12 rearrangements rescued both the spermatogenic arrest phenotype and the male sterility associated with canmutants when introduced into flies by P-element-mediated germ-line transformation (Fig. 1B). We identified a transcript encoded in the 5.7-kb rescue fragment by a combination of cDNA library screening, RT–PCR, and 5′ and 3′ RACE (see Materials and Methods).
Figure 1.
Molecular cloning of can. (A) Genetic map of the can region. The can1 mutation was mapped to a 2.5-cM interval between P-element insertions P1959 and l(3)j4A5 (inverted triangles). Rf#1 and Rf#2: RFLPs used to further define the can interval to a 0.8-cM region. (B) Restriction map of the can genomic region deduced from P1 phage clone DS69 (N) NdeI; (R) EcoRI . The can1 mutation mapped distal but close (1/129 recombinants) to a polymorphic AvaII site (Rf#3) and was not separated from a polymorphic NdeI site (Rf#4) in 124 recombinants between the flanking P-element insertions l(3)j4A5 and P1959. Small filled rectangle: 198-bp deletion in the predicted protein coding region in can1. Inverted triangles: insertions in the can12,can8 and can9 alleles. Arrow: can transcript as determined by 5′ RACE and RT–PCR from testis RNA. Thick horizontal line: predicted transcript coding region. Horizontal bracket: rescue fragment. (*) EcoRI fragment used as probe in Figs. 3 and 4.
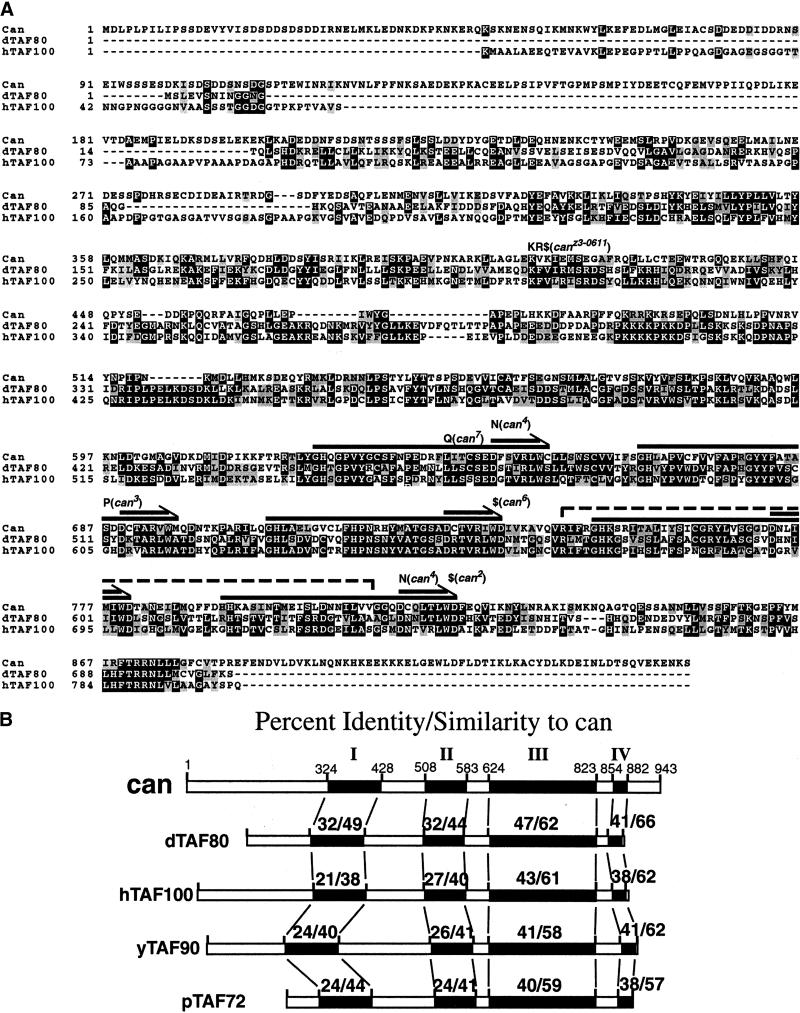
Sequence analysis revealed that the transcript from the 5.7-kb genomic DNA fragment encoded a predicted 943-amino-acid protein with five WD40 repeat motifs clustered near the carboxyl terminus (Fig. 2A). Sequence analysis of genomic DNA derived from homozygous mutant flies identified mutations within the ORF for several can alleles, confirming the identity of the predicted protein as can (Table 1). The can1 strain had a 198-bp deletion removing one and a half of the conserved WD40 repeats (Fig. 2A, dashed line). The can2 and can6 alleles each had a point mutation that changed a conserved tryptophan to a stop codon in the fifth or third WD40 repeat, respectively. The can3 mutation changed a conserved serine to proline in the second WD40 repeat. The can4 allele had two missense mutations: the first changed a signature-conserved aspartic acid to asparagine in the first WD40 repeat, and the second caused a similar conserved aspartic acid to asparagine substitution in the fifth WD40 repeat. The can7 allele had a missense mutation changing a conserved leucine to glutamine in the first WD40 repeat. Finally, the can13 line contained a single base pair deletion resulting in a frame shift and subsequent premature stop codon. The strong loss of function phenotype associated with the single amino acid substitutions in the can3, can4, and can7 missense alleles indicates that the WD40 repeat motifs are stringently required for can function. can3, can4, and can7 all cause the same male sterile phenotype as the null alleles.
