Abstract
The chromosome of Mycobacterium tuberculosis (Mtb) encodes forty seven toxin-antitoxin modules belonging to the VapBC family. The role of these modules in the physiology of Mtb and the function(s) served by their expansion are unknown. We investigated ten vapBC modules from Mtb and the single vapBC from M. smegmatis. Of the Mtb vapCs assessed, only Rv0549c, Rv0595c, Rv2549c and Rv2829c were toxic when expressed from a tetracycline-regulated promoter in M. smegmatis. The same genes displayed toxicity when conditionally expressed in Mtb. Toxicity of Rv2549c in M. smegmatis correlated with the level of protein expressed, suggesting that the VapC level must exceed a threshold for toxicity to be observed. In addition, the level of Rv2456 protein induced in M. smegmatis was markedly lower than Rv2549c, which may account for the lack of toxicity of this and other VapCs scored as ‘non-toxic’. The growth inhibitory effects of toxic VapCs were neutralized by expression of the cognate VapB as part of a vapBC operon or from a different chromosomal locus, while that of non-cognate antitoxins did not. These results demonstrated a specificity of interaction between VapCs and their cognate VapBs, a finding corroborated by yeast two-hybrid analyses. Deletion of selected vapC or vapBC genes did not affect mycobacterial growth in vitro, but rendered the organisms more susceptible to growth inhibition following toxic VapC expression. However, toxicity of ‘non-toxic’ VapCs was not unveiled in deletion mutant strains, even when the mutation eliminated the corresponding cognate VapB, presumably due to insufficient levels of VapC protein. Together with the ribonuclease (RNase) activity demonstrated for Rv0065 and Rv0617 – VapC proteins with similarity to Rv0549c and Rv3320c, respectively – these results suggest that the VapBC family potentially provides an abundant source of RNase activity in Mtb, which may profoundly impact the physiology of the organism.
Introduction
The feature of Mtb that presents the most significant impediment to developing treatment-shortening therapies for tuberculosis (TB) is its remarkable ability to persist in the face of the threats imposed by host immunity and bactericidal drug action [1], [2], [3]. This ability is thought to account, at least in part, for the protracted duration of treatment required to cure TB with short-course chemotherapy. Thus, understanding the physiology of the persistent state/s in Mtb represents one of the most important areas of mycobacterial research today [1], [3], [4]. Much emphasis has been placed on defining the physiology of non-replicating persister cells of Mtb formed under the conditions of nutritional, hypoxic, nitrosative and acidic stress thought to be encountered during infection [5], [6]. This has led to the identification of pathways that are required to maintain mycobacterial viability under such conditions and whose components are being pursued as novel drug targets [7], [8], [9], [10], [11]. Attention has also focused on the mechanisms underlying the formation of persister cells that arise through the stochastic expression of proteins that can affect the physiology and growth rate of the cell by interfering with key cellular processes such as macromolecular synthesis [2], [12], [13], [14], [15], [16], [17], [18], [19], [20]. These mechanisms are the subject of considerable interest having been implicated in phenotypic drug tolerance/indifference in E. coli [2], [14], [15], [16], [17], [18], [21], [22], [23].
Of the genes implicated in such processes, most attention has been paid to toxin-antitoxin (TA) modules, which are bicistronic operons widely distributed in the genomes of free-living prokaryotes [24], [25]. Although their contemporary role in microbial physiology remains the subject of debate [26], [27], there is evidence that chromosomal TA modules may act in stress physiology by serving as metabolic regulators of growth [13], [25], [28], [29]. When bound in a complex, the antitoxin neutralizes the activity of the toxin [30], [31], [32]. In the absence of continued expression of the operon, which regulates its own expression [33], dissociation of the complex and degradation of the relatively unstable antitoxin unveils the biological activity of the toxin.
Mtb possesses an unusually large and diverse complement of TA modules which belong to the MazEF, RelBE, ParDE, HigBA and VapBC families [24], [25]. A systematic analysis of Mtb TA module function revealed that Mtb also possesses a number of novel systems with no similarity to known modules [34]. Within this repertoire, the paralogous expansion of the VapBC family is particularly noteworthy [24], [25], [35], [36] and is a feature that Mtb shares with a small number of unrelated organisms [37]. In stark contrast, the genomes of mycobacteria other than those belonging to the Mtb complex (M. africanum, M. microti, M. canetti and M. bovis) are virtually devoid of VapBC and other TA modules [34]. The TA modules of Mtb have been the subject of intense investigation [31], [34], [38], [39], [40], [41], [42], [43], [44], [45]. Of the 47 VapBC modules (Fig. S1), one has been structurally and biochemically characterized [31], and this and 22 others tested for toxicity in E. coli [40], [45]. Twenty one of 45 VapCs tested were found to be toxic in M. smegmatis and four of these were shown to inhibit translation [34]. VapC toxins belong to the PIN (PilT N-terminus) domain family of proteins whose members have been associated with nuclease activity [36]. Most recently, enteric VapCs were shown to act as site-specific endonucleases that inhibit translation by cleavage of initiator tRNA [46]. PIN domains have RNase-H-like fold in which four conserved acidic residues are located in close proximity to form a negatively charged pocket, as illustrated in the structures of the VapC from Pyrobaculum aerophilum [47], [48], VapC5 from Mtb [31] and FitB from Neisseria gonorrhoeae [32]. Dissociation of the toxin-antitoxin complex is thought to allow binding of divalent metal ion in this acidic pocket of VapC thereby creating an active site for metal-ion-dependent nuclease activity [32], [48].
To investigate vapBC function in mycobacteria, we focused on a subset of 10 modules from Mtb H37Rv and the single vapBC from M. smegmatis mc2155. We found that some, but not all of the VapC proteins confer growth inhibition following inducible over-expression in both mycobacterial species. The toxic activity of these VapCs could be neutralized by the cognate, but not non-cognate antitoxins, indicating that these loci encode functional VapBC modules. A correlation between the expression levels of the VapC proteins and its ability to confer a toxic phenotype was observed. Analysis of mycobacterial vapBC deletion mutants failed to yield observable phenotypes under standard growth conditions. However, toxic VapCs showed enhanced toxicity when expressed in deletion mutants lacking antitoxic VapCs, providing further evidence of the specificity of interaction between VapCs and their cognate antitoxins, as revealed by yeast two-hybrid analyses of VapB-VapC interactions. Finally, the VapCs, Rv0065 and Rv0617, which share ∼50% sequence similarity to the toxic VapCs, Rv0549c and Rv3320c, respectively, were shown to have sequence-selective, Mg2+-dependent RNase activity, further confirming an association between VapC toxicity, translational inhibition and RNA cleavage. We discuss the implications of these findings for the physiology of Mtb.
Results
VapBC modules selected for study
A subset of VapBC modules in Mtb was selected for study with the choice being guided, in part, by information on transcriptional responsiveness and/or essentiality of vapBC gene function available at the time (Table S1). The selected modules are also widely distributed among the main branches of the Mtb VapC phylogenetic tree (Fig. S2). Certain vapB-encoded antitoxins were reported to be induced during infection of human macrophages (Rv0550c, Rv2009, Rv2547 and Rv3321c) [34], [49]. The toxin Rv0627 and antitoxin Rv2830c were identified as essential for growth in vitro [50], with the latter induced by hypoxia [51]. The cluster of three vapBC modules located contiguously on the chromosome (Rv2545-Rv2546, Rv2547-Rv2548 and Rv2550c-Rv2549c) was selected based on the transcriptional responsiveness of most of the genes under in-vivo-relevant conditions [52], [53], [54], [55], [56], [57] and the requirement of Rv2548 for intracellular fitness [58]. The C-terminally truncated Rv1953 was included as a negative control as it lacks residues comprising part of the PIN domain and, as such, is predicted to lack nuclease activity. In addition to the ten Mtb modules, we also included the M. smegmatis vapBC, MSMEG_1283-MSMEG_1284, in our analysis. During the course this study, new information on these modules became available from other studies [31], [34], [40], [59].
