Abstract
The post-transcriptional export of spliced and unspliced HIV-1 (human immunodeficiency virus type 1) RNAs from the nucleus to the cytoplasm is a complex process. Part of the complexity arises from the fact that eukaryotic cells normally retain unspliced RNAs in the nucleus preventing their exit into the cytoplasm. HIV-1 has evolved a protein, Rev, that participates in the export of unspliced/partially spliced viral RNAs from the nucleus. It has been documented that several cellular factors cooperate in trans with Rev, and certain cis-RNA motifs/features are important for transcripts to be recognized by Rev and its co-factors. Here, the post-transcriptional activities of Rev are discussed in the context of a recent finding that an RNA cap methyltransferase contributes to the expression of unspliced/partially spliced HIV-1 transcripts.
Key words: PIMT, TGS1, HIV-1, Rev, CRM1, RNA export
Introduction
Retroviruses express both unspliced and spliced RNAs. HIV-1 employs intricate mechanisms to splice its 9 kb RNA to various 4 kb and 1.8 kb transcripts.1 Over 40 different HIV-1 mRNAs are produced by alternate splicing2 of the primary 9 kb HIV-1 transcript which contains 5 identified splicing donor sites and 8 to 9 splicing acceptor sites. Several cellular factors including arginine/serine rich (SR) proteins [SC35, ASF/SF2 (alternative splicing factor/splicing factor 2), SRp40 (SR protein 40) and 9G8] and heterogenous ribonucleoproteins (hnRNPs; hnRNP A1, hnRNP A2 and hnRNP A3) regulate the expression of different HIV-1 mRNA.3–5 The efficiency of HIV-1 splicing is sub-optimal,6,7 and some of the unspliced and partially spliced viral transcripts appear to be retained in the nucleus;8 however, unspliced and partially spliced HIV-1 RNAs are needed to produce the genomic RNA for progeny viruses as well as for Gag (group-specific antigen), Pol (polymerase) and Env (envelope) proteins. A challenge for the virus is to overcome the cellular mechanisms which normally prevent the nuclear exit of unspliced or incompletely spliced RNAs. A simple view of how HIV-1 solves this challenge is that it encodes a viral “export” protein that recognizes a cis-viral-RNA motif contained in unspliced/partially spliced viral transcripts.9–11 Thus, the HIV-1 Rev protein serves to export from the nucleus into the cytoplasm unspliced/partially spliced genomic, Gag, Pol, Env RNAs that contain a cis-RRE (Rev-responsive element) motif.12
The simple picture of Rev—RRE interaction belies a more complex process that involves many host cell co-factors. Initial efforts to identify the cellular co-factor(s) that mediate Rev nuclear export function revealed cellular factors CRM1 (chromosome region maintenance 1), Ran (Ras-related nuclear protein), FG (phenylalanine-glycine)-repeat nucleoporins, including RIP (Rev-interacting protein)/RAB (Rev/Rex activation domain-binding protein).13–18 In particular, Rev directly interacts with nuclear export receptor CRM1, and CRM1 is required for Rev-mediated export of HIV RNAs.14–16 Additionally, it has been shown that RNA helicases like DDX3 [DEAD (Asp-Glu-Ala-Asp) box 3],19 DDX1,20 and RNA helicase A (RHA),21 also contribute to Rev function. More recently, it has become clear that many other cellular factors22 including those involved in RNA capping,11,23–25 alternative splicing,5 RNA stability,26 and intracellular localization of the viral transcripts may also participate in the cytoplasmic expression of unspliced/partially spliced HIV-1 RNAs.27
Consistent with the view of additional complexities to the expression of unspliced/partially spliced viral RNAs, it was recently reported that a human cellular RNA methyltransferase, PIMT (peroxisome proliferator-activated protein with methyltransferase domain), enhances Rev-mediated export of HIV-1 RNA.23 Indeed, PIMT was demonstrated to be a nuclear-cytoplasmic shuttling protein which binds to Rev, and PIMT overexpression augmented Rev-dependent, but not Rev-independent, expression of viral RNAs. When PIMT was knocked down in cells or when PIMT activity was inhibited by small chemical inhibitors, HIV-1 replication in human cells was reduced. These collective findings implicate a role for the PIMT RNA methyltransferase in HIV-1 post-transcriptional regulation.
What does PIMT do for HIV-1 RNA?