Figure 2.
can encodes a TAFII homolog. (A) Alignment and comparison of the predicted can encoded amino acid sequence (GenBank accession no. AF346730) with dTAFII80 and hTAFII100. Long arrows: WD40 repeats. Dashed line: amino acids deleted in can1. Amino acid changes in other can alleles are indicated above the can protein sequence. ($) Stop codon. (B) Percent identity/similarity of can protein with dTAFII80 (GenBank accession no. S33263) hTAFII100 (AAC50902), yTAFII90 (1091232), and pTAFII72 (T41454). Solid boxes: homology domains. Region III contains the five consensus WD40 repeats. Region II contains a possible degenerate WD40 repeat motif.
Table 1.
Mutations in can alleles
|
can
Allele
|
Base pair change
|
Amino acid change
|
Amino acid
position
|
|---|---|---|---|
| can1 | deletion bp 2281 to 2479 | in frame deletion of 66 amino acid | 746–811 deleted |
| can2 | G(2508) to A | Stop | 822 |
| can3 | T(2105) to C | S to P | 687 |
| can4 | G(1985) to A and | D to N | 667 |
| G(2489) to A | D to N | 815 | |
| can6 | G(2257) to A | Stop | 737 |
| can7 | T(1968) to A | L to Q | 641 |
| canz3-0611 | T(1279) deleted | EK to KR substitution-Stop | 412 |
The position of the base pair change is given relative to the start of the 5′ RACE product. Positions of amino acid changes are relative to the predicted start of translation. All can alleles listed are male sterile.
The cannonball gene encodes a homolog of dTAFII80; a protein identified biochemically from Drosophila embryo extracts as a component of the general transcription factor TFIID (Dynlacht et al. 1993; Kokubo et al. 1993). The predicted can protein has four distinct regions of significant homology to Drosophila dTAFII80 and its human (hTAFII100) and yeast (Saccharomyces cerevisiae; yTAFII90 and Schizosaccharomyces pombe; pTAFII72) homologs (Fig. 2B) (Apone et al. 1996; Dubrovskaya et al. 1996; Tanese et al. 1996; Tao et al. 1997; Yamamoto et al. 1997). Interspersed between the conserved regions are variable segments where the proteins share little or no homology. Overall, the predicted can protein was more related to dTAFII80 than to hTAFII100, but dTAFII80 and hTAFII100 were more related to each other than either was to can. The predicted can protein had a carboxy-terminal extension not present in dTAFII80 or its yeast and human homologs (Fig. 2A). Secondary structure predictions indicated that the carboxy-terminal extension of the can protein was likely to fold into an extended α-helix. The predicted can protein had a high degree of sequence identity/similarity to dTAFII80 and hTAFII100 within each WD40 motif in addition to the GH-X21–40-D-X5-WD backbone of the WD40 motif (Fig. 2B) (Neer et al. 1994). The conserved spacing of the repeats, and the presence of three other conserved domains outside of the WD40 repeat region indicated that the can predicted protein is much more related to dTAFII80 and its homologs than to other WD40-containing proteins.
can expression is tissue specific
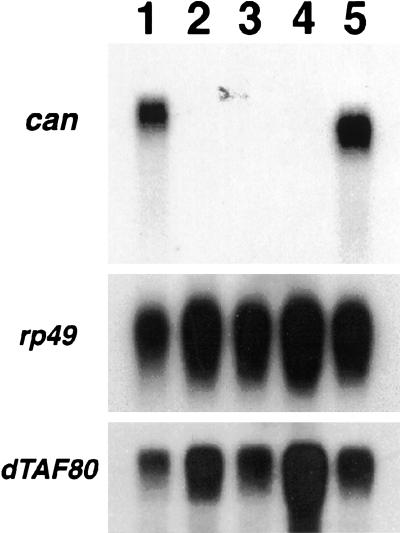
Expression of the can TAFII homolog was stage and tissue specific (Fig. 3). A 3.2-kb can transcript was detected in poly(A)+ mRNA from adult males but not in mRNA from adult females, embryos, or adult males lacking a germ line. In contrast, the can homolog dTAFII80 was widely expressed (Fig. 3). The cantranscript was present but reduced in size to 3.0 kb in males homozygous for the internal deletion allele can1, confirming that the mRNA corresponded to the can gene product. The male-specific and germ-line-dependent expression of can is consistent with the can phenotype; male can mutants are male sterile but viable and female fertile (Lin et al. 1996).
Figure 3.
Expression of can is male specific and germ line dependent. Northern blot of poly(A)+ RNA probed sequentially with sequences from the can open reading frame, rp49 cDNA loading control, and dTAFII80. Poly(A)+ selected RNA from: (lane 1) wild-type adult males; (lane 2) wild-type adult females; (lane 3) adult males lacking germ line; (lane 4) 0- to 24-h embryos; (lane 5) can1 homozygous adult males.