Differential growth inhibitory effects of VapC toxins in M. smegmatis
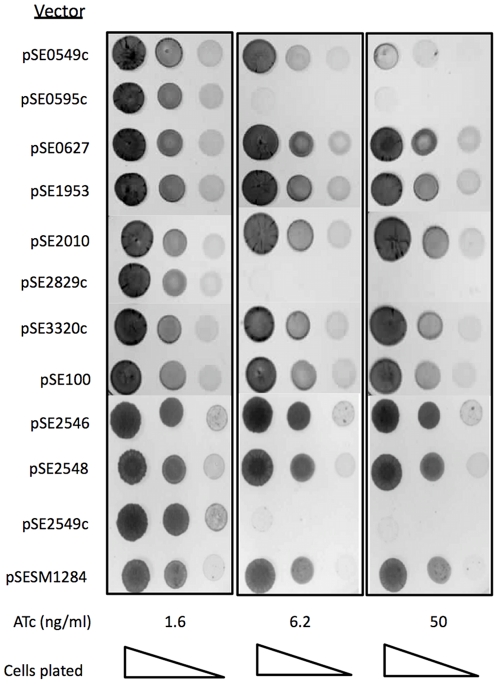
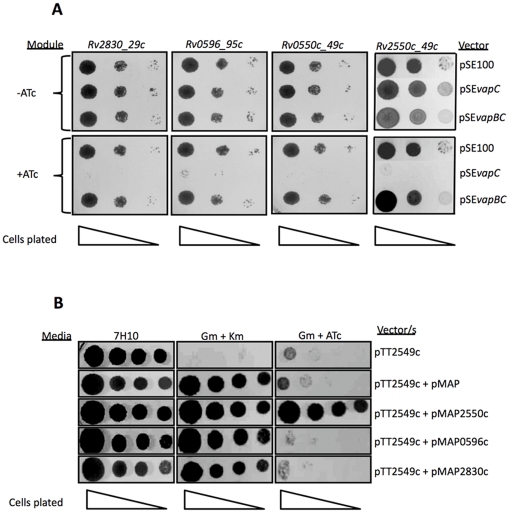
VapC toxicity was initially assessed in M. smegmatis by conditional expression of their encoding genes using an uncoupled system in which the toxin was expressed from a tetracycline (Tet)-regulated promoter (Pmyc1 tetO) contained on the episomal plasmid, pSE100 [60]. In this system, repression from the Pmyc1 tetO promoter in the absence of Tet inducer was mediated by the wild type Tet-repressor (TetR) constitutively expressed from the strong Psmyc promoter contained on an L5-based integration vector (pMC1s) [60]. The resulting vector pairs were co-electroporated into M. smegmatis, and in all cases, were found to be stable in the absence of anhydrotetracycline (ATc) inducer, suggesting that vapC expression was sufficiently repressed to avoid plasmid loss or mutation as a result of toxic gene expression. The toxicity of the VapC proteins was assessed by spotting dilutions of cultures on solid media containing ATc at a concentration of 0–50 ng/ml. Of the ten VapCs assessed, only four, namely, Rv0549c, Rv0595c, Rv2549c and Rv2829c, were growth inhibitory in M. smegmatis at ATc concentrations >3 ng/ml; the toxic effect of Rv0549c expression was, however, noticeably less than that of the other VapCs (Fig. 1). No toxicity was observed following induction of the others VapC, including MSMEG_1284, with ATc at concentrations up to 200 ng/ml.
Figure 1. Effect of mycobacterial VapC expression on growth of M. smegmatis on solid media.
Ten-fold serial dilutions of cells were spotted on 7H10 agar without or with ATc (1.6, 6.2 and 50 ng/ml) and incubated for 24–48 h.
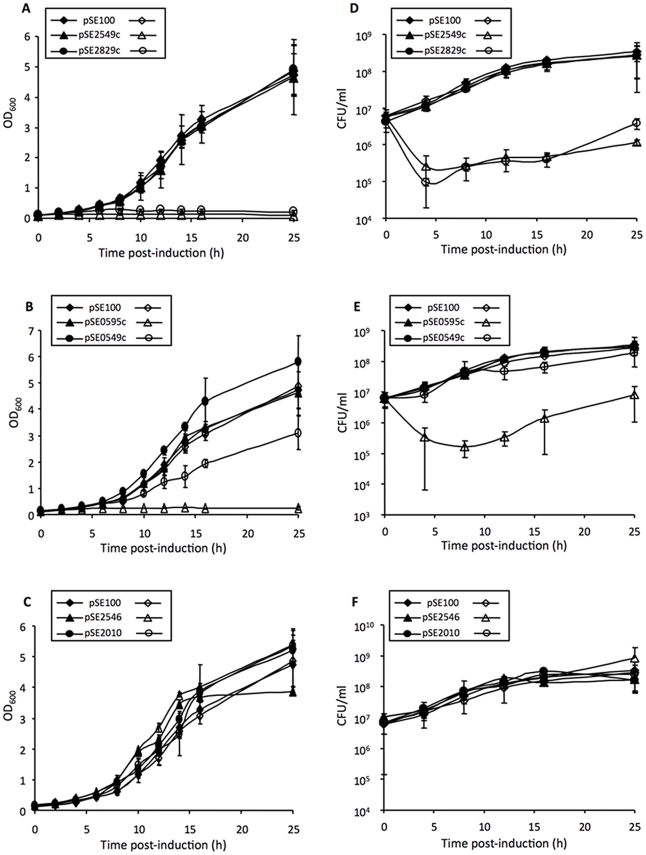
The effect of ectopic VapC expression on the viability of M. smegmatis was then assessed by inducing toxin expression during growth in liquid media (Fig. 2). For three VapCs that were inhibitory in the spotting assay (Rv0595c, Rv2549c and Rv2829c), vapC induction resulted in a ca. 2-log10 reduction in CFUs after 3 h of induction; thereafter, the CFU values stabilized before increasing again between 10 and 24 h post-induction (Fig. 2, panels D and E). Six randomly selected colonies from the Rv2829c-expressing strain that grew on plates after 16 h exposure to ATc in liquid culture were picked and phenotypically analyzed alongside colonies recovered from an ATc-free control. All colonies from the control were severely growth-impaired when plated on ATc confirming that inducible VapC expression was retained in these cells. In contrast, four clones derived from the ATc-induced sample were no longer resistant to hygromycin (Hyg) or had lost responsiveness to ATc. Similar results were obtained in the case of Rv0595c and Rv2549c: all colonies recovered after prolonged ATc exposure had undergone plasmid loss or rearrangement with concomitant loss of ATc responsiveness of growth (data not shown). Therefore, high-level expression of toxic VapCs exerts a selective pressure that promotes plasmid loss or instability in M. smegmatis.
Figure 2. Variable effects of Mtb VapC over-expression on growth and viability of M. smegmatis in liquid culture.
Growth and viability were assessed spectrophotometrically (A, B, C) and by CFU enumeration (D, E, F). Open symbols represent ATc-induced samples and filled symbols represent uninduced controls. The results represent the average and standard deviations from one of three independent experiments.
Since use of a strong promoter to drive expression of tetR may limit the ability to de-repress vapC expression by ATc treatment and thus mask the activity of VapCs for which no growth inhibition was observed, we replaced pMC1s with pMC2m, in which tetR is expressed from an intermediate strength promoter. However, in this configuration, no toxicity was observed for Rv0627, Rv1953, Rv2010, Rv2546, Rv2548, Rv3320c and MSMEG_1284, even at the highest concentration of ATc tested (200 ng/ml) (data not shown). The effect of fully de-repressing vapC expression was then assessed by measuring the transformation efficiencies in M. smegmatis of the expression vectors in the absence of TetR. The transformation efficiencies of pSE0549c, pSE0595c, pSE2549c, pSE2829c and pSE3320c were ≥3-log10 lower than the empty vector control, pSE100 (Table S2). Of these, four were previously scored as toxic when conditionally expressed (Rv0549c, Rv0595c, Rv2549c and Rv2829c), whereas toxicity of Rv3320c was only revealed when this VapC was constitutively expressed. In contrast, the transformation efficiencies of pSE0627, pSE1953, pSE2010, pSE2546, pSE2548 and pSESM1284 were comparable to that of pSE100 (0.08-1.3-fold), confirming that no significant toxicity was conferred by these VapCs, even when constitutively expressed (Table S2). We tested whether Rv2546, Rv2548 and MSMEG_1284 may have been rendered non-toxic through plasmid mutation/rearrangement by sequencing the promoter-operator region and insert of plasmid recovered from transformants obtained with and without the tetR-expressing vector. However, in all cases, the recovered plasmid was unaltered (data not shown).
