Abstract
The mechanisms of the control and activity of the autophagy-lysosomal protein degradation machinery are emerging as an important theme for neurodevelopment and neurodegeneration. However, the underlying regulatory and functional networks of known genes controlling autophagy and lysosomal function and their role in disease are relatively unexplored. We performed a systems biology-based integrative computational analysis to study the interactions between molecular components and to develop models for regulation and function of genes involved in autophagy and lysosomal function. Specifically, we analyzed transcriptional and microRNA-based post-transcriptional regulation of these genes and performed functional enrichment analyses to understand their involvement in nervous system-related diseases and phenotypes. Transcriptional regulatory network analysis showed that binding sites for transcription factors, SREBP1, USF, AP-1 and NFE2, are common among autophagy and lysosomal genes. MicroRNA enrichment analysis revealed miR-130, 98, 124, 204 and 142 as the putative post-transcriptional regulators of the autophagy-lysosomal pathway genes. Pathway enrichment analyses revealed that the mTOR and insulin signaling pathways are important in the regulation of genes involved in autophagy. In addition, we found that glycosaminoglycan and glycosphingolipid pathways also make a major contribution to lysosomal gene regulation. The analysis confirmed the known contribution of the autophagy-lysosomal genes to Alzheimer and Parkinson diseases and also revealed potential involvement in tuberous sclerosis, neuronal ceroidlipofuscinoses, sepsis and lung, liver and prostatic neoplasms. To further probe the impact of autophagy-lysosomal gene deficits on neurologically-linked phenotypes, we also mined the mouse knockout phenotype data for the autophagy-lysosomal genes and found them to be highly predictive of nervous system dysfunction. Overall this study demonstrates the utility of systems biology-based approaches for understanding the autophagy-lysosomal pathways and gaining additional insights into the potential impact of defects in these complex biological processes.
Key words: systems biology, autophagy, lysosome, transcription factors, microRNA
Introduction
The autophagy-lysosomal pathway is an important mechanism for regulating homeostasis of intracellular long-lived proteins and organelles.1,2 Macroautophagy delivers damaged proteins and organelles to the lysosome via double-membrane vesicles called the autophagosomes, which then fuse with lysosomes to complete the process.3 Failure at any of the steps in this process may lead to accumulation of toxic, structurally disruptive and energy-draining structures and cellular components. Ultimately, cells in which damaged proteins or organelles accumulate die because of bioenergetic and metabolic dysfunction. This autophagy-lysosomal degradation process is particularly important for nervous system function and neuronal survival because of the nondividing nature of postmitotic neurons. Aging brains exhibit reduced lysosomal function and, in neurodegenerative diseases, increased accumulation of macroautophagic vesicles has been observed in postmortem patient brains compared to normal controls.4–7
Because of the importance of macroautophagy and lysosomal activities to cellular functions, it is expected that the two processes are coordinately regulated in many situations to ensure high efficiency of clearance of excessive and damaged proteins and organelles. A candidate gene approach to identify such targets has clear limitations given the very large number of proteins involved in both pathways. For example, both macroautophagy and lysosomal function are induced under nutrient deprivation and stress to degrade proteins, lipids, nucleic acids and pathogens.8–11 For this reason, even at this early stage in autophagy/lysosomal research a computational systems biology approach may reveal new and unpredicted interactions which cannot be identified by manual inspection of the overwhelming number of potential candidate proteins. How autophagy and lysosomal genes functionally interact, and how they are regulated to achieve synergy and flexibility in response to diverse physiological and environmental stimuli is not well understood. Here we describe the insights obtained from a computational data analysis of the literature and mining of the relevant databases. An integrative systems biology approach can improve the understanding of the molecular basis of disease. For instance, how does dysfunction of lysosomal genes affect multiple steps of the integrated autophagy-lysosomal pathway and contribute to disease susceptibility and pathogenesis? As an example, mutation of lysosome-associated membrane protein-2 (LAMP2) causes abnormalities in autophagosome-lysosome fusion and accumulation of autophagic vesicles,12 which leads to Danon disease with accelerated tissue degeneration in heart, muscle and brain.12 Likewise, mutations in the lysosomal enzyme cathepsin D lead to Batten disease, characterized by lysosomal storage of lipofuscin.13,14 Genetic models of cathepsin D-deficient animals accumulate alpha-synuclein and autophagic vesicles.15–17 Both LAMP2 and cathepsin D mutations affect not only the lysosomes but also autophagy and lipid metabolism,18–20 further emphasizing the potential value of an integrated analyses of genes, biological pathways and disease connections. Furthermore, loss-of-function mutations in NPC1 and NPC2 that encode lysosomal acid sphingomylenases lead to Niemann-Pick Type C (NPC) disease, with accumulation of cholesterol and other lipids in late endosomes and lysosomes in virtually every tissue including the brain. In addition, increase of Beclin 1, autophagic vacuoles and LC3II were prominently associated with the tissue pathologies associated with NPC.21 Under certain conditions, macroautophagy and lysosomal activities may be independently regulated because in addition to macroautophagy, substrates for lysosomal degradation also arrive from endocytosis and phagocytosis.1,3 In short, the complexity of the physiological and pathological pathways calls for a systems biology approach to better understand the interconnections of genes, gene regulation, signaling pathways and disease pathogenesis.
