Abstract
The misfolding of α-synuclein (αS) to a cross-β-sheet amyloid structure is associated with pathological conditions in Parkinson's and other neurodegenerative diseases. Using pulse electron paramagnetic resonance spectroscopy combined with a cross-labeling strategy involving four double mutants, we were able to determine the intramolecular distance between the extremal β-strands. The distance of 4.5 ± 0.5 nm is in good agreement with the dimensions of a protofilament reported by other low-resolution techniques, such as x-ray scattering and atomic force microscopy.
α-Synuclein (αS) fibrils are a major component of the intracellular Lewy body inclusions that are present in the brains of patients with Parkinson's disease (1). αS is a small 140-amino-acid protein characterized by three different regions: an N-terminal domain (residues 1–60) containing seven KTKEGV repeats, a central hydrophobic NAC domain (residues 61–95), and a highly negatively charged C-terminal domain (residues 96–140) (2). The protein monomer is intrinsically disordered, with some residual structure in solution. Solution NMR studies have revealed intramolecular long-range contacts between the NAC and the C-terminal regions as well as the N- and C-termini. This residual structure is autoinhibitory for aggregation (3,4). In the aggregated fibrillar form, the protein arranges in a cross-β-sheet structure, characterized by x-ray and electron diffraction (5,6), in which individual β-strands stack perpendicularly to the fibril axis. Solid-state (SS)-NMR and continuous wave (CW) electron paramagnetic resonance (EPR) measurements have revealed that amino-acid residues 38–96 form the β-sheet-rich core region of the fibrils (7–11). In particular, the EPR data (8) suggested that α-synuclein stacks in register along the fibril axis. However, the detailed folding and arrangement of αS monomers in the fibrils is still unresolved. Recently, a folding model consisting of five β-sheets per monomer in the fibril was proposed based on SS-NMR data (10), although long-range restraints at the molecular level in support of this model were lacking.
In the past decade, pulse EPR spectroscopy techniques, such as PELDOR and DEER, have emerged as powerful tools for determining distances in the range between 2 and ∼8 nm by measuring the dipolar coupling between two paramagnetic centers in proteins and nucleic acids (12–14). These methods provide a means of obtaining long-range distance restraints in structural studies of amyloid fibrils and their intermediate aggregates if suitable labeling schemes can be established. In diamagnetic proteins, such as αS, the technique requires the incorporation of two spin labels, such as MTSL, which can be covalently attached to a cysteine in the protein backbone via formation of a disulfide bond. The labeling procedure was successfully demonstrated in CW EPR studies of fibrils containing single cysteine mutations (8), in which the dynamics of the label and the protein backbone were investigated in detail. EPR distance measurements were recently employed to investigate the interaction of αS with lipids and micelles (15–17). However, to our knowledge, no pulse-EPR distance measurements on amyloid fibrils have been reported to date.
Here we present an investigation of αS fibrils by PELDOR/DEER. The labeling positions for distance measurements were selected based on the reported observations of β-strand regions in the fibrils by magnetic resonance methods. Fig. 1 summarizes the αS amino-acid sequence and the corresponding reported β-strands in fibrils. To probe the distance between the two extremal strands, we chose to use residues 41/42 and 90/91, respectively. We prepared a set of four double cysteine mutants (G41C/A90C, G41C/A91C, S42C/A90C, S42C/A91C) and labeled them with MTSL (see the Supporting Material for details regarding the materials and methods used). From the set of four mutants, we were able to infer the orientation of the specific residue with respect to the plane of the β-strand, as discussed further below. The EPR distance measurements necessitated dilution of the labeled proteins with wild-type (wt) protein to reduce the likelihood of intermolecular electron-electron couplings. Several dilution ratios from 1:4 to 1:40 (double-labeled protein versus wt ratio) were tested to optimize the aggregation conditions and to ensure an adequate intramolecular PELDOR signal and an adequate ratio of the intra- and intermolecular PELDOR signals (both increasing with concentration of labeled αS). The aggregation protocol is described in the Supporting Material. In all samples, the aggregation kinetics was followed by means of a thioflavin-T fluorescence assay and CW EPR spectroscopy (Fig. S1 and Fig. S2). Samples with dilution of ≤1:10 showed aggregation characteristics (lag-phase ∼40 h, t1/2 of aggregation 60–80 h) comparable to those of wt protein, and the EPR spectra of the fibrils displayed EPR line shapes close to the rigid limit of immobilized, isolated labels (Fig. S2). Our results are consistent with the line shape analysis of Heise et al. (7), and with the residues being buried and in tertiary contact. The overall morphology of the fibrils was characterized by electron microscopy (EM) (Fig. S3). Twisted and straight filaments were observed in all samples. This structural heterogeneity was previously reported in fibrils prepared from wt αS for SS-NMR measurements (7,10). Based on all characterizations, the 1:20 dilution was used for EPR at Q-band (34 GHz) and the 1:10 dilution was employed for X-band (9 GHz) due to the lower EPR sensitivity at this frequency.
Figure 1.