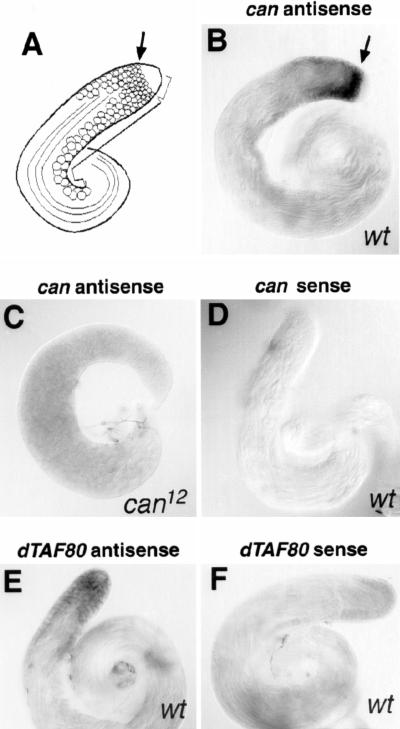
Within the testis, expression of can mRNA was restricted to primary spermatocytes (Fig. 4). In wild-type Drosophila testis, male germ-line stem cells and mitotically dividing spermatogonia are localized to the testis apical tip. The germ cells make the transition to the primary spermatocyte stage just below this region (Fig. 4A, arrow). can mRNA was detected in male germ cells by in situ hybridization beginning at the onset of the primary spermatocyte period (Fig. 4B, arrow). The can signal was highest in early spermatocytes but persisted at lower levels in more mature primary spermatocytes until approximately the time of the meiotic divisions. can message was not detected in mitotically dividing gonial cells or stem cells at the apical tip of the testis, or in postmeiotic germ cells and elongating spermatids. The can message was not detected in testes from flies homozygous for the can12 insertion allele (Fig. 4C). The cell type in which can mRNA is expressed in the testis is consistent with the defects observed in flies containing can loss of function mutations. All target genes identified to date that require wild-type can function for normal levels of transcription are expressed in male germ cells beginning early in the primary spermatocyte growth and gene expression period (White-Cooper et al. 1998).
Figure 4.
can mRNA is expressed in primary spermatocytes, beginning early in the growth and gene expression phase. In situ hybridization to whole adult Drosophila testes with single-stranded riboprobes. (A) Diagram of a whole adult testis with position of crucial stages indicated: (short bracket) mitotically dividing germ cells before the primary spermatocyte stage, (arrow) start of the primary spermatocyte period, (long, bent bracket) primary spermatocytes, (lines) elongating spermatid bundles. (B) Wild-type testis probed with antisense can. Cells entering the primary spermatocyte growth and gene expression program (arrow) express high levels of can mRNA. (C) The can signal was greatly reduced in testes from males homozygous for the can12 allele, which has an insertion in the can open reading frame (antisense can riboprobe). (D) Control: wild-type testis probed with can sense riboprobe. (E,F) Wild-type testis probed with dTAFII80 (E) antisense and (F) sense.
It is possible that primary spermatocytes have one complex containing can protein and a second alternative complex containing dTAFII80. In situ hybridization to wild-type testis revealed dTAFII80 message in both primary spermatocytes and spermatogonia (Fig. 4E).
Transcription of target genes is reduced in can mutants
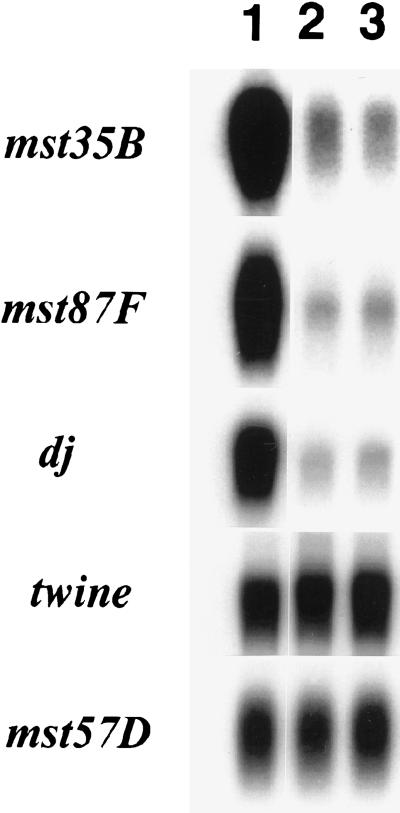
Wild-type function of can is required for normal accumulation of transcripts encoded by a suite of spermatid differentiation genes (White-Cooper et al. 1998). Early characterizations of TAFII function using in vitro transcription assays indicated that TBP supported basal transcription, whereas TBP with TAFIIs supported activated transcription in the presence of transcriptional activators (Pugh and Tjian 1990; Dynlacht et al. 1991; Zhou et al. 1992). The level of expression of can target genes in can mutant testes was reminiscent of this quantitative effect. Null mutations in can significantly reduced but did not completely eliminate transcription of target gene messages relative to wild type (Fig. 5). Nevertheless, in each case, a low level of transcript was detected in the mutants. As each of these target genes is expressed for the first time during development in primary spermatocytes, the residual message is not likely to be due to transcripts expressed at earlier stages.