We are fully aware that the incomplete state of current gene annotations raises two critical questions: first, how do the existing gaps in current knowledge affect our ability to develop effective computational approaches focusing on knowledge integration, and second, how can we draw any conclusions or assess the performance of our approach when much of the ground truth is either obscure or yet to be established? We believe that the strength of our approach is in the manually compiled list of autophagy-lysosomal gene sets derived by mining existing annotations in the Gene Ontology, Pathways and literature. Previous studies have shown that incompleteness of gene annotation repositories does not necessarily impair computational function prediction. However, it can hamper comparative performance evaluation of different techniques.22 There are several reports of systems biology-based approaches to study the autophagy-lysosomal system. For example, proteomics studies have identified a complex network of interacting proteins in the autophagy system requiring a complementary transcriptional regulatory mechanism that is currently poorly understood.23 Previous studies24 analyzed lysosomal genes and identified a transcription factor, TFEB, that is clearly one of the master regulators of lysosomal gene regulation, but the connections with autophagy have not yet been fully defined. This is important because it has been shown that TFEB can reduce mutant huntingtin level24 in a cell model and may be a novel therapeutic target for other neurodegenerative diseases. Studies of aging and Alzheimer disease brains also found transcriptional regulation of autophagy genes.11
In this study we found a number of novel and interesting interactions between regulatory and functional components of the autophagy-lysosomal pathway. Most striking was the role of SREBP1 as a potential regulatory link between the known autophagy-lysosomal pathway and lipid metabolism. The emergence of microRNAs as regulatory elements in a broad range of biological processes is now challenging our ideas of biological regulation in a number of fields.25 Our analysis revealed a potentially novel role for the microRNA, miR-124, in calpain regulation, which is a calcium-regulated protease important in autophagy.26 The extension of our analysis to pathology identified diseases for which a connection to the autophagy-lysosomal pathway has been suspected. Among these pathophysiological processes are Alzheimer and Parkinson diseases, tuberous sclerosis, neuronal ceroid-lipofuscinoses, sepsis and lung, liver and prostatic neoplasms. Extending these studies to the 43 reported murine knockouts of the autophagy-lysosomal genes identified in our analysis, we found that they all exhibited multiple abnormalities in the central nervous system. The implications of these findings for understanding the role of the autophagy-lysosomal system in the pathogenesis of neuronal and developmental disease are discussed.
Results and Discussion
Overview of mammalian autophagy-lysosomal genes and their functional interactions.
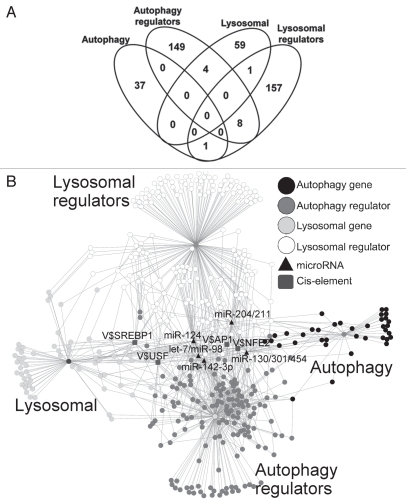
As a first step in examining the functional and regulatory interactions modulating the autophagy-lysosomal pathway, we compiled relevant genes from the established databases, including Gene Ontology,27 an autophagy database (http://autophagy.lu), KEGG pathways28 and literature3,8,29,30 for both human and murine data. This resulted in a total of 416 genes that included 38 genes with functions in autophagy, 161 autophagy regulators, 64 genes with a role in the lysosomal function or pathways, and 167 lysosomal regulators (Sup. Materials Table 1). The intersection of these four categories of genes (autophagy, autophagy regulators, lysosome and lysosomal regulators) is shown in the Venn diagram in Figure 1A. Interestingly, the four groups do not have extensive overlap based on existing literature and Gene Ontology, indicating that biological regulation is integrated at relatively few key steps. In Figure 1B the putative transcription factors and microRNAs with potential functional involvement in the autophagy-lysosomal pathway are shown with their potential interactions. We found the most prominent putative transcription factors and microRNAs that may coordinately regulate many of these autophagy-lysosomal genes included SREBP1, USF, AP-1, NFE2 and miR-130, 98, 124, 204 and 142 (Fig. 1B). We placed the symbol “V$” before the IDs of transcription factor binding sites in the diagram in this figure.
Figure 1.
An overview of functional and physical interactions among autophagy-lysosomal genes and the most common binding sites for transcription factors and microRNA. (A) Venn diagram shows the total of 416 autophagy-lysosomal genes we included in the analysis. We assigned the genes according to their most relevant and best known functions based on Gene Ontology and extensive literature review to four groups that include 38 autophagy genes, 64 lysosomal genes, 161 autophagy regulators and 167 lysosomal regulators. Fourteen of these 416 genes are shared between two groups, as shown in the Venn diagram. (B) Network representation of the autophagy-lysosomal genes showing the physical interactions as well as genes with enriched cis-acting elements for binding of transcription factors, and microRNA regulators that may be important in regulating expression of these genes. The nodes represent genes, cis-elements, or microRNAs, while the edges denote either a protein-protein interaction or a regulatory connection (TFs or microRNAs). In the diagram, the symbol “V$” is placed before the IDs of transcription factor binding sites. The most enriched TFBSs include those recognized by E-box transcription factors SREBP1, USF, AP-1 and NFE2, while the putative microRNA regulators include miR-130, 98, 124, 204 and 142.