PIMT is the human homolog of the yeast RNA cap hypermethylase TGS1 (trimethylguanosine synthase 1). TGS1 hypermethylates the m7G RNA cap to a trimethylguanosine (TMG; m2,2,7 guanosine) cap.28–33 A nuclear isoform of human PIMT has been shown to have methyl transferase activity.28,34,35 In the new finding, PIMT was found to hypermethylate selectively the RNA-cap of unspliced and partially spliced HIV-1 transcripts and to increase their cytoplasmic distribution23 (Fig. 1). It was shown that PIMT, a nucleocytoplasmic shuttling protein, is recruited by Rev to RRE containing transcripts (i.e., unspliced and partially spliced HIV-1 RNA). Rev-recruited PIMT then hypermethylates the m7G-cap on these RNAs to a TMG-cap. How the TMG-cap then acts to facilitate the cytoplasmic expression of the RNA is not clearly understood, but one suggestion is that the TMG-cap may be recognized by CRM1 which then directs the transcript for nuclear export. The activity of PIMT appears to be selective for only RRE-containing HIV-1 RNAs; thus, the expression of fully spliced viral transcripts (which do not contain the RRE-sequence and are not TMG-capped) does not appear to be regulated by PIMT. Rather than using the CRM1-associated route, fully spliced HIV-1 RNAs are known to exit the nucleus through the NFX1 (nuclear transcription factor X-box binding 1)/TAP (tyrosine kinase interacting protein associated protein) dependent pathway.36,37
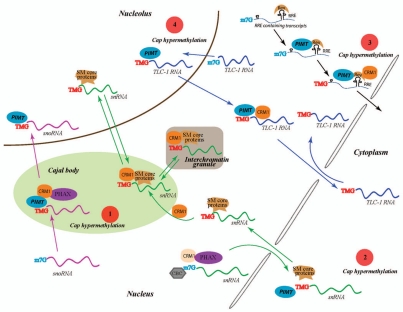
Figure 1.
Schematic representation of some of the CRM1 dependent nuclear and cytoplasmic events that influence the expression and function of hypermethylated cellular and viral RNAs. The hypermethylation of RNA caps is a complex process which occurs in different regions of the cells and appears to determine RNA trafficking inside the cell. Some proposed CRM1 dependent mechanisms that regulate post-transcriptional expression of TMG containing transcripts are indicated as follow. (1) m7G capped snoRNAs are targeted to “Cajal bodies” by CBC (cap binding complex) and PHAX (phosphorylated adapter for RNA export) where they are hypermethylated by PIMT and then bound by CRM1 through recognition of the TMG caps and transported to the nucleolus; (2) snRNAs are exported to the cytoplasm in a m7G-cap dependent manner. snRNAs bound by CBC, CRM1 and PHAX are exported to cytoplasm. The association of Sm core proteins with snRNA in cytoplasm recruits PIMT which hypermethylates the snRNA cap. The snRNA with TMG-cap are then re-imported into the nucleus in a complex with Sm core proteins. This snRNP interacts with CRM1 and localizes to Cajal bodies from where they travel to the nucleolus and interchromatin granules. (3) Rev binds to the cis-RRE motif in unspliced and partially spliced HIV-1 RNA and recruits PIMT which then hypermethylates the m7G-capped RNA. CRM1 binds to Rev and the TMG cap and transports the RNA into the cytoplasm. (4) Telomerase TLC-1 RNA (Telomerase component-1 RNA) also has been reported to acquire a TMG cap in the nucleus before being export through a CRM1 route into the cytoplasm.32
So where does the recently published report lead us? An important next question is which cellular RNAs are targets of PIMT for TMG cap hypermethylation. Currently, RNA Pol II transcribed snRNA (small nuclear RNA) and snoRNA (small nucleolar RNA) are two types of RNAs subjected to additional methyl modifications to their m7G cap structures to form TMG-caps.38–41 Both types of TMG-capped RNAs are transported within the nucleus or between the nucleus and the cytoplasm by the CRM1 protein42–44 (Fig. 1). Interestingly, PIMT-mediated TMG-hypermethylation occurs in the nucleus for snoRNA45 and in the cytoplasm for snRNA38–41 (Fig. 1). It remains unclear why one type of RNA requires nuclear PIMT TMG-activity while the other needs a cytoplasmic PIMT TMG-activity. This question warrants further resolution; however, the finding that PIMT is a nuclear-cytoplasmic shuttling protein23 suggests that it can serve both types of activity. A bigger question is whether human cellular RNAs, other than snRNA and snoRNA, also have TMG-cap modification. While many human mRNAs are CRM1 dependent for expression,14,46,47 there is yet no evidence that any of these mRNAs are TMG-capped. The technology is available to address this question which seems certain to be answered soon.