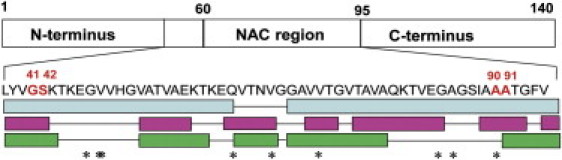
Schematic sequence of αS and amino acids proposed in β-strand regions. Marked are residues G41, S42, A90, and A91, which were selected for labeling with MTSL in this study. Bottom: β-strand regions as identified by EPR (light blue: Chen et al. (8)) and SS-NMR (cyan: Heise et al. (7); green: Vilar et al. (10). Stars denote flexible residues observed by high-resolution NMR (11).
Fig. 2 shows representative dipolar oscillation (PELDOR/DEER) traces of the fibrils containing the four doubly labeled mutants. Because the dipolar frequency is not dependent on the EPR frequency, a comparison of the traces at the two frequencies distinguishes reproducible distances given by intramolecular spin pairs from other undesirable physical effects such as electron spin echo envelope modulation, and measurement perturbations such as baseline distortions or processing artifacts (e.g., from Tikhonov regularization). Two doubly labeled mutants, G41C/A90C and S42C/A90C, showed time traces with distinct modulation, yielding distances of 4.5± 0.5 nm and 4.0 ± 0.5 nm, respectively. The experiment on mutant S42C/A91C did not display a modulation discernable by eye, but the analysis of both X- and Q-band traces suggested a distance in the range of ∼5 nm. The analysis of both latter traces also showed some peaks at shorter distances, which we attribute to artifacts of the Tikhonov regularization due to the weak signal. Finally, mutant G41C/A91C lacked a detectable PELDOR effect, probably because the distance was too long to allow for detection of the first oscillation maximum within 2 μs. Indeed, the short T2 relaxation time of the samples limited the time acquisition window to ∼2 μs, corresponding to a maximal detectable distance of ≤5 nm. All observed distances did not depend on the used dilution ratio within the error.
Figure 2.
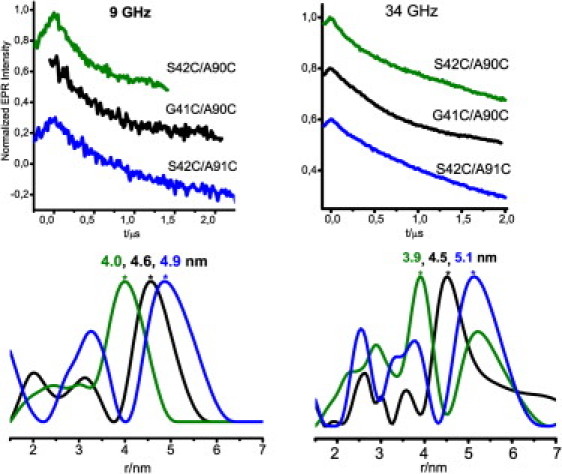
Normalized four-pulse PELDOR/DEER traces of αS double mutants diluted in wt protein at 9 GHz (left) and 34 GHz (right) and T = 10 K. Traces were plotted in stack. The typical acquisition time per trace was 48 h. Data were analyzed with Deer Analysis (18). See the Supporting Material for more details.
The distances shown in Fig. 2 cannot be rationalized independently because the N-O group of MTSL, which carries the electron spin density, is displaced by ∼0.5–0.7 nm from the Cα in the β-strand. Note that the three observed distance distributions are centered at discrete values, i.e., 4, 4.5, and 5 nm, with a distance increment of ∼0.5 nm. The data reflect the different orientations of the spin label and the displacement at each residue. In a β-strand, two neighboring residues are oppositely oriented with respect to the β-strand plane. Consequently, our cross-labeling scheme should lead to four distances, two of which might be similar, the third shorter and the fourth longer, if the considered strands are parallel, according to the currently most widely accepted model based on SS-NMR data (10). A schematic view of the orientation of the spin labels with respect to their β-strand and to each other is given in Fig. 3. Based on this scheme, we assign the shortest distance of 4 nm to two spin labels pointing toward each other with side chains in opposite directions. The intermediate distances of 4.5–5.0 nm between spin-label pairs are assigned to side chains pointing in the same direction. Accordingly, the fourth distance should be considerably larger than 5.0 nm and would not be detectable under our experimental conditions.
Figure 3.
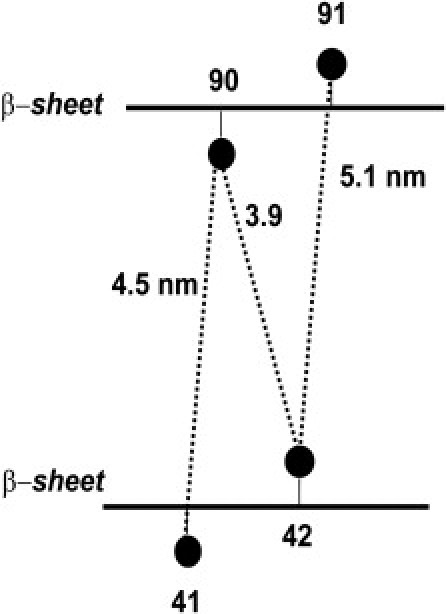
Labeling scheme employed to extract the distance between the two β-strands containing residues 41/42 and 90/91. Assignment of the spin-label orientation was based on the observed distances. The circles denote the position of the spin labels.