Transcriptional regulation of autophagy-lysosomal genes.
The computational analysis of the autophagy-lysosomal gene promoters identified several putative transcription factor binding sites (from the approximately 400 known transcription factor binding sites). Figure 2A illustrates in more detail the relationship between the binding sites for specific transcription factors—SREBP1, USF, AP-1 and NFE2—and the autophagy-lysosomal genes they potentially regulate. We again placed the symbol “V$” before the IDs of transcription factor binding sites in the diagram in this figure. In this analysis autophagy and lysosomal genes and regulators are potentially controlled by transcription factors SREBP1 (26 genes), USF1 (16 genes), AP-1 (16 genes) and NFE2 (17 genes) (Sup. Materials Table 2). Recent studies also suggested that transcription factor FoxO3 may be important in regulation of autophagy genes.31 We searched for FoxO3 target genes using MatInspector and found 64 autophagy genes that had at least one putative FoxO3 binding site within 1 kb upstream region (Sup. Material Table 3). However, FoxO3 was not among the statistically significant enriched TFBSs in autophagy-lysosomal genes, suggesting that FoxO3 may not be specific to just autophagy-class genes and that it may regulate transcription of many nonautophagy, nonlysosomal genes.
Figure 2.
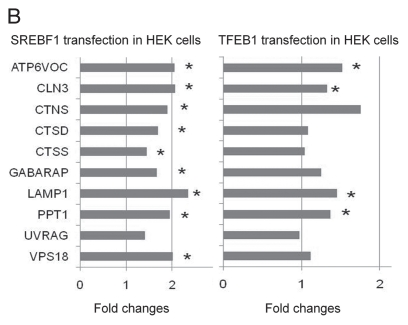
A close-up view of the autophagy and lysosomal genes containing cis-acting elements of the E-box transcription factors SREBP1 and USF, AP-1 and NFE2. In the diagram, the symbol “V$” is placed before the IDs of transcription factor binding sites to distinguish them from the autophagy-lysosomal genes. Validation of regulation of autophagy-lysosomal genes by SREBP1. HEK293 cells were transfected by SREBP1 or TFEB cDNA. RNA was harvested by Trizol reagent at 24 hours post-transfection and passed quality control. Quantitative RT-PCR was performed in triplicate. SREBP1 and TFEB mRNA level were increased by 21- and 343-fold, respectively, by real-time RT-PCR assay. The mRNA levels of target genes were plotted in the bar graph. LAMP1, GABARAP, CLN3, CTSS , CTSD, CTNS, ATP6VOC, VPS 18 and PPT1 are upregulated by SREBP1. LAMP1, CLN3, ATP6VOC and PPT1 are upregulated by TFEB. *p < 0.05 by Student t-test compared to control empty vector transfection.
USF-1, AP-1 and NFE2.
Upstream stimulating factor-1 (USF-1) is another E-box-binding transcription factor found enriched. Knockouts of USFs indicate that they are key regulators of stress and immune response, cell cycle and proliferation, as well as lipid metabolism.32 The autophagic system is altered during stress, cell cycle changes and immune challenges as well as intracellular lipid metabolism. The process of autophagy plays an integral role in all the above cellular functions. Stress response kinase p38 can phosphorylate and activate USF-1.33 Previous studies have shown that p38 can regulate autophagy by localization to Golgi membranes and binding to Atg9 via an intermediate protein.33 This physical interaction triggers Atg9 re-localization from the Golgi and endosomal membranes to MAPLC3 upon initiation of autophagy during starvation via a ULK-1-dependent process.34–36 Our observation that USF-1 binding sites are common in autophagy-lysosomal genes therefore suggests that USF-1 may also regulate stress and immune response, cell cycle and proliferation, as well as lipid metabolism via regulation of autophagy-lysosomal genes. Transcriptional regulation of autophagy-lysosomal genes by USF-1 may be an additional mechanism for the observed p38 activation of autophagy, in addition to the reported p38 function in regulating Atg9 re-localization.
AP-1 proteins JunB and c-Jun, but not JunD, c-Fos or Fra-1, have been shown to inhibit autophagy.37 Starvation induces transient induction of JunB and c-Jun and downregulation of their mRNA and proteins after one hour. Overexpression of JunB inhibits autophagy induced by starvation, as assessed by both LC3-GFP puncta accumulation and by p62 clearance. JunB inhibition of autophagy is independent of mTOR activation.37 JunB and c-Jun inhibition of starvation-induced autophagy are dependent on both the dimerization domain and the DNA binding domain, implicating a requirement of their transcription regulation activities for their inhibition of autophagy. Here we found that GABARAPL1, SQSTM1, SLC11A1 ABCA2, WDFY3, CAPN6, CAPN12, MAPK3 and PPP1R15A, may be regulated by AP-1, thus providing transcriptional inhibition of autophagy.