Translation of TMG-Capped RNAs
If some mRNAs do have TMG caps how might this modification affect their translation? Normally the recognition of m7G cap by eIF-4F is the first step in mRNAribosome association and the translation of the mRNA. Thus, the presence of a TMG cap might have repercussions for the initiation of translation.48,49 Of note, there are published data that during the replication of Togaviruses, Semliki Forest virus and Sindbis virus,50,51 late viral mRNAs acquire hypermethylated guanosine caps and are expressed into viral proteins. Moreover, in nematode C. elegans and A. lumbricoides where mRNAs acquire trimethylguanosine caps by trans-splicing (TMG-capped 5′ leader sequence from snRNA are spliced to the 5′ ends of mRNAs) virtually all of the mRNAs coding for actin and ribosomal proteins have trimethylguanosine caps, and are apparently efficiently translated.52–55 These findings together with the HIV-1 report23 suggest that TMG-capping per se may not be a barrier to ribosome-translation of RNAs.
PIMT as an Anti-HIV-1 Drug Target
If TMG-capping of HIV-1 RNA is a significant biological process in viral replication, then it stands to reason that the PIMT enzymatic activity could be a potential drug target for inhibiting viral pathogenesis. Targeting PIMT, a cellular protein, avoids the inherent problem posed by rapid HIV-1 mutation to all currently utilized chemotherapeutics targeted to virus-encoded proteins. There is some early evidence that treatment of cells with RNA methylation inhibitors suppresses HIV-1 replication.23 There are also reports which indicate that RNA methylation inhibitors can be used successfully to inhibit the replication of Herpes simplex virus, Vesicular Stomatitis Virus and other viruses.56–59 Future studies on whether these inhibitions correlate with a requirement for hypermethylated capping of viral RNAs will be needed.
Post-transcriptional Role of Rev on RNA Packaging?
Full length (unspliced) HIV-1 RNA functions as a template for translation of Gag-Pol proteins and also serves as the genomic RNA that is packaged into virions. Recently a new post-transcriptional activity for Rev in the packaging of genomic RNA was reported. It was shown that Rev influenced viral RNA encapsidation and subsequent HIV-1 infectivity by more than 1,000 fold.60–63 Interestingly, this latter activity of Rev was in part linked to the nuclear export of RNA suggesting that this apparent activity of Rev may lie in its ability to modulate nuclear RNP (ribonuclear protein) assembly on RRE containing RNAs.60–62 It was observed that in cells expressing similar amounts of different forms of HIV-1 RNA, Rev/RRE dependent viral RNA encapsidation was 100 fold higher than similar viral RNA engineered to be Rev and RRE-independent by codon optimization of the Gag or by substituting the CTE (constitutive transport element, which acts independently of Rev),64 in place of the RRE. It should be noted that viral RNA which is codon optimized in Gag and viral RNA that substitutes CTE for RRE are exported through NXF1/TAP pathway and are not CRM1 substrates.36,37 Additionally, evidence suggests that the presence of Rev alone is not sufficient for efficient encapsidation of HIV-1 RNA that has been engineered to be exported via a CRM1 independent pathway. These collective results indicate that direct CRM1/Rev—RRE RNA interaction is a critical event needed for optimal viral RNA encapsidation. Previously it was also reported that the major HIV-1 packaging signal (SL3; stem loop 3) though important was not sufficient for RNA packaging.65 The above observations on RNA-packaging agree with a SL3-independent role contributed by Rev in viral RNA encapsidation.60–62 It remains to be determined if a TMG-PIMT/Rev interaction may also influence the encapsidation of viral genomic RNA. Preliminary analyses of HIV-1 virion RNAs suggest the presence of TMG-capped moieties (Yedavalli VS, unpublished results).
Concluding Remarks
Several different transcripts synthesized by RNA polymerase I [rRNAs (ribosomal RNA)], RNA polymerase II [mRNAs (messenger RNA) and snRNAs] and RNA polymerase III [tRNAs (transfer RNA) and 5S rRNA], use distinct and independent pathways to exit the nucleus. Efforts to identify the critical trans-acting protein components that define each of these distinct nuclear RNA export pathways have produced insights into several functional factors and their pathways. To date, the study of HIV-1 has contributed to the understanding of many important RNA transport proteins. Going forward it will be important to explore how cis-RNA features such as RNA-cap and interstitial RNA modifications in viral transcripts could determine RNA distribution.