Figure 5.
can mutants show lowered but detectable expression of target genes. RNA blots containing poly(A)+ selected RNA isolated from testis and accessory glands from wild-type and can mutant flies were probed with labeled DNA from known can-dependent or -independent genes. (Lane 1) wild type (red e); (lane 2) can2/can12; (lane 3) can6/can12. Similar results were obtained using can2/ can6 males (data not shown). can dependent genes: mst35B, mst87F, don juan: can independent gene: twine. Loading control: mst57D (expressed in male accessory glands but not in testes).
The effects of can mutations on transcript levels in primary spermatocytes were gene specific. The twine cell cycle regulatory phosphatase is normally transcribed in primary spermatocytes, starting at the onset of the primary spermatocyte growth and gene expression stage (Alphey et al. 1992; Courtot et al. 1992). Expression of twine mRNA was not can dependent in primary spermatocytes, as similar levels of the twine message were detected in can mutant and wild-type male reproductive tracts (Fig. 5).
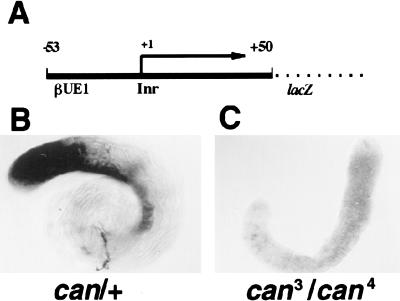
To explore whether wild-type can influences the levels of target gene expression by regulating transcription or message stability, we assayed the effect of can mutations on expression of a promoter lacZ fusion construct in vivo. Sequences from −53 to +50 of the β2t promoter fused to lacZ and introduced into flies allowed lacZ reporter mRNA expression in primary spermatocytes (Fig. 6A,B) (Michiels et al. 1989). Expression of the lacZ mRNA in primary spermatocytes was greatly reduced in can homozygotes compared to their can/+ siblings, indicating that can is likely to regulate expression of this reporter construct at the level of transcription (Fig. 6B,C).
Figure 6.
can-dependent transcription of a reporter construct. (A) Reporter construct carrying genomic sequences from −53 to +50 (+1 start of transcription) of the β2t gene fused to lacZ (provided by F. Michiels and R. Renkawitz-Pohl [Philipps Universität, Germany]). (B,C) In situ hybridization to whole adult testes showing expression of β-galactosidase RNA from the reporter construct at high levels in primary spermatocytes from +/can males (B) but only at low levels in their can3/can4 mutant brothers (C).
Discussion
Regulation of differential gene expression by different TAF isoforms
Our results demonstrate that a tissue-specific homolog of a more widely expressed TAFII is required in vivo for a developmentally regulated gene expression program in a metazoan. The can gene of Drosophila encodes a homolog of dTAFII80 expressed only in primary spermatocytes and required for the stage- and tissue-specific transcription of target genes needed for terminal differentiation of male gametes. These findings strongly suggest that substitution of tissue-specific TAF isoforms may adapt the general transcription machinery for expression of specific sets of developmentally regulated target genes.
Although similar ideas have been proposed for tissue-specific TAFIIs, our results constitute the first demonstration of in vivo function. The human hTAFII130 has a cell type-specific homolog hTAFII105 (Dikstein et al. 1996). Likewise, two Drosophila genes encoding homologous proteins dTAFII24 and dTAFII16 are differentially expressed during embryogenesis in certain cells (Georgieva et al. 2000). However, in neither case was a developmental gene expression program dependent on the different TAFII isoforms described. In addition to tissue-specific TAFIIs, homologs of TBP may be used to adapt the general transcription machinery. For example, the ubiquitously expressed Caenorhabditis elegans TBP homolog TLF (CeTLF) is required for multiple events during embryogenesis, although the specific developmental pathways affected are not known (Dantonel et al. 2000; Kaltenbach et al. 2000).
There are several mechanisms by which the can protein might adapt the general transcription machinery to regulate testis-specific transcription. The can protein might simply replace dTAFII80 in a protein complex that otherwise contains the same subunits as TFIID from Drosophila embryos. Alternatively, the can protein might act as part of a specialized TFIID-like complex consisting of a number of tissue-specific TAFIIs or a different TBP. Genes predicted to encode second homologs of dTAFII110, dTAFII30α, and dTAFII60 are present in the Drosophila genome (Adams et al. 2000). In addition, the Drosophila TBP homolog TRF1 is expressed in primary spermatocytes (Crowley et al. 1993).
It is also possible that the can protein regulates transcription of spermatid differentiation genes as a component of a tissue-specific HAT complex. HAT complexes are thought to regulate the transcription of certain genes by locally altering nucleosome acetylation and chromatin structure to allow the general transcription machinery access to the promoter (Georgakopoulos and Thireos 1992; Grant et al. 1997). The PCAF complex of humans includes a protein with homology to can, whereas both the SAGA and TFIID complexes of S. cerevisiae contain the can homolog yTAFII90 (Grant et al. 1998; Ogryzko et al. 1998).