NFE2 is a heterodimeric transcription factor and consists of p45 and small Maf proteins. P45 knockout mice exhibit neonatal massive bleeding due to defects in megakaryocyte maturation.38,39 P45 belongs to the Cap'N'Collar (CNC) family of transcription factors, with Nrf2 as the most studied stress response gene regulated by oxidative stress.40 Both p45 and Nrf2 activate cytoprotective genes including NAD(P) H:quinone oxidoreductase (NQO1), with p45 competing for binding to the same cis-acting elements but a weaker activator. It was proposed that p45 modulates reactive oxygen species accumulation.38 Accumulation of reactive oxygen species can in turn regulate autophagy by modifying ATG4.41 Among NFE2 target genes, CAPN6, BNIP3 and GABARAPL1 are also AP-1 target genes. Additional NFE2 target genes include WDFY3, a WD repeat and FYVE domain containing protein that may be downstream of class III PI3K (VPS34) and mediate beclin/VPS34 complex-induced autophagy induction.
SREBP1.
SREBP1 (sterol-regulatory element binding protein1) is a basic-helix-loop-helix protein belonging to the family of transcription factors including TFE, Myc and USF1 that bind to the CACGTG E-box.42 SREBP1 has three isoforms: SREBP1a and SREBP1c play a role in fatty acid synthesis while SREBP2 plays a role in cholesterol synthesis.43 Lipid breakdown and degradation within the cell has been shown to require the autophagy-lysosomal pathway.44 Recent studies also report that lipids can regulate the autophagic process, which in turn can regulate lipid metabolism and adipocyte differentiation.45–52 Thus, SREBP1 may be important for lipid homeostasis not only by regulating lipid synthesis and signaling but also by regulating lipid degradation via the autophagy-lysosomal pathway. Maintenance of intracellular lipid metabolism and homeostasis is important in diseases such as diabetes, atherosclerosis, lipid storage disorders and neurodegeneration.48,53,54 Increased renal accumulation of lipids leads to an increased expression of SREBP1 in a rat model of diabetes.43 Cultured rat mesangial cells grown in high glucose media have increased SREBP1 expression.43 Hyperglycemic stress has been shown to induce autophagy.55 Prior studies have found that from 11,000 transcripts analyzed, 214 showed significant differences between lean and obese mice, including those important for adipocyte differentiation, lysosomal cathepsins and transcription factor SREBP1.56 Altered lipid metabolism has commonalities with lysosomal storage disorders such as Fabry's disease,53 type IA glycogen storage disease (von Gierke's disease),48 and neurodegeneration.54
Prior studies by Sardiello et al. have reported that another E-box family of transcription factor, TFEB, is important for lysosomal biogenesis. In our initial search for enriched TFBSs within the autophagy-lysosomal gene promoters, we did not find TFEB because the binding site library used to scan the promoters did not have the position weight matrix (PWM) for TFEB. To address this, we used the CLEAR PWM (from Sardiello et al.) and scanned the 230 lysosomal gene promoters using the RSAT application (http://rsat.ulb.ac.be/rsat/) as described by Sardiello et al. We found 76 lysosomal genes that had at least one CLEAR site within the promoter region. Of these, 13 genes also had SREBP1 sites within their promoter region (Table 1).
Table 1.
Common TFEB and SREBP1 targets, and newly identified potential SREBP1 targets from the selected 416 autophagy-lysosomal genes
| Potential SREBP1 targets that are in common with TFEB targets | SREBP1 targets that were not identified previously as TFEB targets |
| ACP2 | APP |
| ATP6V0C | BAX |
| CTSD | CAPN3 |
| CTSS | CLN3 |
| GABARAP | CTNS |
| GLA | DDIT3 |
| IGF2R | EIF2S1 |
| LAMP1 | EIF4G1 |
| MANBA | HEXA |
| MCOLN1 | PPT1 |
| PSAP | TYR |
| SLC36A1 | UVRAG |
| VPS11 | VPS18 |
Because of the potential importance of SREBP1 in coordinating autophayg-lysosomal activities and lipid metabolism, we performed experiments to validate transcriptional regulation of autophagy-lysosomal genes by SREBP1. We have performed quantitative RT-PCR analyses of potential target genes identified in our study, after transfection of full length human SREBP1 into HEK cells. Twenty-four hours after transfection of SREBP1, mRNA levels of LAMP1, GABARAP, CLN3, CTSS, CTSD, CTNS, ATP6VOC, VPS18 and PPT1 are all increased (Fig. 2B). LAMP1 and CTSD have both been extensively used as lysosomal markers that largely correlate with lysosomal numbers, sizes and activities. LAMP1 and CTSD expression is regulated by a variety of growth factors, as well as other intracellular and extracellular signaling molecules.3 TFEB upregulated LAMP1, CLN3, ATP6VOC and PPT1 in HEK cells after transfection (Fig. 2B). Our data indicate that SREBP1 is also an important transcription factor in regulating the autophagy-lysosomal pathway. Taken together this small sampling of genes lends credence to our analysis predicting that the time course and expression levels of SREBP1 and TFEB play a role in affecting the mRNA levels of target genes in the autophagy-lysosomal pathway.
Regulation of autophagy-lysosomal genes by microRNA.