Differential growth inhibitory effects of VapC toxins in wild type Mtb
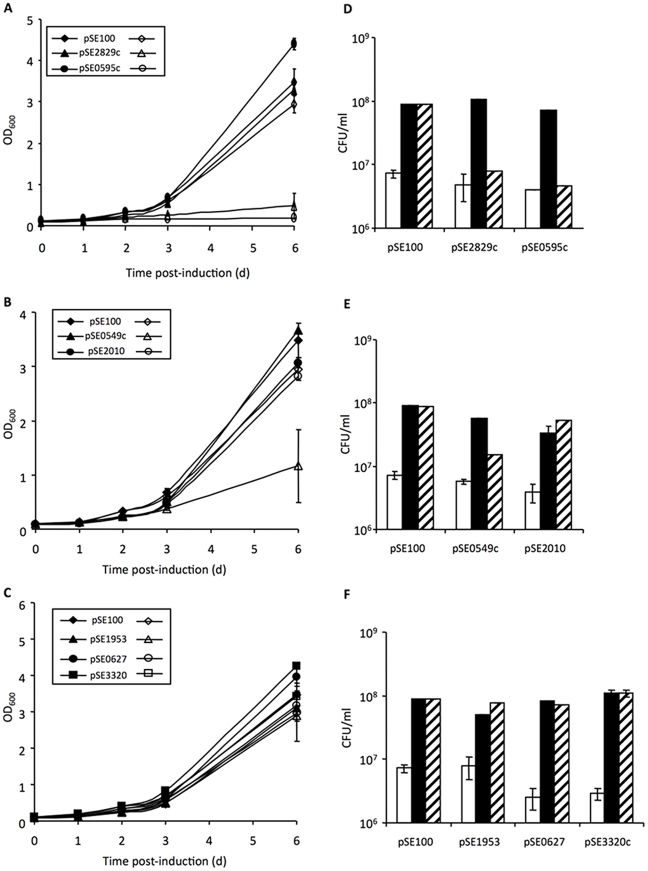
The effect of regulated expression of a subset of VapCs was analyzed in wild type Mtb (Fig. 3 and data not shown). Rv0627, Rv1953, Rv2010, Rv2546, Rv2548 and Rv3320c had no effect on the growth of H37Rv in liquid culture (Fig. 3C, Fig. 3F and data not shown) even though corresponding vapC transcript was detected (Fig. 4A and data not shown). However, as observed in M. smegmatis, only Rv0549c, Rv0595c, Rv2549c and Rv2829c were growth inhibitory when conditionally expressed in Mtb, with Rv0549c being the least toxic (Fig. 3, panels A, B, D and E). Moreover, while expression of the toxic VapCs in M. smegmatis resulted in a marked decline in CFUs after ATc induction (Fig. 2), no reduction in CFUs was observed over a 3–4-day induction period in Mtb (Fig. 3D, Fig. 3E and data not shown) with CFUs remaining unchanged over this period. The effect of constitutive VapC expression in Mtb was then assessed by measuring the transformation efficiencies of the VapC expression vectors in the absence of a tetR-expressing partner (Table S3). Toxicity of Rv0549c, Rv0595c, Rv2549c and Rv2829c was revealed by reduced transformation efficiency relative to the empty vector control. Moreover, as in M. smegmatis (Table S2), toxicity of Rv3320c in Mtb was only revealed when this VapC was constitutively expressed (Table S3).
Figure 3. Variable effects of Mtb VapC expression on growth and viability of wild type Mtb in liquid culture.
A, B, C: Growth was assessed spectrophotometrically for a period of 6 days after induction of gene expression, as described under Materials and Methods. Open symbols represent ATc-induced samples and filled symbols represent uninduced controls. The results show the data from one of three independent experiments. D, E, F: Viability was assessed by enumerating CFU. Open bars, CFU prior to induction of gene expression (day 0); black bars, untreated control (day 3); and striped bars, ATc-treated culture (day 3). The results show the average CFU values from duplicate platings at one serial dilution from one of three independent experiments.
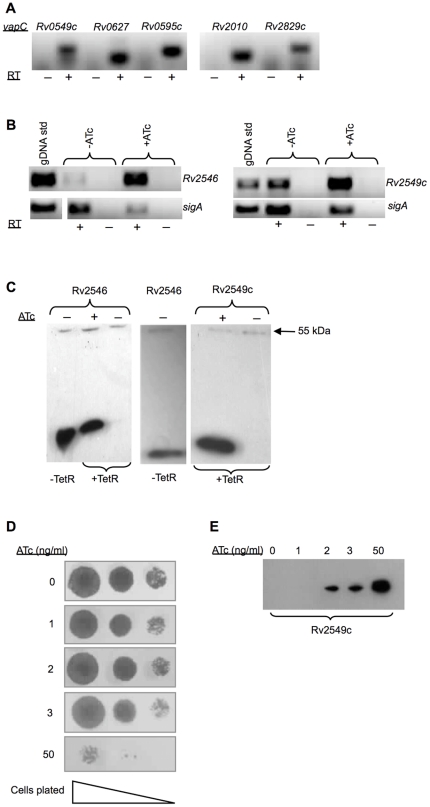
Figure 4. Analysis of ATc-regulated vapC expression in mycobacteria.
Expression was analyzed by RT-PCR (A and B) and detection of expressed proteins by epitope tagging (C). RT-PCR was carried out using mRNA samples obtained from (A) induced cultures of wild type Mtb (6 h treatment with ATc at 25 ng/ml); and (B) ATc-induced and uninduced cultures of M. smegmatis (1 h treatment with ATc at 50 ng/ml) over-expressing various vapC toxins under the control of the Pmyc1 tetO promoter. Samples obtained with (+) and without (-) reverse transcription (RT) were compared to distinguish cDNA from genomic DNA contamination. (C) Cellular fractions isolated from M. smegmatis were subjected to Western blot analysis using the anti-FLAG M2 antibody to detect the epitope-tagged VapC Rv2546 and Rv2549c proteins. Cultures were grown in either the presence (+) or absence (-) of 50 ng/ml ATc for 3 h. (D) The effect of epitope tagged Rv2549c expression on the growth of M. smegmatis on solid media. Ten-fold serial dilutions of cells were spotted on 7H10 agar without or with the ATc (1, 2, 3 and 50 ng/ml) and incubated for 24–48 h. (E) Comparison of the relative abundance of Rv2549c induced with varying concentrations of ATc. Cultures were grown in either the presence (+) or absence (−) of 50 ng/ml ATc for 3 h. Equal amounts (3 µg) were subjected to Western blot analysis using the anti-FLAG M2 antibody to detect the epitope-tagged Rv2549c protein.