The requirement of can for normal levels of transcription is gene specific. Certain genes transcribed in primary spermatocytes require can, whereas transcription of many other genes in primary spermatocytes is independent of can function (White-Cooper et al. 1998). At least three mechanisms could account for this specificity. (1) Particular core promoter sequences within target genes might bind a protein complex containing can, but not a similar dTAFII80-containing complex. Promoter swapping studies in yeast have demonstrated that dependence of activated transcription on the largest TAFII subunit yTAFII145 is conferred by core promoter sequences rather than upstream enhancer elements (Shen and Green 1997). Likewise, hTAFII250-dependent activated transcription depended on both core promoter sequences and upstream enhancer elements in vivo (Wang et al. 1997). In addition, several TAFIIs have been shown to physically interact with core promoter elements (Burke and Kadonaga 1997; Chalkley and Verrijzer 1999). (2) Certain genes may require can to activate transcription because one or more spermatocyte-specific transcriptional activators require can protein instead of dTAFII80 to recruit the Pol II transcription machinery to their promoters. The can protein could either bind such a transcriptional activator or its coactivators directly, or participate in a protein complex that does so. The proposed spermatocyte-specific transcriptional activators would thus work in concert with a can protein complex to achieve full transcription of target genes. (3) Spermatid differentiation genes dependent on can function for normal levels of transcription could be enveloped in a special chromatin structure that requires an activity associated with can or its partner proteins, but not with dTAFII80 or its partners to achieve full levels of activated transcription. Consistent with this hypothesis, chromatin may be in a different overall state in primary spermatocytes than in many cell types, as the chromosomes in primary spermatocytes appear as partially condensed, discrete entities when stained with the DNA dye Hoechst 33342 (Lin et al. 1996). These models are not mutually exclusive and the transcription of spermatid differentiation genes may involve any two or even all three of these mechanisms.
Understanding TAFII function through the study of tissue-specific TAFIIs
Phenotypic analysis of the can mutants may shed light on the in vivo mechanisms of the action of TAFIIs in general. Wild-type can function is not required for viability or any earlier stages of somatic or germ-line development (Lin et al. 1996). Thus, we were able to assess the effect of null alleles on transcription in primary spermatocytes, circumventing problems of interpretation due to the need to use partial loss of function alleles, temperature-sensitive alleles, or possible incomplete depletion of pre-existing protein.
We have exploited this strength to assess the effects of the loss of can function on target gene expression in vivo. Although expression levels for several testis-specific can-dependent target transcripts were much reduced in can mutant testis compared to wild type, low levels of testis-specific message were nonetheless detected. Our results suggest that tissue-specific transcription in primary spermatocytes may involve at least a two-step activation mechanism. For example, the low level of target gene transcripts expressed in can mutant testis may result from the function of a testis-specific transcriptional activator, whereas the full wild-type transcript levels may require the function of a TFIID- or HAT-like complex containing the can protein. Alternatively, residual target gene expression in the absence of can protein function could reflect action of a different TAFII or TBP alone.
The primary spermatocyte transcription program
Many can-dependent target genes are expressed for the first time during development in primary spermatocytes, in preparation for meiosis and spermatid differentiation. Other can-dependent target genes are more widely expressed, but use testis-specific promoters for expression in primary spermatocytes. Our results suggest that the general Pol II transcription machinery may be different in primary spermatocytes than in many other cell types. The use of testis-specific forms of TFIID or other components of the general transcription machinery could explain why many generally expressed genes use new, testis-specific promoters during male germ line development in both Drosophila and mammals (Fuller 1993; Hecht 1993). Indeed, a testis-specific TFIIA homolog has recently been described in humans (Upadhyaya et al. 1999; Ozer et al. 2000). In addition, the human TBP homolog hTRF2 is expressed predominantly in the testis (Teichmann et al. 1999). For several can-dependent genes, DNA sequences sufficient for primary spermatocyte expression lie very near the start site of transcription. Testis-specific promoters with cis-acting sequences near the start of transcription have also been observed for genes expressed in spermatocytes in mammals. For example, <200 bp of promoter region are sufficient for expression of the lactate dehydrogenase C and protamine 1 genes in testis extracts or transgenic mouse testes (Zambrowicz et al. 1993; Goldberg 1996). These parallels evoke the possibility that testis-specific components of the general transcription machinery may also play an important role in the male germ cell-specific gene expression programs in mammals.
Materials and methods
Drosophila strains and husbandry
Drosophila were raised on standard cornmeal and molasses medium at 25°C, unless otherwise noted. can alleles 1–12 were as described (Lin et al. 1996). Three new alleles, canz3–0611, canz3–2824, and canz3–0246, were isolated in a large-scale screen for EMS-induced viable but male sterile mutations by B. Wakimoto, D. Lindsley, and C. Zuker (unpubl.). osk8 (FlyBase ID FBal0013310) and oskce3 (FBal0032324) mutant flies were a gift of R. Lehmann (New York University Medical School) and described in FlyBase (http://flybase.bio.indiana.edu/). Recombination mapping indicated that the can1 mutation was not associated with the single marked P-element in the line (inserted at 86°C). Although the can8 and can9 alleles were induced by hybrid dysgenesis using the Birm2 chromosome (Robertson 1988), no P-element inserts in the can interval were detected by in situ hybridization to polytene chromosomes and none of the P-element sequences detected by Southern blot analysis of these strains segregated with the can mutation during recombination mapping.