The extent of microRNA regulation of autophagy is of great interest, particularly in the context of how the autophagy-lysosomal pathway is altered in nervous system diseases. MicroRNAs are important for brain function and neuronal survival.57,58 Knockout of Dicer, which is responsible for microRNA processing, in dopaminergic neurons leads to cell death and reduced locomotion.59 Dicer knockout in the hippocampus, Purkinje cells or striatum caused neuronal phenotypes with reduction of dendritic branches and size and increase of degeneration in respective brain areas.60–62 Out of all known mammalian microRNAs, about half of them have been found in different brain regions.63 We thus focused on microRNAs that have high nervous system expression in mammals. Supplemental Table 2 lists the microRNAs we analyzed and their target genes. We found autophagy and lysosomal genes contain conserved 3′UTR binding sequences recognized by miR-130, 98, 124, 204 and 142 (Fig. 3A). One of the enriched microRNAs, the miR-130/301/454 cluster, putatively regulates at least nine genes of the mTOR pathway that are involved in autophagy-lysosomal functions, including signaling genes IGF1, MAPK1, PRKAA1, PRKAA2, PRKAB1, PTEN, TSC1, ULK1 and ULK2, as well as autophagy mediators WDFY3, ATG2B and ATG16L1. Presumably, the coordinated regulation of the entire pathway helps improve efficiency. In addition, this coordinated regulation can provide new targets for up or downregulation of the autophagy-lysosomal pathway in disease therapies. Of the other enriched microRNAs, miR-142-3p targets the mTOR pathway genes RHEB and RICTOR, multiple autophagy regulators and mediators including TP53INP2, CAPN6, ATG16L1, GABARAPL1 and the gene encoding aggregation-prone protein in Alzheimer disease, APP. This property may allow miR-142-3p to serve as a converging point for multiple signaling inputs to regulate the autophagy-lysosomal activities.
Figure 3.
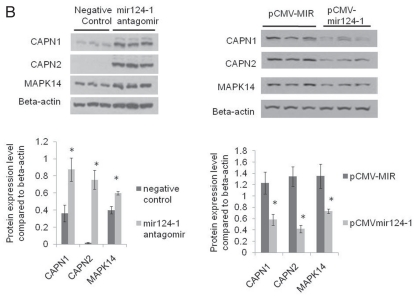
Autophagy and lysosomal genes contain sequences recognized by miR-130, 98, 124, 204 and 142. Validation of regulation of CAPN1 and CAPN2 by miR-124. Western blot analyses after transfection of control RNA and mir124 antagomir into HEK cells indicated that mir124 antagomir led to increased MAPK14, CAPN1 and CAPN2 protein levels. Western blot analyses after transfection into HEK cells of pCMV-MIR control vector and pCMV-mir-124 vector that expresses miR-124 indicated that miR-124 expression led to decreased MAPK14, CAPN1 and CAPN2 protein levels. Different ECL developing conditions were used in the left and right panels to allow sufficient dynamic range of signal for comparison of both upregulated and downregulated proteins in the two different experiments. β-actin was used as a loading control. n = 6 for each group. DATA = mean ± SEM. *p < 0.05 by Student t-test compared to control.
Of interest, we identified that miR-124 potentially regulates 52 target genes in the autophagy-lysosomal pathway. The following nine have been validated in the literature: ITGB1, MAPK14, KLHL24, GNAI3, TOM1L1, SLC17A5, LITAF, ANKRD27 and DRAM1. Interestingly, we found four calpains (CAPN1, CAPN2, CAPNS1 and CAPN6) that could be targeted by miR-124. To validate, we transfected a plasmid expressing miR-124 and an antagomir RNA of mir-124 into HEK cells. We used western blot analyses to examine CAPN1 and CAPN2 expression because microRNA can regulate both mRNA degradation and translation from the mRNA. MAPK14 western blot analyses were used as a positive control64 (Fig. 3B). We found that CAPN1 and CAPN2 are altered by the miR-124 expression and by miR-124 antagomir RNA transfection. This result validated our approach and suggested a novel regulatory mechanism for calpains by miR-124 microRNA.
Analyses of autophagy-lysosomal genes for their participation in intracellular signaling, human diseases and nervous system phenotypes in murine genetic models.
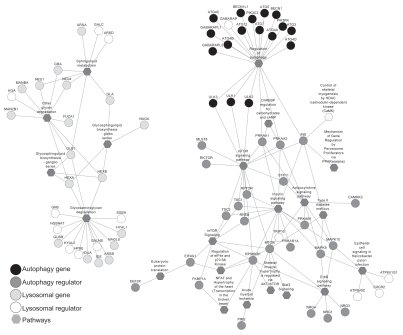
Using the ToppGene knowledgebase65,66 and Cytoscape,67 we examined and displayed functional networks based on target gene interactions and control of cell signaling pathways.67–69 In this analysis, mTOR and insulin signaling pathways were shown to play a key role in autophagy regulation, whereas glycosaminoglycan degradation, glycosphingolipid biosynthesis, sphingolipid metabolism and glycan degradation pathways were involved in lysosomal gene regulation (Fig. 4 and Sup. Material Table 4). Again, regulation of lipid metabolism seems to be an important function of lysosomal gene products and lysosomal regulators.
Figure 4.