Detection of VapC toxicity is influenced by the level of expressed protein
To investigate the reasons underlying the apparent lack of toxicity of some VapCs, transcript levels of the genes encoding the toxic Rv2549c and non-toxic Rv2546 VapCs were compared by RT-PCR analysis of mRNA from M. smegmatis cells cultured with or without ATc (Fig. 4B). Expression of both genes was significantly induced by ATc. Similar results were obtained for the other vapCs investigated in this study (data not shown) suggesting that the lack of toxicity was not due to a defect in gene expression from the Tet-regulated promoter. To compare the levels of VapC proteins induced in M. smegmatis, we tagged Rv2549c and Rv2546 at their C-termini with the 3×FLAG epitope. Epitope-tagged Rv2549c displayed toxicity equivalent to its native counterpart, confirming that the 3×FLAG tag did not affect the biological activity of this VapC (data not shown). Immuno-reactive bands of the size predicted for epitope-tagged Rv2546 and Rv2549c proteins (∼17 kDa) were observed in Western blots of cell-free extracts from M. smegmatis strains carrying the tagged VapC expression vectors that were cultured in the presence ATc (Fig. 4C, “+ATc”). A 55-kDa immuno-cross-reactive band detected in the cell-free extract of M. smegmatis provided an internal control that allowed the levels of FLAG-tagged VapCs to be compared between strains. The level of Rv2546 protein was noticeably higher in cells lacking TetR than in ATc-induced cells carrying TetR (Fig. 4C; left-hand panel, “-TetR” vs. “+TetR” lanes). However, the level of Rv2546 under fully de-repressed conditions was still lower than that of Rv2549c in TetR-containing cells induced with ATc (Fig. 4C, middle and left-hand panels). These results suggest that the level of Rv2546 might have been insufficient to cause a growth inhibitory effect in M. smegmatis.
To further investigate the effects of VapC levels on cell viability, we analysed the effect of epitope-tagged Rv2549c over-expression on the growth of M. smegmatis at various concentrations of ATc, and compared this with the level of protein expressed in cells cultured under the same conditions. As shown previously (Fig. 1), the toxic effect of Rv2549c expression only became evident at ATc concentrations >3ng/ml, whereas lower concentrations of inducer had little or no impact on the growth or viability of M. smegmatis (Fig. 4D). Western blot analysis of cell-free extracts of M. smegmatis cultures exposed to the same concentrations of inducer revealed an ATc dose-dependent increase in the level of epitope-tagged Rv2549c (Fig. 4E). Therefore, although Rv2549c is expressed at the lower concentrations of inducer (≤3 ng/ml), these results suggest that a specific threshold must be breached in order for the toxic activity of this VapC to become apparent in M. smegmatis.
Abrogation of VapC toxicity by cognate antitoxin expression
To determine whether the activity of the toxic VapC proteins could be neutralized by expression of their cognate antitoxins, we compared the growth of M. smegmatis strains in which the vapC gene was expressed either individually from the Pmyc1 tetO promoter, or together with its cognate vapB gene in its native configuration as a vapBC operon. As observed previously (Fig. 1), ATc-induced expression of the VapCs encoded by Rv0549c, Rv0595c, Rv2549c and Rv2829c resulted in growth inhibition in the absence of its cognate antitoxin (Fig. 5A). However, in all cases, co-expression of the toxin together with its cognate antitoxin rescued the growth of the strains in the presence of ATc (Fig. 5A). The finding that VapC toxicity is abrogated by cognate VapB antitoxin co-expression supports the notion that these proteins form functional TA pairs in mycobacteria, and that the growth inhibition observed in the absence of antitoxin was not due to generalized toxicity due to protein over-expression.
Figure 5. Neutralization of Mtb VapC toxicity in M. smegmatis by cognate antitoxin expression.
A: Cognate toxin-antitoxin modules were co-expressed as an operon under control of the Tet-regulated promoter. Serial dilutions were plated on 7H10 agar alone (-ATc) or with ATc at 25 ng/ml (+ATc). B: Toxin and antitoxin genes were expressed separately on Tweety (vapC) or L5-based integration vectors (vapB) under control of Tet- or acetamide-regulated promoters, respectively. Serial dilutions were plated on 7H10 agar alone or supplemented with Gm and Km, or Gm and ATc (50 ng/ml).
We then investigated whether the ability to neutralize VapC activity is restricted to cognate antitoxins using an uncoupled system in which the genes encoding toxic VapCs were expressed from the ATc-inducible promoter on a Tweety-based integration vector [61] carrying tetR, and the antitoxins constitutively expressed from the acetamidase promoter [62] on a L5-based integration vector. In this configuration, growth inhibition of M. smegmatis following ATc-induced over-expression of the Rv2549c VapC protein was retained (Fig. 5B). The toxicity was, however, neutralized by co-expression of its cognate VapB, Rv2550c, when its encoding gene was integrated at a chromosomal locus distal from that of the toxin (Fig. 5B). In contrast, expression of the non-cognate antitoxins, Rv0596c and Rv2830c, which had previously been shown to abolish toxicity of their cognate toxins (Fig. 5A), had no effect on the growth inhibition caused by induction of Rv2549c (Fig. 5B). Similarly, expression of its cognate VapB, Rv0596c, alleviated the toxicity of Rv0595c in the presence of ATc whereas expression of Rv2550c or Rv2830c had no effect (data not shown). Together, these data confirm that the activity of VapC toxins can only be neutralized by their cognate antitoxins.
Specificity of VapC interaction with cognate VapB confirmed by yeast-two hybrid (Y2H) analysis
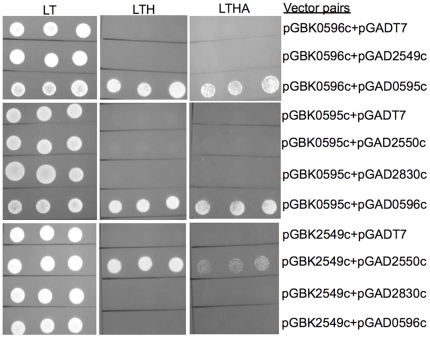
The interaction between cognate vs. non-cognate VapC and VapB proteins was further analyzed by Y2H analysis (Fig. 6). In this assay system, the VapC, Rv0595c, was found to interact specifically with its cognate VapB, Rv0596c as evidenced by growth of the corresponding yeast strains on high-stringency media. In contrast, no interaction was observed between Rv0595c and the non-cognate VapBs, Rv2550c and Rv2830c, or between Rv0596c and the non-cognate VapC, Rv2549c. Similar results were obtained for Rv2549c, which interacted specifically with Rv2550c, but showed no detectable interaction with the non-cognate VapBs, Rv2830c and Rv0596c. These results independently confirm the specificity of cognate VapB-VapC interaction deduced from the toxicity neutralization data and suggest that spurious interactions between non-cognate VapB-VapC pairs may not be functionally relevant in the native host.
Figure 6. Y2H analysis of VapC-VapB interactions.
Interactions between VapCs and either cognate or non-cognate VapBs were tested by co-transformation and scoring for growth on selection (LT) vs. media of differing stringency. Three independent colonies of each strain were re-suspended in sterile water, the cell density adjusted to an OD600 of 1 and aliquots spotted on plates. LT, Leu-Trp; LTH, Leu-Trp-His; LTHA, Leu-Trp-His-Ade dropout-supplemented media.
Effect of vapBC loss on susceptibility to cognate vapC-mediated toxicity
To investigate the role of the vapBC TA modules in the physiology of mycobacteria, unmarked Mtb deletion mutants lacking the vapC, Rv0595c, the vapBC, Rv2009-Rv2010, or the three contiguous vapBC modules, Rv2545-Rv2546, Rv2547-Rv2548 and Rv2550c-Rv2549c, were generated by allelic exchange mutagenesis. In addition, a M. smegmatis mutant lacking the MSMEG_1283-MSMEG_1284 module was constructed. None of the strains displayed a growth defect relative to the wild type when cultured under standard conditions in either liquid medium or on agar plates (data not shown). The susceptibility of the M. smegmatis vapBC mutant to cell wall, oxidative, nitrosative, genotoxic and heat stress was also assessed and found to be indistinguishable from wild type under all conditions tested (data not shown).