P-element-mediated germ-line transformation was carried out using the indicated 5.7-kb KpnI–SacII fragment subcloned into pCaSpeR-4 (Rubin and Spradling 1983). Five independent transformed lines were tested. In each case the clones rescued both the meiotic arrest and sterility phenotypes of can3/can4 trans-heterozygotes.
Molecular cloning of can
The can locus was identified by fine structure recombination mapping using RFLP markers. Two P-element insertions, P1959 (Lin et al. 1996) in 67C10 and l(3)j4A5 (FlyBase ID FBti0004953) in 67E5, were selected for their disparate origins so that polymorphisms between the two parental chromosomes might be readily detectable. First, can1 was recombined onto the P1959 chromosome, then a total of 135 recombinants were generated between the two w + inserts by screening the progeny of P1959 can/l(3)j4A5 females crossed to w− males for either w− or a darker w+ (indicating the double insert) eye color. Recombination between the two flanking P-element insertions occurred at a frequency of 290 in 11,523 flies, placing them 2.5 cM apart. Scoring for inheritance of can1 among the recombinants placed the mutation ∼0.8 cM proximal to P1959 and 1.7 cM distal to l(3)j4A5. RFLPs between the two parental chromosomes were first identified by Southern blot analysis of genomic DNA from P1959 can1/P1959 can1 compared to P1959 can1/l(3)j4A5 flies digested with a panel of restriction enzymes and probed with end fragments of P1 clones (Berkeley Drosophila Genome Project) in the region. Segregation of the RFLPs was then scored in the recombinants. Initially, the can1 mutation mapped 0.3 cM proximal to a RFLP identified by the proximal end fragment of the insert in P1 DS69 (Rf#1) and 0.5 cM distal to a RFLP identified by the distal end of the P1 DS1778 insert (Rf#2). The proximal end of the P1 DS7440 insert identified two RFLPs tightly linked to can1: one caused by a polymorphic AvaII site just proximal to can1 based on 1/129 recombinants (Rf#3), the other caused by a polymorphic NdeI site not separated from can1 in 124 recombinants (Rf#4) (Fig. 1B). This P1 end fragment was used to screen the Tamkun λEMBL3 genomic phage library to isolate overlapping bacteriophage clones carrying inserts covering 23.5 kb from the can region.
Sequence analysis
DNA fragments spanning the can region were isolated from genomic phage clones, subcloned into Bluescript, and sequenced fully on both strands by dideoxy chain termination (Sanger et al. 1977) using T3 and T7 primers and gene-specific oligonucleotides (PAN facility, Stanford) to fill remaining gaps. Point mutations of EMS induced can alleles and the can1 deletion were identified by sequencing bulk PCR products amplified from genomic DNA from can homozygous mutant flies using gene-specific oligonucleotides. Sequences were aligned and analyzed using Sequencher (Gene Codes Corp.) and MacVector (Oxford Molecular Group plc) DNA analysis software.
The structure of the can transcript was deduced from a combination of testis cDNAs, RT–PCR, 5′ and 3′ RACE. The 5′ end of the can mRNA was determined by 5′ RACE using the GeneRacer system (Invitrogene) in two independent reactions. The sequence of the can transcript (GenBank accession no. AF346730) was deduced from the sequence of the 5′ RACE product plus the sequence of a separate, overlapping RT–PCR product generated from testis poly(A)+ selected mRNA using can gene-specific primers. The sequence of the 5′ RACE product differed from the sequence of genomic DNA at 4 bases resulting in four predicted amino acid changes in the nonconserved amino-terminal region of the predicted can protein. The amino acid sequence in Figure 2 was predicted from the genomic sequence of the exons using the intron/exon structure indicated from the sequence of the 5′ RACE product. The predicted initiating methionine had in-frame stop codons upstream. One additional 5′ RACE reaction using a different system (Life Technologies) detected a possible shorter RNA initiating in the fourth intron and encoding a predicted protein starting at methionine 186. However, Northern blot analysis revealed only one mRNA that most closely matched the larger transcript in size. For PCR from the testis cDNA library (gift of T. Hazelrigg, Columbia University), oligonucleotides complementary to sequences on the phage arms of the Hazelrigg λZAP testis cDNA library were used in combination with gene-specific primers to amplify the 3′ region of the can cDNA by nested PCR. To bias the PCR reaction toward the cDNA of interest, the first round of PCR was carried out with 10-fold of the gene-specific primer compared to the primer recognizing the phage arm sequence.
The predicted can protein sequence was used to search nucleotide sequence databases translated in all reading frames (tBLASTn). Sequence alignment was generated using the ClustalW Multiple Sequence Alignment (Thompson et al. 1994) and Boxshade programs for the three proteins shown. Percents identity/similarily in Figure 1B were calculated based on pairwise comparisons from a ClustalW alignment of all five proteins.