The intermediate distance between spin labels pointing in the same direction, i.e., 4.5–5 nm, is expected to correspond to the distance between the two extremal β-strands. The distance between β−sheets that are stacked perpendicular to the fibril axis was reported to be 1.1 nm by x-ray diffraction methods (6). Our distance of 4.5–5 nm (with an error of ± 0.5 nm) is a factor of 4 larger, suggesting that three other β-sheets could lie in a parallel stack between the two extremal ones, with a total of five sheets per αS monomeric unit. We point out that this result is consistent with the folding model proposed by Vilar et al. (10) assuming that the residues are more or less in the middle of the respective β-strand. However, we cannot exclude other, more complicated packing models such as that found in the HET-s prion protein (19). Furthermore, our data are consistent with the size of a protofilament being ∼4 nm as measured by lower-resolution techniques such as atomic force microscopy (6) and cryo-EM (10).
In conclusion, we have demonstrated that it is possible to obtain intramolecular EPR distance measurements between spin labels site-specifically inserted into αS β-strands with dilutions between 10- and 20-fold with wt protein. In combination with a specific labeling scheme, and assuming some prior knowledge about the secondary structure, the known directions of the spin label can provide new information at the molecular level on the distance between β-strands in fibrils. The method should be useful in future structural studies of amyloid-forming proteins by providing long-range distance constraints, and complements well-established techniques such as NMR, x-ray diffraction, and atomic force microscopy.
Acknowledgments
We thank Shyamala Thirunavukkurasu and Dmytro Yushenko for initial help with the aggregation assays, and Giuseppe Sicoli and Markus Zweckstetter for fruitful discussions.
This work was supported by the Max Planck Society (project “Toxic protein conformation” to M.B., C.G., and T.J.).
Supporting Material
References and Footnotes
- 1.Spillantini M.G., Schmidt M.L., Goedert M. α-Synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.Fink A.L. The aggregation and fibrillation of α-synuclein. Acc. Chem. Res. 2006;39:628–634. doi: 10.1021/ar050073t. [DOI] [PubMed] [Google Scholar]
- 3.Bertoncini C.W., Jung Y.-S., Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured α-synuclein. Proc. Natl. Acad. Sci. USA. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dedmon M.M., Lindorff-Larsen K., Dobson C.M. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 5.Conway K.A., Harper J.D., Lansbury P.T., Jr. Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 6.Serpell L.C., Berriman J., Crowther R.A. Fiber diffraction of synthetic α-synuclein filaments shows amyloid-like cross-β conformation. Proc. Natl. Acad. Sci. USA. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heise H., Hoyer W., Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M., Margittai M., Langen R. Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 2007;282:24970–24979. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 9.Kloepper K.D., Zhou D.H., Rienstra C.M. Temperature-dependent sensitivity enhancement of solid-state NMR spectra of α-synuclein fibrils. J. Biomol. NMR. 2007;39:197–211. doi: 10.1007/s10858-007-9189-z. [DOI] [PubMed] [Google Scholar]
- 10.Vilar M., Chou H.T., Riek R. The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M.K., Kim H.-Y., Zweckstetter M. Conserved core of amyloid fibrils of wild type and A30P mutant α-synuclein. Protein Sci. 2011;20:387–395. doi: 10.1002/pro.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milov A.D., Ponomarev A.B., Tsvetkov Y.D. Electron-electron double resonance in electron spin echo: model biradical systems and the sensitized photolysis of decalin. Chem. Phys. Lett. 1984;110:67–72. [Google Scholar]
- 13.Martin R.E., Pannier M., Spiess H.W. Determination of end-to-end distances in a series of TEMPO diradicals of up to 2.8 nm length with a new four-pulse double electron electron resonance experiment. Angew. Chem. Int. Ed. 1998;37:2833–2837. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2833::AID-ANIE2833>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Schiemann O., Prisner T.F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 15.Drescher M., van Rooijen B.D., Huber M. A stable lipid-induced aggregate of α-synuclein. J. Am. Chem. Soc. 2010;132:4080–4082. doi: 10.1021/ja909247j. [DOI] [PubMed] [Google Scholar]
- 16.Rao J.N., Jao C.C., Ulmer T.S. A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins. J. Am. Chem. Soc. 2010;132:8657–8668. doi: 10.1021/ja100646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgieva E.R., Trudy F.R., Eliezer D. The lipid binding domain of wild type and mutant α-synuclein: compactness and interconversion between the broken and extended helix forms. J. Biol. Chem. 2010;285:28261–28274. doi: 10.1074/jbc.M110.157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeschke G., Chechik V., Jung H. Deer Analysis—a comprehensive software package for analyzing pulse ELDOR data. Appl. Magn. Reson. 2006;230:473–498. [Google Scholar]
- 19.Wasmer C., Lange A., Meier B.H. Amyloid fibrils of the HET-s (218–289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