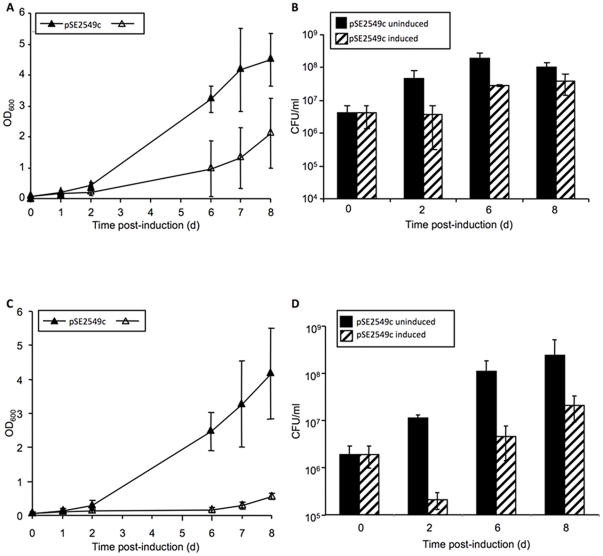
To assess the effect of loss of vapB function on the susceptibility of Mtb to growth inhibition by cognate or non-cognate vapC expression, we compared the transformation efficiencies of the vapC expression vectors in the ΔRv2545-Rv2550c mutant and wild type strains (Table S3). Neither Rv2546 nor Rv2548 was toxic when expressed in the mutant strain, which lacks their cognate vapB genes. Similarly, no toxicity was observed when MSMEG_1284 was conditionally or constitutively expressed in wild type M. smegmatis or the deletion mutant lacking the MSMEG_1283-MSMEG_1284 module. In contrast, an exacerbation of Rv2549c toxicity in the absence of its cognate antitoxin was revealed by comparing its effects on growth and viability in Mtb when conditionally expressed in liquid cultures of the wild type vs. ΔRv2545-Rv2550c strains (Fig. 7). Expression of Rv2549c was growth inhibitory in wild type Mtb (Figs. 7A and B); however, a 10-fold reduction in CFUs was observed within two days of induction of Rv2549c expression in the mutant strain (Figs. 7C and D). As in M. smegmatis, the outgrowth observed subsequent to the early killing of the ΔRv2545-Rv2550c mutant upon induction of Rv2549c expression was due to abrogation of toxicity by plasmid rearrangement or loss (data not shown).
Figure 7. Rv2549c displays increased toxicity in the ΔRv2545-Rv2550c mutant strain of Mtb.
Growth and viability were assessed spectrophotometrically (A, C) and by CFU enumeration (B, D). Open symbols represent ATc-induced samples and filled symbols represent uninduced controls. The results represent the average and standard deviations from three independent experiments.
The VapCs Rv0065 and Rv0617 display sequence-selective RNase activity
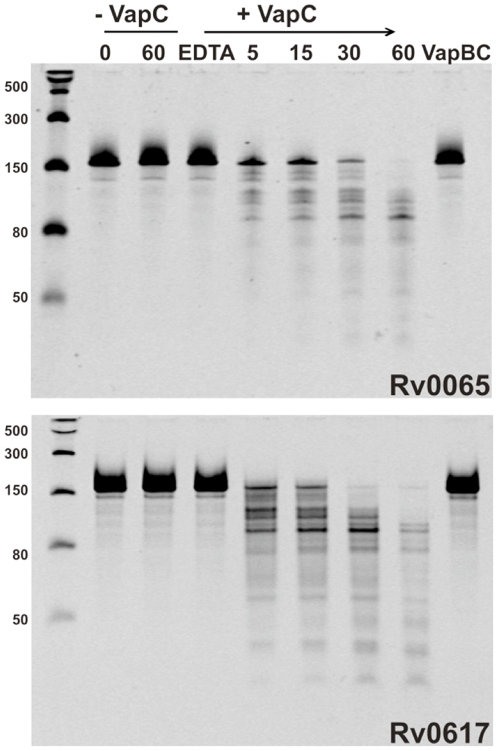
To investigate the mechanism underlying VapC toxicity in mycobacterial hosts, we sought to biochemically characterize five VapCs analyzed in this study (Rv0549c, Rv0595c, Rv2546, Rv2549c and Rv2829c). In general, mycobacterial VapC proteins cannot be expressed in E. coli due to a combination of their insolubility and toxicity (J.L.M & V.A., unpublished observations). Thus, the VapCs were expressed in M. smegmatis [59], [63] as tagged fusion protein complexes with their cognate VapBs. The Rv0596c-Rv0595c and Rv2830c-Rv2829c complexes did not express in M. smegmatis, and the other three complexes did express but the proteins were insoluble after cell lysis (Rv0550c-Rv0549c, Rv2545-Rv2546 and Rv2550c-Rv2549c). In contrast, the related VapBC complexes Rv0065A-Rv0065 and Rv0616A-Rv0617 expressed in a soluble form and could be purified. Since Rv0065 is closely related to Rv0549c, sharing 36% amino acid identity and 50% amino acid similarity with Rv0549c (Fig. S2) and Rv0617 shares 38% sequence identity and 55% sequence similarity with Rv3320c, we reasoned that these VapCs could serve as useful models for analyzing the biochemical function of a toxic mycobacterial VapCs. Limited proteolysis of the VapBC complex resulted in selective degradation of the unstable VapB allowing purification of Rv0065 and Rv0617 alone. As shown in Fig. 8, these purified proteins show Mg2+-dependent sequence-selective RNase activity on a single-stranded RNA substrate. This RNase activity is completely inhibited when the VapC proteins are in complex with their cognate VapBs.
Figure 8. The VapCs Rv0065 and Rv0617 have RNase activity.
VapC proteins Rv0065 and Rv0617 display sequence-selective RNase activity against an RNA substrate of ∼150 bases in the presence of 6 mM MgCl2. Activity is Mg2+-dependent as addition of 12 mM EDTA abolishes activity (lane 4). Ribonuclease activity is also inhibited in the presence of VapB (lane 9). Addition of VapC (+VapC) results in degradation of the RNA substrate over a period of 5 – 60 minutes (lanes 5–8). RNA only controls (-VapC) show no contaminating ribonuclease activity (lanes 2 & 3). The molecular weight marker shows molecular masses of single-stranded RNA in number of bases (lane 1).
Discussion
In this study, the function of a representative subset of mycobacterial VapBC modules was investigated in M. smegmatis and Mtb. VapC toxicity was initially assessed using a modular system in which vapC expression was either constitutive (in the absence of TetR) or conditionally regulated by TetR as a function of inducer concentration or the level of tetR expression. Of the VapCs tested, five were growth inhibitory in M. smegmatis, with Rv0549c being noticeably less toxic than Rv0595c, Rv2549c and Rv2829c when conditionally expressed, and Rv3320c only displaying toxicity when constitutively expressed. The five VapCs that were growth inhibitory in M. smegmatis demonstrated the same differential toxicity in wild type Mtb, with Rv0549c displaying only modest growth inhibition when expressed conditionally or constitutively, and Rv3320c being most toxic, but only when expressed constitutively.
Conditional expression of the three Mtb VapCs that were most toxic in M. smegmatis (Rv0595c, Rv2549c and Rv2829c) resulted in a rapid initial decline in bacterial CFUs, suggestive of VapC-mediated cell death or protracted bacteriostasis. Selection against VapC toxicity in M. smegmatis was revealed by loss or rearrangement of the expression vector to eliminate vapC expression in cells that survived toxic VapC exposure. Although the same three VapCs were growth inhibitory in Mtb, their effects were bacteriostatic in this host. However, a bactericidal effect in Mtb was observed by conditional expression of Rv2549c in a mutant strain that lacks the cognate Rv2550c antitoxin, confirming that VapC toxicity in Mtb can be tempered by expression if its cognate antitoxin from the corresponding vapBC module on the chromosome. In line with this observation, co-expression of the cognate vapB on an operon with vapC abrogated toxicity of all VapCs that were growth inhibitory in M. smegmatis. The specificity of toxin neutralization by the cognate VapB was demonstrated using an uncoupled system in which vapC and vapB genes were conditionally expressed under control of different regulatable promoters and from distinct chromosomal loci. In this assay, VapC-induced growth inhibition was specifically neutralized in M. smegmatis expressing the cognate vapB; in contrast, expression of non-cognate vapBs was ineffective. Together with the enhanced toxicity of Rv2549c in a mutant of Mtb that lacks the cognate antitoxin (Rv2550c), and the specificity of interaction between cognate VapB-VapC pairs revealed by Y2H analysis, our results argue against (functionally relevant) interactions between the toxin and antitoxin components of different VapBC modules in Mtb, consistent with findings from another study [34].