Unbiased bioinformatics analyses identified that mTOR and insulin signaling pathways play a key role in autophagy regulation, whereas glycosaminoglycan degradation, glycosphingolipid biosynthesis, sphingolipid metabolism, glycosphingolipid biosynthesis and glycan degradation pathways are involved in lysosomal gene regulation.
Next we asked whether our findings of transcriptional and post-transcriptional regulators of autophagy and lysosomal genes could be linked to known pathologies. A list of known lysosomal proteins with involvement of diseases is presented in Supplemental Table 5. We used OMIM, AlzGene, PDGene, SZGene and the ToppGene knowledgebase65,66 and built disease by gene binary matrices using CIMminer (cluster method Jaccard) (http://discover.nci.nih.gov/cimminer). To represent this analysis we used a heat map, as shown in Figure 5A. This unbiased analysis revealed two clusters of genes highly enriched for a wide range of diseases including tuberous sclerosis and neuronal ceroid-lipofuscinoses. Interestingly, our gene sets significantly overlap with those associated with neurodegenerative diseases including Alzheimer and Parkinson diseases as well as neoplasms including lung, liver and prostatic neoplasms and sepsis (Fig. 5A and Sup. Table 6). The top five genes that have been identified as involved in four out of the eight human diseases encode estrogen receptor, TGFβ, cathepsin D, TNF and p53. Whether autophagy has beneficial or adverse effects in sepsis warrants further investigation. Interestingly, a recent study reported a marked increase in hepatocyte autophagocytosis in both patients with sepsis and in a clinically relevant animal model of sepsis.70
Figure 5.
(A) Unbiased bioinformatics analyses also found that autophagy and lysosomal genes are most implicated in Alzheimer and Parkinson diseases, tuberous sclerosis, neuronal ceroid-lipofuscinoses, sepsis and lung, liver and prostatic neoplasms, implicating a critical role of autophagy-lysosomal pathway in these diseases. Shown are heat maps of relevance of genes and diseases. (B) A summary of the extent that knockout mice deficient of these genes exhibit nervous system phenotypes. Shown are heat maps of relevance of genes and knockout phenotypes.
Finally, we analyzed the autophagy-lysosomal genes for nervous system and behavior-related phenotypes using the mammalian phenotype ontology and associated genes from MGI (Fig. 5B). Interestingly, we found highly enriched motor and motor coordination phenotypes along with abnormal brain morphology. Other enriched phenotypes included abnormal neuron apoptosis, brain size, glia cell morphology and gliosis, suggesting that disruptions of the autophagy-lysosomal genes may play a role in these phenotypes. Forty-three genes have been shown to exhibit 10 or more nervous system phenotypes out of the 29 phenotypes that constitute the top hits in the database (Sup. Table 7).
Summary.
In summary, we have analyzed 416 key genes involved in the autophagy-lysosomal pathway, identified common transcription factor binding sites and microRNA binding sites for these genes, and identified their involvement in mTOR, insulin and glycosphingolipid synthesis pathways and nervous system and neoplasm pathogenesis. Interestingly, their potential association with nervous system pathologies and behavior-related phenotypes in mouse models supports the utility of the systems biology approach we have adopted. We envision our findings will help in our understanding of the regulation of the autophagy-lysosomal activities and shed light on their role in human nervous system-related abnormalities and other diseases.
Methods
Gene lists compilation.
We mined Gene Ontology27 annotations, an autophagy database (http://autophagy.lu/) and KEGG pathways28 to compile a preliminary data set of autophagy and lysosomal genes and their regulators. The compiled lists were further enhanced by manual curation of published literature.3,8,29,30 The final list comprises 38 autophagy genes, 161 autophagy regulator genes, 64 lysosomal genes and 167 lysosome regulator genes. Although these gene lists may not be exhaustive and extensive, we reasoned that they represented the best available current knowledge related to autophagy and lysosome regulation in the mammals and can provide key insights into the biology of autophagy-lysosomal pathway.
Functional enrichment analysis.
In order to assess the relative functional enrichment of the four groups of genes (autophagy, autophagy regulators, lysosome and lysosomal regulators), we first grouped the four lists together into a single list (416 unique genes) and examined this list for relative enrichment of genes associated with Gene Ontology, pathways, mouse phenotypes, human diseases, transcription factor binding sites (TFBS) and microRNAs. We used the ToppGene server for this purpose.65,66 In the case of TFBS, we mined the catalog of human, mouse, rat and dog conserved regulatory motifs in promoters,71 again using ToppGene server.65 The microRNA targets were based on TargetScan (Release 5.1),72 predictions. The gene ontology annotations were obtained from NCBI while the pathway annotations were based on several resources compiled and stored in the ToppGene knowledgebase (reviewed in ref. 65 and 66). The mouse phenotype was obtained from MGI, and for this study we limited it to nervous system and behavior-related phenotypes only. The disease-gene data was from multiple sources including OMIM and specialized databases related to neurodegenerative diseases (AlzGene, PDGene and SZGene) and stored in the ToppGene knowledgebase (reviewed in ref. 65 and 66). For the enrichment analysis, a p-value cut-off of 0.05 (with Bonferroni correction) was used for all categories except for microRNA. In the case of microRNA, enrichment analysis using a p-value <0.05 (Bonferroni) did not yield any results. We therefore used a false discovery rate (FDR <0.1; represents the rate of potential falsely identified genes in the enrichment results).