In situ hybridization
In situ hybridization to whole adult Drosophila testes was carried out as described in White-Cooper et al. (1998). In each well, mutant testes were mixed with testes from wild-type or heterozygous sibling males as a positive control. The color reaction was allowed to develop until the control testes were well stained. Mutant testes could be easily distinguished from the controls by their smaller size and lack of elongating spermatids. In all cases, mutant and control testes were processed for the same time in the same well so that signal levels could be compared directly. Single-stranded riboprobes were generated using the Genius System (Boehringer Mannheim) per the manufacturer's instructions. Probes were: can: a 1.2-kb EcoRI fragment (* in Fig. 1A) of can coding sequence cloned into Bluescript; dTAFII80: PCR product corresponding to base pairs 902 to 2079 of the cDNA cloned into pCR2.1 (Invitrogen). lacZ–pBSlac (Lessing and Nusse 1998).
RNA and DNA blot analysis
Southern blot analysis was carried out as described by Sambrook et al. (1989) modified as follows: genomic DNA was extracted from whole adult flies (Steller and Pirrotta 1984) and DNA from ∼5–10 flies were loaded per lane. All prehybridizations and hybridizations were done in Church Buffer (7% SDS, 1% BSA, 1 mM EDTA, 250 mM sodium phosphate buffer at pH 7.2). Probes used for DNA or RNA blots were labeled according to the manufacturer's instructions using the Megaprime DNA labeling system (Amersham) or using Rediprime II (Amersham Pharmacia Biotech) from gel-purified DNA fragments.
RNA from whole adult flies was isolated by homogenization in TRIzol Reagent (Life Technologies) according to the manufacturer's instructions. Adult males lacking germ line were generated by crossing oskarCE3/oskar8females to males. Testis mRNA was isolated from dissected testis of 1-day-old (or younger) males from indicated genotypes using the Micro-FastTrack 2.0 kit according to the manufacturer's instructions (Invitrogen). Approximately 5 μg of poly(A)+ RNA selected using PolyATtract mRNA isolation systems (Promega) was loaded per sample, separated on a 1.2% agarose gel with formaldehyde, transferred onto Hybond nylon membrane (Amersham) in 10× SSPE, and fixed to the membrane by UV cross-linking (Stratagene, Stratalinker model 2400). Prehybridization and hybridization was done in Church buffer. Probes as for in situ hybridizations above, plus mst35B: PCR product corresponding to base pairs 220 to 728 from the mst35Ba cDNA sequence (Ashburner et al. 1999); mst87F: PCR product corresponding to the entire 3′ untranslated sequence (Kuhn et al. 1988); don juan: 900-bp EcoRI sequence from the don juan cDNA (Santel et al. 1997); mst57D: 666-bp PCR fragment corresponding to the mst57Dc transcript from base pair 3607 to 2941 (Simmerl et al. 1995); rp49: PCR product using T7 and T3 universal primers from pBluescript containing rp49.
Acknowledgments
We are grateful to F. Michiels, R. Renkawitz-Pohl, and R. Lehmann for fly strains, B. Wakamoto and D. Lindsley for can alleles, R. Tjian for the dTAFII80 cDNA clone, the Berkeley Drosophila genome project for P1 clones and P-element insert markers, and the Stanford PAN facility for oligonucleotide synthesis and sequencing. We thank members of the Fuller laboratory and R. Kornberg, A. Villeneve, and C. Thut for discussions and critical comments on the manuscript. This work was supported by an NICHD research grant to M.T.F., a National Institute of Child Health and Human Development Reproductive Biology Training Grant, a National Research Service Award (HD082778), and a Stanford Medical School Dean's Fellowship to M.A.H., and the National Institute of General Medical Sciences Medical Scientist Training Program (T-Y.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL fuller@cmgm.stanford.edu; FAX (650) 725-7739.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.869101.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–988. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- Apone LM, Virbasius CA, Reese JC, Green MR. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes & Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Misra S, Roote J, Lewis SE, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adhregion. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A J. TBP-like factors come into focus. Cell. 2000;103:5–8. doi: 10.1016/s0092-8674(00)00098-2. [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. DrosophilaTFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- ————— The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: A TAFII250–TAFII150 complex recognizes the Initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Attardi LD, Verrijzer CP, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- Courtot C, Frankhauser C, Simanis V, Lehner C. The Drosophila cdc25 homolog twineis required for meiosis. Development. 1992;116:405–416. doi: 10.1242/dev.116.2.405. [DOI] [PubMed] [Google Scholar]
- Crowley TE, Hoey T, Liu JK, Jan YN, Jan LY, Tjian R. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature. 1993;361:557–561. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- Dantonel J C, Quintin S, Lakatos L, Labouesse M, Tora L. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell. 2000;6:715–722. doi: 10.1016/s1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- Dubrovskaya V, Lavigne A, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIFβ (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Weinzierl ROJ, Admon A, Tjian R. The dTAFII80 subunit of DrosophilaTFIID contains β-transducin repeats. Nature. 1993;363:176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The development of Drosophila. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 71–147. [Google Scholar]
- Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S, Kirschner DB, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B, Tora L. Two novel DrosophilaTAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol Cell Biol. 2000;20:1639–1648. doi: 10.1128/mcb.20.5.1639-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E. Transcriptional regulatory strategies in male germ cells. J Andrology. 1996;17:628–632. [PubMed] [Google Scholar]
- Goodrich J, Cutler G, Tjian R. Contacts in context: Promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, 3rd, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Hecht NB. Gene expression during male germ cell development. In: Desjardins C, Ewing LL, editors. Cell and molecular biology of the testis. New York, NY: Oxford University Press; 1993. pp. 400–432. [Google Scholar]
- Holstege FC, Jennigs EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegansembryogenesis. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Kokubo T, Takada R, Yamashita S, Gong D-W, Roeder RG, Horikoshi M, Nakatani Y. Identification of TFIID components required for transcriptional activation by upstream stimulatory factor. J Biol Chem. 1993;268:17554–17558. [PubMed] [Google Scholar]
- Kuhn R, Schafer U, Schafer M. Cis-acting regions sufficient for spermatocyte-specific transcriptional and spermatid-specific translational control of the Drosophila melanogaster gene mst(3)gl-9. EMBO J. 1988;7:447–454. doi: 10.1002/j.1460-2075.1988.tb02832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- Lessing D, Nusse R. Expression of wingless in the Drosophila embryo: A conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development. 1998;125:1469–1476. doi: 10.1242/dev.125.8.1469. [DOI] [PubMed] [Google Scholar]
- Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- Lin T, Visvanathan S, Wood C, Wilson PG, Wolf N, Fuller MT. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophilamales. Development. 1996;122:1331–1341. doi: 10.1242/dev.122.4.1331. [DOI] [PubMed] [Google Scholar]
- Michiels F, Gasch A, Kaltschmidt B, Renkawitz-Pohl R. A 14 bp promoter element directs the testis specificity of the Drosophilaβ2-tubulin gene. EMBO J. 1989;8:1559–1565. doi: 10.1002/j.1460-2075.1989.tb03540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Ogryzko V, Kotani T, Xiaolong Z, Schiltz RL, Howard T, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Ozer J, Moore PA, Lieberman PM. A testis-specific transcription factor IIA (TFIIAtau) stimulates TATA-binding protein–DNA binding and transcription activation. J Biol Chem. 2000;275:122–128. doi: 10.1074/jbc.275.1.122. [DOI] [PubMed] [Google Scholar]
- Poon D, Bai Y, Campbell AM, Bjorklund S, Kim Y-J, Zhou S, Kornberg RD, Weil PA. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: Evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Robertson H, Preston C, Phillis R, Johnson-Schlitz D, Benz W, Engels W. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Vectors for P element-mediated transformation in Drosophila. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. New York, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanders SL, Weil PA. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Winhauer T, Blümer N, Renkawitz-Pohl R. The Drosophila don juan (dj) gene encodes a novel sperm specific protein component characterised by an unusual domain for repetitive amino acid motif. Mech Dev. 1997;64:19–30. doi: 10.1016/s0925-4773(97)00031-2. [DOI] [PubMed] [Google Scholar]
- Shen W, Green MR. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- Simmerl E, Schafer M, Schafer U. Structure and regulation of a gene cluster for male accessory gland transcripts in Drosophila melanogaster. Insect Biochem Mol Biol. 1995;25:127–137. doi: 10.1016/0965-1748(94)00034-f. [DOI] [PubMed] [Google Scholar]
- Steller H, Pirrotta V. Regulated expression of genes injected into early Drosophila embryos. EMBO J. 1984;3:165–173. doi: 10.1002/j.1460-2075.1984.tb01778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N, Pugh BF, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes & Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- Tanese N, Saluja D, Vassallo M, Chen J-L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci. 1996;93;:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Guermah M, Martinez E, Oelgeschlager T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder RG. Specific interactions and potential functions of human TAFII100. J Biol Chem. 1997;272:6714–6721. doi: 10.1074/jbc.272.10.6714. [DOI] [PubMed] [Google Scholar]
- Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci. 1999;96:13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya AB, Lee SH, DeJong J. Identification of a general transcription factor TFIIAalpha/beta homolog selectively expressed in testis. J Biol Chem. 1999;274:18040–18048. doi: 10.1074/jbc.274.25.18040. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- Wang EH, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes & Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H, Schafer MA, Alphey LS, Fuller MT. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development. 1998;125:125–134. doi: 10.1242/dev.125.1.125. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Poon D, Weil PA, Horikoshi M. Molecular genetic elucidation of the tripartite structure of the Schizosaccharomyces pombe72 kDa TFIID subunit which contains a WD40 structural motif. Genes to Cells. 1997;2:245–254. doi: 10.1046/j.1365-2443.1997.1180316.x. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Harendza CJ, Zimmermann JW, Brinster RL, Palmiter RD. Analysis of the mouse protamine 1 promoter in transgenic mice. Proc Natl Acad Sci. 1993;90:5071–5075. doi: 10.1073/pnas.90.11.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Lieberman PM, Boyer TG, Berk AJ. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes & Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]