The lack of toxicity of Rv1953 under all conditions tested was consistent with the predicted abrogation of nuclease activity in this C-terminally truncated protein. In contrast, the reason for the lack of toxicity of Rv0627, Rv2010, Rv2546 and Rv2548 following expression in both Mtb and M. smegmatis was less clear. During the course of our study, the effects of toxin expression on the growth of E. coli, M. smegmatis and/or Mtb hosts were reported by other groups [38], [40], [43], [44]. Although some concordance exists, there are many differences between studies, even when the same host organism was employed for assessing toxicity. For example, Rv0627 was non-toxic in M. smegmatis and Mtb in our study, and in M. smegmatis in that of Ramage et al. [34], but this VapC was reported by Miallau et al. [31] as toxic in Mtb and M. smegmatis. Similarly, we found Rv0595c, Rv2549c and Rv3320c to be toxic in M. smegmatis, but Ramage et al. [34] did not, whereas the converse was true for Rv2010 and Rv2548 which were scored as toxic in that study, but not in the present one. Moreover, while Robson et al. reported growth inhibition of M. smegmatis by conditional expression of MSMEG_1284 [59], no such effect was observed in either wild type M. smegmatis or a deletion mutant lacking the MSMEG_1283-MSMEG_1284 module in this study. These discrepancies may be due to differences in expression vectors, translation initiation signals, growth conditions and/or host strains used for VapC over-expression, which could result in assay-dependent biases. One or more of these factors may preclude the detection of toxin-induced growth inhibition if insufficient levels of the protein are produced following over-expression, leading to the incorrect classification of a subset of VapC as ‘non-toxic’. Our observations suggest that the reduced level of Rv2546 relative to Rv2549c might explain why this was non-toxic in our assay system, a notion substantiated by the finding that the toxicity of Rv2549c was only evident once the level of induced protein exceeded a certain threshold. Such factors clearly complicate distinguishing ‘functional’ TA modules from others, and suggest that the list of 30 modules defined as ‘toxic’ is likely an under-representation, biased by assay-dependent effects, as postulated [34].
The five VapCs identified as toxic in this study proved to be refractory to expression in a soluble form when complexed with their cognate VapCs, and as such, could not be isolated in a form suitable for biochemical analysis. Nonetheless, we were able to isolate and purify the Rv0549c homolog, Rv0065 and the Rv3320c homolog, Rv0617, respectively, and demonstrate that they have sequence-selective RNase activity. This finding strongly implies an association between VapC toxicity and translational inhibition as a result of RNA cleavage in mycobacteria, and moreover, suggests that the vapBC family provides a potentially abundant source of RNase activity in Mtb that might vary as a function of regulated expression of individual modules, and/or the rates of antitoxin degradation. The effects of extensive and variable nuclease activity on the biology of Mtb are likely to be profound and underscore the importance of identifying the cellular target(s) of VapCs in Mtb. Such studies are likely to provide valuable insights into the physiological role(s) that this family of proteins plays in the biology of this major human pathogen.
Materials and Methods
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are described in Table S4. Unless otherwise indicated, M. smegmatis strains were grown in Difco Middlebrook 7H9 media (BD) supplemented with 0.085 % NaCl, 0.2 % glucose, 0.2 % glycerol and 0.05 % Tween 80 or on solid Difco Middlebrook 7H10 media (BD) supplemented with 0.085 % NaCl, 0.2 % glucose and 0.5 % glycerol. Mtb strains were grown in Middlebrook 7H9 media supplemented with 0.2 % glycerol, Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment (Merck) and 0.05 % Tween 80. Hygromycin (Hyg), kanamycin (Km) and gentamycin (Gm) were used in mycobacterial cultures at final concentrations of 50, 25 and 5 µg/ml, respectively. ATc (Sigma) inducer was used at concentrations up to 200 ng/ml.
Construction of deletion mutant strains
A 6097 bp EcoRI/Asp718 fragment carrying Rv2009-Rv2010 was excised from BacD8 [64] and inserted in pGEM3Zf(+). An upstream homologous fragment was obtained by subcloning a 2455 bp PstI fragment in pGEM3Z(+)f. A 3143 bp downstream homologous fragment was excised from the BacD8 subclone by digestion with BsDRI and PstI and cloned in the XbaI site of the vector carrying the upstream fragment. The resulting deletion allele was excised as a 5654 bp HindIII/Asp718 fragment and inserted in p2NIL before introducing the lacZ-sacB-hyg cassette from pGOAL19 to produce p2Δ2009_10KO. This vector was used to construct a mutant of Mtb in which the last 78 amino acids of the antitoxin (Rv2009, containing the DNA binding motif) and all except the C-terminal 33 amino acids of the toxin (Rv2010) were removed. Suicide plasmids for generating other deletion mutants were constructed by PCR amplification from genomic DNA of upstream and downstream homologous sequences including the 5′ and 3′ termini of the gene of interest using primer pairs described in Table S5. Amplicons were cloned in pGEM3Z(+)f and sequenced before sub-cloning the corresponding upstream and downstream fragments in p2NIL to create deletion alleles. The lacZ-sacB-hyg cassette from pGOAL19 was inserted in the p2NIL subclones to create p2Δ0595cKO, and p2Δ2545_50cKO as suicide substrates for introducing unmarked deletions in Mtb Rv0595c and Rv2545-Rv2550c, and p2ΔSM1283_84KO as a substrate for generating an unmarked deletion in MSMEG_1283-MSMEG_1284 (Table S4). Suicide vectors were electroporated into Mtb H37Rv or M. smegmatis mc2155 and allelic exchange mutants recovered by two-step selection, as described [65]. Mutant genotypes were confirmed by Southern blot analysis.
Construction of vectors for conditional expression of toxins and antitoxins in mycobacteria
VapC toxin-encoding sequences were amplified by PCR from Mtb H37Rv chromosomal DNA using the primers listed in Table S6. In all cases, ribosome binding sites were changed to a standardized consensus sequence (GGAAG/A) in order to optimize the yield of expressed protein [66]. For cloning downstream of the Pmyc1 tetO promoter-operator element and ATc-dependent regulation of expression [60], Rv0549c, Rv0595c, Rv2829c, Rv1953 were expressed as BamHI/PstI fragments, Rv2546, Rv2548, Rv2549c were expressed as BamHI/HindIII fragments, and Rv0627 was expressed as a PstI-HindIII fragment and cloned in pSE100. The MSMEG_1284 ORF was cloned in the PvuII site of pSE100. Derivatives of Rv2546 and Rv2549c containing a C-terminal 3×FLAG epitope tag were expressed using the same forward primer as for the native version, and a modified reverse primer containing the 3× FLAG peptide-encoding sequence (Table S6). To ensure efficient translation of the expressed VapC proteins following ATc-induction, a consensus RBS (GGAAG/A) [66] was included upstream of the start codon of the vapC genes in those instances where an endogenous ribosome-binding site (RBS) was not clearly discernible. Other than this modification, the promoter architecture and start codons of the native vapC genes were preserved.
Vectors for expression of the vapBC modules Rv0550c-Rv2549c, Rv0596c-Rv0595c, Rv2550c-Rv2549c and Rv2830c-Rv2829c were generated using a vapB-specific forward primer and the reverse primer used for expression of the corresponding vapC (Table S6). All expression constructs were confirmed by DNA sequencing, and electroporated into M. smegmatis or Mtb alone or with an integrative tetR-containing plasmid in which tetR is expressed under control a strong (pMC1s) or intermediate strength promoter (pMC2m) [60]. Transformants were selected on 7H10 agar supplemented with Hyg or Hyg and Km respectively.