Comparative network and heat map-based visualization of enrichment analysis.
For obtaining a comparative network view of enriched pathways, phenotypes, diseases and putative shared conserved TFBSs and microRNAs and the corresponding four categories of gene sets we used Cytoscape,67 a JAVA-based bioinformatics software package for visualizing, modeling and analyzing molecular and genetic interaction networks. Cytoscape provides a unique in silico approach to examine and display functional and transcriptional networks based on putative TFBS and target gene interactions characterized from either previous studies or computational predictions.67–69 The enrichment results for each category (e.g., Pathways, TFBS, etc.) were converted into individual Cytoscape-compatible files and loaded into Cytoscape along with attribute files for node size and color to visualize the enrichment results as networks. Additionally, when building the networks we also included protein-protein interaction data for the 416 genes. For the mammalian phenotype and disease enrichment results we used heatmap views. Phenotype by gene and disease by gene binary matrices were built using the enriched phenotype and disease terms, and the candidate genes and the heat map were built using CIMminer (cluster method Jaccard) (http://discover.nci.nih.gov/cimminer).
Cell culture and transfection.
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum (FBS). Cells were seeded in six-well plates at 10% confluence before transfection. Transfection was performed by using Lipofectamine Reagent (Invitrogen 11668-019) according to the manufacturer's protocols. At 24 hours after transfection, RNA was harvested using Trizol reagent (Invitrogen 15596-026). pcDNA3.0-TFEB and control vector pcDNA3.0 are the gifts from Dr. Marco Sardiello, Baylor College. pCMV-SREBP1 and control vector pCMVXL5 are from Origene (SC117502, PCMV6XL5). miR-124 antagomir and negative control from Dharmacon (IH-300592-06-0005, IN-001005-01-05) were transfected to HEK cells and protein extracts were made 24 hrs after transfection. miR-124 expression vector and negative controls from Origene (SC400060, PCMVMIR) were transfected to HEK cells and protein extracts were made 48 hrs after transfection.
Quantitative real-time PCR.
RNA was isolated from cells using TRIzol (invitrogen 15596-026) according to the manufacturer's protocol. Three micrograms of RNA was used to convert to cDNA using iScriptTM cDNA Synthesis Kit (Bio-Rad 170-8891) according to the manufacturer's protocol. Quantitative real-time PCR was performed with the SYBR Green Mastermix (Invitrogene 4364346) with the following conditions: 95°C, 5 min; (95°C, 10 s; 60°C, 10 s; 72°C, 15 s) ×40. Real-time quantitative RT-PCR results were normalized against an internal control (GAPDH). Oligonucleotide sequences are listed in Table 2.
Table 2.
Real-time RT-PCR primers
| Genes | NCBI reference sequence ID | Primers | |
| UVRAG | NM_003369.3 | F | TGT ATG CGG TGT CAA GTT GCC T |
| R | TCA TGG AGA CCA GAT GTG CAG T | ||
| LAMP1 | NM_005561.3 | F | CCA GTT CGG GAT GAA TGC AAG T |
| R | TTG GCA GCT TTA AAG GCA GGG T | ||
| GABARAP | NM_007278.1 | F | AGC TGT ACC AGG AAC ACC ATG A |
| R | AGC TTC ACA GAC CGT AGA CAC T | ||
| CTSS | NM_004079.3 | F | TGG CTT CAT GAC AAC GGC TT |
| R | TGA ACA TGT GGC AGC ACG AT | ||
| CTSD | NM_001909.3 | F | TTC CCG AGG TGC TCA AGA ACT ACA |
| R | TGT CGA AGA CGA CTG TGA AGC ACT | ||
| CTNS | NM_004937.2 | F | AAT GTG AGT CAA GCG TCA GCC T |
| R | TGC ATT TAA TGG TGG CCG CA | ||
| CLN3 | NM_000086.2 | F | AAG AAG AAG CAG AGA GCG CA |
| R | ACA ATG TAC CAC AGC AGA CCC T | ||
| ATP6V0C | NM_001694.2 | F | TCA GCC TCT ACA AGA GCT TCC T |
| R | TCA GGA TCA TGC CCA CGA AT | ||
| VPS18 | NM_020857.2 | F | TCC AGC AAT CAG CTG TGC AT |
| R | TGG TTG GGC TCA TTT GCC TT | ||
| PPT1 | NM_001142604.1 | F | TGG CCA AGC CAA GGA AAC CAT T |
| R | AGC TGT CCT GCA TTG TCC AT | ||
| DRAM1 | NM_018370.2 | F | AAC AGT CTC TCG ACA TGC CAC A |
| R | TGG ATT CCA CTC CAG CTT GGT T | ||
| CAPN1 | NM_005186.2 | F | AGC AAA CAC AAA GAC CTG CGG A |
| R | TTC CCA TTG CCA TCA CGA TCC A | ||
| CAPN2 | NM_001146068.1 | F | TCA GGA GAC TGT TTG CCC AGT T |
| R | TTG GCG CTT TGC TAG AAC CCT T | ||
| CAPNS1 | NM_001003962.1 | F | AAC TTC ATC AGC TGC TTG GTC AGG |
| R | ACC TGG ATT TGT CCA GTG CCA T | ||
| CAPN6 | NM_014289.2 | F | TGC AAA GCT GCT AGG CTG TT |
| R | TTT CAG CCA ATG TGC CCG TGA A |
Western blot analysis.