Acknowledgements
Work in our laboratory is supported in part by NIAID, NIH, intramural funding and by the Intramural AIDS Targeted Antiviral Program (IATAP) from the office of the Director, NIH.
References
- 1.Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowling D, Nasr-Esfahani S, Tan CH, O'Brien K, Howard JL, Jans DA, et al. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology. 2008;5:18. doi: 10.1186/1742-4690-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saliou JM, Bourgeois CF, yadi-Ben ML, Ropers D, Jacquenet S, Marchand V, et al. Role of RNA structure and protein factors in the control of HIV-1 splicing. Front Biosci. 2009;14:2714–2729. doi: 10.2741/3408. [DOI] [PubMed] [Google Scholar]
- 4.Exline CM, Feng Z, Stoltzfus CM. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J Virol. 2008;82:3921–3931. doi: 10.1128/JVI.01558-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- 6.Madsen JM, Stoltzfus CM. A suboptimal 5′ splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication. Retrovirology. 2006;3:10. doi: 10.1186/1742-4690-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology. 2006;3:89. doi: 10.1186/1742-4690-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang DD, Sharp PA. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 9.Nakielny S, Fischer U, Michael WM, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 10.Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- 11.Felber BK, Zolotukhin AS, Pavlakis GN. Posttranscriptional control of HIV-1 and other retro-viruses and its practical applications. Adv Pharmacol. 2007;55:161–197. doi: 10.1016/S1054-3589(07)55005-2. [DOI] [PubMed] [Google Scholar]
- 12.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 16.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 17.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 18.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 19.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Fang J, Kubota S, Yang B, Zhou N, Zhang H, Godbout R, et al. A DEAD box protein facilitates HIV-1 replication as a cellular co-factor of Rev. Virology. 2004;330:471–480. doi: 10.1016/j.virol.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, et al. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nekhai S, Jeang KT. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Future Microbiol. 2006;1:417–426. doi: 10.2217/17460913.1.4.417. [DOI] [PubMed] [Google Scholar]
- 23.Yedavalli VS, Jeang KT. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci USA. 2010;107:14787–14792. doi: 10.1073/pnas.1009490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cougot N, van DE, Babajko S, Seraphin B. ‘Captabolism’. Trends Biochem Sci. 2004;29:436–444. doi: 10.1016/j.tibs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 26.Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, et al. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff H, Hadian K, Ziegler M, Weierich C, Kramer-Hammerle S, Kleinschmidt A, et al. Analysis of the influence of subcellular localization of the HIV Rev protein on Rev-dependent gene expression by multifluorescence live-cell imaging. Exp Cell Res. 2006;312:443–456. doi: 10.1016/j.yexcr.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 29.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, et al. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res. 2007;35:1411–1420. doi: 10.1093/nar/gkl1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke J, Gehlen J, Ehrenhofer-Murray AE. Hypermethylation of yeast telomerase RNA by the snRNA and snoRNA methyltransferase Tgs1. J Cell Sci. 2008;121:3553–3560. doi: 10.1242/jcs.033308. [DOI] [PubMed] [Google Scholar]
- 32.Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. TLC1 RNA nucleo-cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J. 2008;27:748–757. doi: 10.1038/emboj.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hausmann S, Zheng S, Costanzo M, Brost RL, Garcin D, Boone C, et al. Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors and RNA decay pathways. J Biol Chem. 2008;283:31706–31718. doi: 10.1074/jbc.M806127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enunlu I, Papai G, Cserpan I, Udvardy A, Jeang KT, Boros I. Different isoforms of PRIP-interacting protein with methyltransferase domain/trimethylguanosine synthase localizes to the cytoplasm and nucleus. Biochem Biophys Res Commun. 2003;309:44–51. doi: 10.1016/s0006-291x(03)01514-6. [DOI] [PubMed] [Google Scholar]
- 35.Girard C, Verheggen C, Neel H, Cammas A, Vagner S, Soret J, et al. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J Biol Chem. 2008;283:2060–2069. doi: 10.1074/jbc.M704209200. [DOI] [PubMed] [Google Scholar]