Vectors for uncoupled, regulated expression of the Rv2549c and either cognate (Rv2550c), or non-cognate VapB antitoxins (Rv2830c, Rv0596c) were constructed as follows. The L5-based integrative vector, pMC1s [60], was digested with NotI and the vector backbone re-ligated to generate pMC1r. The acetamide-inducible acetamidase promoter from M. smegmatis [62] was cloned in pMC1r to produce pMAP. The antitoxin-encoding genes, Rv2550c, Rv0595c and Rv2830c, were PCR-amplified (Table S6) and cloned in pMAP to produce pMAP2550c, pMAP0595c and pMAP2830c, respectively. The Tweety-based integration vector [61], pTTP1BG, was prepared by digesting pTTP1B [61] with HindIII, re-ligation of the vector and insertion of a GmR cassette [67] in the PstI site. A NotI fragment from pMC1s carrying the Psmyc-tetR element was cloned in the SmaI site of pTTP1BG followed by cloning of the 883-bp SpeI/ClaI fragment from pSE2549c which carries the Pmyc1 tetO::Rv2549c element in the EcoRI site of the resulting vector to produce pTT2549c. This vector was electroporated into M. smegmatis alone or together with each of the antitoxin-expressing pMAP subclones.
Effect of conditional overexpression of VapCs on mycobacterial growth and viability
VapC toxicity was assessed both on solid medium and in liquid culture in M. smegmatis and in liquid culture only in Mtb. For the M. smegmatis plating assay, transformants were grown in Middlebrook 7H9 media containing Hyg and Km to an OD600 of approximately 1.0. The cultures were diluted in a 10-fold series and spotted on 7H10 plates with or without varying concentrations of ATc. The plates were incubated at 37°C and the growth checked after 24 and 48 h. For toxicity assessment in liquid culture, M. smegmatis transformants were grown in Middlebrook 7H9 media containing Km and Hyg to an OD600 of 0.1 - 0.4 and diluted into fresh warm media to an OD600 of 0.1. The cultures were split and either treated with ATc (25 ng/ml; induced) or left untreated (uninduced). The OD600 of each culture was measured every 2 h and viability scored by plating, in duplicate, serial dilutions of samples taken every 4 h over a period of 25 h.
Mtb transformants were grown in Middlebrook 7H9 media containing Hyg and Km to an OD600 of 0.1. The cultures were diluted to an OD600 of 0.04 and left to grow overnight before being split into two equal aliquots which were treated with ATc (25 or 50 ng/ml – induced) or left untreated (uninduced). Growth and viability were assessed by periodically monitoring OD600 and CFUs over 6 d.
Stress sensitivity testing
The susceptibility of the M. smegmatis vapBC mutant to various stresses was tested using previously described methods [52], [68], [69], [70].
Analysis of gene expression by RT-PCR
RNA was extracted from early log-phase cultures, as described [71]. Samples were treated twice with Turbo DNase (Ambion) and RT-PCR performed with the Phusion RT-PCR kit (Finnzymes) according to the manufacturer's instructions. Primers for RT-PCR analysis of expression of Mtb vapCs were designed using Primer 3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and are described in Table S6. Expression of M. smegmatis sigA was detected as described previously [72].
Detection of FLAG-tagged proteins by Western blot analysis
M. smegmatis cells containing 3×FLAG-tagged VapC fusion proteins were grown in 90 ml cultures to mid log-phase (OD600∼0.4–0.5) and split equally. One 45-ml aliquot was treated with ATc (50 ng/ml) and the other served as the uninduced control. After 3 h induction, 20 ml of cells were harvested and resuspended in 250 µl of Bacterial Protein Extraction Reagent (B-PER II Reagent, Thermo Scientific) containing complete mini protease inhibitor cocktail (Roche). The cells were lysed three times for 20 s at speed 6 using the Savant Fastprep FP120 (BIO101), with 5 min intervals between pulses when the cells were cooled on ice. The protein concentration was determined using a Bradford assay and equivalent amounts of soluble and insoluble fractions of each induced and uninduced sample were resolved on an SDS-PAGE gel and the proteins transferred to a PVDF membrane (Amersham). The membrane was incubated with the HRP-conjugated mouse Anti-FLAG M2 antibody (Sigma) and the FLAG-tagged proteins proteins were detected using the ProteoQwest™ chemiluminescent Western blotting kit (Sigma).
Y2H analysis
Y2H analysis was performed using the Clontech Matchmaker Y2H system and vectors carrying the VapCs, Rv0595c and Rv2549c, or VapBs Rv0596c, Rv2550c, and Rv2830c, cloned as GAL-4 Activation Domain (AD) and/or Binding Domain (BD) fusions (Table S4). Interactions between selected VapC toxins and cognate vs. non-cognate VapB antitoxins were assessed according to the manufacturer's instructions.
Expression, purification and biochemical analysis of recombinant Rv0065 and Rv00617
The ORFs encoding the vapBC operons Rv0065A-Rv0065 and Rv0616A-Rv0617 were amplified from Mtb H37Ra genomic DNA. Importantly, there are no differences in sequence between H37Rv and H37Ra in these regions. Moreover, the antitoxins for Rv0065 and Rv0617, which are designated herein as Rv0065A and Rv0617A, are not annotated in the H37Rv genome. However, these antitoxins are annotated in the H37Ra, BCG and CDC5115 genomes: Rv0065A corresponds to BCG0095A and Rv0617A corresponds to MT0645.2. The amplicons were digested with NcoI/HindIII restriction enzymes, purified and inserted into the pYUB28b shuttle vector [63] enabling expression of a C-terminal His-tag on VapC. The pYUBRv0065A-5 and pYUBRv0616A-6 constructs were then transformed into M. smegmatis mc24517 cells. Rv0065A-Rv0065 and Rv0616A-Rv0617 VapBC complexes were expressed and purified as previously described for the VapBC complex from M. smegmatis [59]. VapBC complexes were digested with trypsin to remove VapB. Digestion reactions were stopped by addition of trypsin inhibitor from soybean and VapC was subsequently purified using anion exchange chromatography. RNA was transcribed from a ‘pentaprobe’ PCR product [73] using the T7 MEGAscript® kit (Ambion, USA) according to manufacturer's instructions. RNase activity assays for VapC proteins contained 1 µg purified VapC protein, 1 µg purified RNA, 12 mM sodium phosphate buffer, 6 mM NaCl and 6 mM MgCl2. Negative controls included addition of 12 mM EDTA and substitution of VapBC for VapC. Time course assay reactions were stopped by the addition of 10 µl formamide loading dye and heated to 70°C before loading onto a 10% urea-denaturing PAGE gel.
Supporting Information
Multiple sequence alignment of mycobacterial VapCs. Sequences were aligned using the ClustalW2 multiple sequence alignment tool at the European Bioinformatics Institute website, http://www.ebi.ac.uk/Tools/clustalw2/index.html.
(PDF)
Phylogenetic tree of VapC proteins from mycobacteria. VapCs were aligned using ClustalW2 multiple sequence and server alignment server (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The tree was generated in Jalview 2.08.1 based on percentage identity between sequences.
(PDF)
Properties of mycobacterial vapBC toxin-antitoxin modules selected for study.
(PDF)
VapC toxicity in M. smegmatis mc2155 assessed by transformation efficiency of VapC expression vector.
(PDF)
VapC toxicity in Mtb assessed by transformation efficiency of VapC expression vector.
(PDF)
Strains and plasmids used in this study.
(PDF)
Oligonucleotides used to create knockout vectors for allelic exchange mutagenesis.
(PDF)
Oligonucleotides used for expression vector construction, yeast two-hybrid (Y2H) vector construction and RT-PCR.
(PDF)
Acknowledgments
We thank Dirk Schnappinger and Sabine Ehrt for providing pSE100, pMC1s and pMC2m, Stewart Cole for providing the cosmid library, Cristina Stallings for advice on immunoblotting, Digby Warner for critically reviewing the manuscript and Bavesh Kana for advice and assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by an International Scholars grant from the Howard Hughes Medical Institute to V.M. (grant no. 55005522; www.hhmi.org), and by grants from the National Research Foundation (www.nrf.ac.za) and the South African Medical Research Council (www.mrc.ac.za). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Young DB, Perkins MD, Duncan K, Barry CE, 3rd Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest. 2008;118:1255–1265. doi: 10.1172/JCI34614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat Rev Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- 4.Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, et al. Spectrum of latent tuberculosis - existing tests cannot resolve the underlying phenotypes: author's reply. Nat Rev Microbiol. 2010;8:242. doi: 10.1038/nrmicro2236-c1. [DOI] [PubMed] [Google Scholar]
- 5.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–570. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell Microbiol. 2009;11:1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryk R, Gold B, Venugopal A, Singh J, Samy R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, Manjunatha U, Boshoff HI, Ha YH, Niyomrattanakit P, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boshoff HI, Xu X, Tahlan K, Dowd CS, Pethe K, et al. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J Biol Chem. 2008;283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryk R, Arango N, Venugopal A, Warren JD, Park YH, et al. Triazaspirodimethoxybenzoyls as selective inhibitors of mycobacterial lipoamide dehydrogenase. Biochemistry. 2010;49:1616–1627. doi: 10.1021/bi9016186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korch SB, Hill TM. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol. 2006;188:3826–3836. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 15.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 17.Spoering AL, Vulic M, Lewis K. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol. 2006;188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, et al. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 20.Hansen S, Lewis K, Vulic M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 22.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 23.Wiuff C, Zappala RM, Regoes RR, Garner KN, Baquero F, et al. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob Agents Chemother. 2005;49:1483–1494. doi: 10.1128/AAC.49.4.1483-1494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 26.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodogai M, Ferenczi S, Bashtovyy D, Miclea P, Papp P, et al. The ntrPR operon of Sinorhizobium meliloti is organized and functions as a toxin-antitoxin module. Mol Plant Microbe Interact. 2006;19:811–822. doi: 10.1094/MPMI-19-0811. [DOI] [PubMed] [Google Scholar]
- 29.Hopper S, Wilbur JS, Vasquez BL, Larson J, Clary S, et al. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect Immun. 2000;68:896–905. doi: 10.1128/iai.68.2.896-905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoo SK, Loll B, Chan WT, Shoeman RL, Ngoo L, et al. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J Biol Chem. 2007;282:19606–19618. doi: 10.1074/jbc.M701703200. [DOI] [PubMed] [Google Scholar]
- 31.Miallau L, Faller M, Chiang J, Arbing M, Guo F, et al. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems: VapBC-5 from Mycobacterium tuberculosis. J Biol Chem. 2009;284:276–283. doi: 10.1074/jbc.M805061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattison K, Wilbur JS, So M, Brennan RG. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J Biol Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- 33.Li GY, Zhang Y, Inouye M, Ikura M. Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J Mol Biol. 2008;380:107–119. doi: 10.1016/j.jmb.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 34.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arcus VL, Rainey PB, Turner SJ. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 2005;13:360–365. doi: 10.1016/j.tim.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Arcus VL, McKenzie JL, Robson J, Cook GM. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng Des Sel. 2011;24:33–40. doi: 10.1093/protein/gzq081. [DOI] [PubMed] [Google Scholar]
- 37.Sevin EW, Barloy-Hubler F. RASTA-Bacteria: a web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korch SB, Contreras H, Clark-Curtiss JE. J Bacteriol; 2008. Three Mycobacterium tuberculosis Rel toxin:antitoxin modules inhibit mycobacterial growth and are expressed in human-infected macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll P, Brown AC, Hartridge AR, Parish T. Expression of Mycobacterium tuberculosis Rv1991c using an arabinose-inducible promoter demonstrates its role as a toxin. FEMS Microbiol Lett. 2007;274:73–82. doi: 10.1111/j.1574-6968.2007.00842.x. [DOI] [PubMed] [Google Scholar]
- 40.Gupta A. Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2009;290:45–53. doi: 10.1111/j.1574-6968.2008.01400.x. [DOI] [PubMed] [Google Scholar]
- 41.Kumar P, Issac B, Dodson EJ, Turkenburg JP, Mande SC. Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein. J Mol Biol. 2008;383:482–493. doi: 10.1016/j.jmb.2008.08.067. [DOI] [PubMed] [Google Scholar]
- 42.Zhao L, Zhang J. Biochemical characterization of a chromosomal toxin-antitoxin system in Mycobacterium tuberculosis. FEBS Lett. 2008;582:710–714. doi: 10.1016/j.febslet.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 43.Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, et al. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem. 2006;281:18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- 44.Singh R, Barry CE, 3rd, Boshoff HI. The Three RelE Homologs of Mycobacterium tuberculosis Have Individual, Drug-Specific Effects on Bacterial Antibiotic Tolerance. J Bacteriol. 2010;192:1279–1291. doi: 10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. Journal of Biological Chemistry; 2010. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem. 2004;279:16471–16478. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 48.Bunker RD, McKenzie JL, Baker EN, Arcus VL. Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon. Proteins. 2008;72:510–518. doi: 10.1002/prot.22048. [DOI] [PubMed] [Google Scholar]
- 49.Dubnau E, Fontan P, Manganelli R, Soares-Appel S, Smith I. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect Immun. 2002;70:2787–2795. doi: 10.1128/IAI.70.6.2787-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 51.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manganelli R, Voskuil MI Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 53.Fisher MA, Plikaytis BB, Shinnick TM. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb) 2004;84:239–246. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Talaat AM, Lyons R, Howard ST, Johnston SA. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez GM, Voskuil MI, Gold B Schoolnik GK, Smith I. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart GR, Patel J, Robertson BD, Rae A, Young DB. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1:269–278. doi: 10.1371/journal.ppat.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J Mol Biol. 2009;390:353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology. 2007;153:2711–2723. doi: 10.1099/mic.0.2007/008904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143 ( Pt. 1997;7):2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 63.Goldstone RM, Moreland NJ, Bashiri G, Baker EN, Shaun Lott J. A new Gateway vector and expression protocol for fast and efficient recombinant protein expression in Mycobacterium smegmatis. Protein Expr Purif. 2008;57:81–87. doi: 10.1016/j.pep.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 64.Brosch R, Gordon SV, Billault A, Garnier T, Eiglmeier K, et al. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146. 2000;1969-1975 doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 66.Ma J, Campbell A, Karlin S. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol. 2002;184:5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Labes M, Puhler A, Simon R. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 68.Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, et al. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol. 2009;191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Firmani MA, Riley LW. Reactive nitrogen intermediates have a bacteriostatic effect on Mycobacterium tuberculosis in vitro. J Clin Microbiol. 2002;40:3162–3166. doi: 10.1128/JCM.40.9.3162-3166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, et al. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 71.Downing KJ, Betts JC, Young DI, McAdam RA, Kelly F, et al. Global expression profiling of strains harbouring null mutations reveals that the five rpf-like genes of Mycobacterium tuberculosis show functional redundancy. Tuberculosis (Edinb) 2004;84:167–179. doi: 10.1016/j.tube.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 72.Mowa MB, Warner DF, Kaplan G, Kana BD, Mizrahi V. Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J Bacteriol. 2009;191:985–995. doi: 10.1128/JB.01409-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwan AH, Czolij R, Mackay JP, Crossley M. Pentaprobe: a comprehensive sequence for the one-step detection of DNA-binding activities. Nucleic Acids Res. 2003;31:e124. doi: 10.1093/nar/gng124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of mycobacterial VapCs. Sequences were aligned using the ClustalW2 multiple sequence alignment tool at the European Bioinformatics Institute website, http://www.ebi.ac.uk/Tools/clustalw2/index.html.
(PDF)
Phylogenetic tree of VapC proteins from mycobacteria. VapCs were aligned using ClustalW2 multiple sequence and server alignment server (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The tree was generated in Jalview 2.08.1 based on percentage identity between sequences.
(PDF)
Properties of mycobacterial vapBC toxin-antitoxin modules selected for study.
(PDF)
VapC toxicity in M. smegmatis mc2155 assessed by transformation efficiency of VapC expression vector.
(PDF)
VapC toxicity in Mtb assessed by transformation efficiency of VapC expression vector.
(PDF)
Strains and plasmids used in this study.
(PDF)
Oligonucleotides used to create knockout vectors for allelic exchange mutagenesis.
(PDF)
Oligonucleotides used for expression vector construction, yeast two-hybrid (Y2H) vector construction and RT-PCR.
(PDF)