- 36.Pasquinelli AE, Ernst RK, Lund E, Grimm C, Zapp ML, Rekosh D, et al. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 38.van ZK, Dickmanns A, Fischer U, Luhrmann R, Fanning E. A cytoplasmically anchored nuclear protein interferes specifically with the import of nuclear proteins but not U1 snRNA. J Cell Biol. 1993;121:229–240. doi: 10.1083/jcb.121.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plessel G, Fischer U, Luhrmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, et al. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 41.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 42.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 44.Sleeman J. A regulatory role for CRM1 in the multidirectional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci. 2007;120:1540–1550. doi: 10.1242/jcs.001529. [DOI] [PubMed] [Google Scholar]
- 45.Terns MP, Grimm C, Lund E, Dahlberg JE. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schutz S, Chemnitz J, Spillner C, Frohme M, Hauber J, Kehlenbach RH. Stimulated expression of mRNAs in activated T cells depends on a functional CRM1 nuclear export pathway. J Mol Biol. 2006;358:997–1009. doi: 10.1016/j.jmb.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 48.Darzynkiewicz E, Stepinski J, Ekiel I, Jin Y, Haber D, Sijuwade T, et al. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai A, Jankowska-Anyszka M, Centers A, Chlebicka L, Stepinski J, Stolarski R, et al. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- 50.HsuChen CC, Dubin DT. Di-and trimethylated congeners of 7-methylguanine in Sindbis virus mRNA. Nature. 1976;264:190–191. doi: 10.1038/264190a0. [DOI] [PubMed] [Google Scholar]
- 51.van Duijn LP, Kasperaitis M, Ameling C, Voorma HO. Additional methylation at the N(2)-position of the cap of 26S Semliki Forest virus late mRNA and initiation of translation. Virus Res. 1986;5:61–66. doi: 10.1016/0168-1702(86)90065-1. [DOI] [PubMed] [Google Scholar]
- 52.Liou RF, Blumenthal T. trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol Cell Biol. 1990;10:1764–1768. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van DK, Hirsh D. mRNAs that mature through trans-splicing in Caenorhabditis elegans have a trimethylguanosine cap at their 5′ termini. Mol Cell Biol. 1990;10:1769–1772. doi: 10.1128/mcb.10.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maroney PA, Denker JA, Darzynkiewicz E, Laneve R, Nilsen TW. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 55.Gordon RK, Ginalski K, Rudnicki WR, Rychlewski L, Pankaskie MC, Bujnicki JM, et al. Anti-HIV-1 activity of 3-deaza-adenosine analogs. Inhibition of S-adenosylhomocysteine hydrolase and nucleotide congeners. Eur J Biochem. 2003;270:3507–3517. doi: 10.1046/j.1432-1033.2003.03726.x. [DOI] [PubMed] [Google Scholar]
- 56.Hong JH, Kim SY, Oh CH, Yoo KH, Cho JH. Synthesis and antiviral evaluation of novel open-chain analogues of neplanocin A. Nucleosides Nucleotides Nucleic Acids. 2006;25:341–350. doi: 10.1080/15257770500544578. [DOI] [PubMed] [Google Scholar]
- 57.De CE. John Montgomery's legacy: carbocyclic adenosine analogues as SAH hydrolase inhibitors with broad-spectrum antiviral activity. Nucleosides Nucleotides Nucleic Acids. 2005;24:1395–1415. doi: 10.1080/15257770500265638. [DOI] [PubMed] [Google Scholar]
- 58.Kim JW, Hong JH. Simple synthesis of novel acyclic (e)-bromovinyl nucleosides as potential antiviral agents. Nucleosides Nucleotides Nucleic Acids. 2006;25:879–887. doi: 10.1080/15257770600793877. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H, Schinazi RF, Chu CK. Synthesis of neplanocin F analogues as potential antiviral agents. Bioorg Med Chem. 2006;14:8314–8322. doi: 10.1016/j.bmc.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J Virol. 2010;84:6598–6604. doi: 10.1128/JVI.02264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brandt S, Blissenbach M, Grewe B, Konietzny R, Grunwald T, Uberla K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007;3:54. doi: 10.1371/journal.ppat.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 2004;23:2632–2640. doi: 10.1038/sj.emboj.7600270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cochrane A. How does the journey affect the message(RNA)? RNA Biol. 2009;6:169–170. doi: 10.4161/rna.6.2.8095. [DOI] [PubMed] [Google Scholar]
- 64.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, et al. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berkowitz RD, Hammarskjold ML, Helga-Maria C, Rekosh D, Goff SP. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]