Total cellular extracts were collected in lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, plus protease inhibitor mixture (Roche 04693132001). Homogenates were centrifuged at 10,000x g for 15 min at 4°C. Protein concentrations were determined by detergent-compatible protein assay (Bio-Rad 500-0013, 500-0114). Thirty micrograms of protein was resolved on 12% SDS-PAGE gel and transferred to nitrocellulose membranes. Membranes were blocked in 5% nonfat dry milk in TBST (50 mM Tris-HCl, 150 mM NaCl, pH 7.4, 0.1% Tween 20) for 30 min at room temperature. The membranes were then incubated overnight at 4°C with primary antibodies: monoclonal anti-β-actin (1:5,000) (Sigma A5441); anti-calpain 1 1:1,000, anti-calpain 2 1:1,000 and anti-MAPK14 1:1,000 (Cell Signaling 2556S, 2539S and 9212S). The membranes were then washed four times with TBST and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. After washing four times with TBST, the membranes were developed using enhanced chemiluminescence (ECL) substrate kit (Pierce 32106). We used NIH imaging software to quantify the western blot band intensity.
Acknowledgements
We thank Dr. Victor Darley-Usmar for critical discussions and for editing the manuscript. We acknowledge the help of Ron Bryson, Technical Writer, Division of Biomedical Informatics, CCHMC, OH USA, in editing the article. This work was partially supported by the grants from National Institutes of Health DK078392 (Gene Expression and Sequencing Core and Bioinformatics Core, Digestive Disease Research Core Center in Cincinnati) (A.J.), and University of Alabama at Birmingham start-up support, NIHR01-NS064090 and a VA merit award (J.Z.).
Supplementary Material
References
- 1.de Duve C. The lysosome turns fifty. Nat Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 2.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider L, Zhang J. Lysosomal function in macro-molecular homeostasis and bioenergetics in Parkinson's disease. Mol Neurodegener. 2010;5:14. doi: 10.1186/1750-1326-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 5.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 6.Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- 7.Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008;13:718–736. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- 8.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 9.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuertes G, Martin de Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci USA. 2010;107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saftig P, Tanaka Y, Lullmann-Rauch R, von FK. Disease model: LAMP-2 enlightens Danon disease. Trends Mol Med. 2001;7:37–39. doi: 10.1016/s1471-4914(00)01868-2. [DOI] [PubMed] [Google Scholar]
- 13.Steinfeld R, Reinhardt K, Schreiber K, Hillebrand M, Kraetzner R, Bruck W, et al. Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am J Hum Genet. 2006;78:988–998. doi: 10.1086/504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siintola E, Partanen S, Stromme P, Haapanen A, Haltia M, Maehlen J, et al. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain. 2006;129:1438–1445. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- 15.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–1728. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen V, Lindfors M, Ng J, Paetau A, Swinton E, Kolodziej P, et al. Cathepsin D expression level affects alpha-synuclein processing, aggregation and toxicity in vivo. Mol Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, et al. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haidar B, Kiss RS, Sarov-Blat L, Brunet R, Harder C, McPherson R, et al. Cathepsin D, a lysosomal protease, regulates ABCA1-mediated lipid efflux. J Biol Chem. 2006;281:39971–39981. doi: 10.1074/jbc.M605095200. [DOI] [PubMed] [Google Scholar]
- 20.Mutka AL, Haapanen A, Kakela R, Lindfors M, Wright AK, Inkinen T, et al. Murine cathepsin D deficiency is associated with dysmyelination/myelin disruption and accumulation of cholesteryl esters in the brain. J Neurochem. 2010;112:193–203. doi: 10.1111/j.1471-4159.2009.06440.x. [DOI] [PubMed] [Google Scholar]
- 21.Pacheco CD, Lieberman AP. The pathogenesis of Niemann-Pick type C disease: a role for autophagy? Expert Rev Mol Med. 2008;10:26. doi: 10.1017/S146239940800080X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttenhower C, Hibbs MA, Myers CL, Caudy AA, Hess DC, Troyanskaya OG. The impact of incomplete knowledge on evaluation: an experimental benchmark for protein function prediction. Bioinformatics. 2009;25:2404–2410. doi: 10.1093/bioinformatics/btp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sardiello M, Palmieri M, di RA, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2010 doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 26.Demarchi F, Bertoli C, Copetti T, Eskelinen EL, Schneider C. Calpain as a novel regulator of autophagosome formation. Autophagy. 2007;3:235–237. doi: 10.4161/auto.3661. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu ZZ, Valencia JC, Huang H, Chi A, Shabanowitz J, Hearing VJ, et al. Comparative Bioinformatics Analyses and Profiling of Lysosome-Related Organelle Proteomes. Int J Mass Spectrom. 2007;259:147–160. doi: 10.1016/j.ijms.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubke T, Lobel P, Sleat DE. Proteomics of the lysosome. Biochim Biophys Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 33.Galibert MD, Carreira S, Goding CR. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced Tyrosinase expression. EMBO J. 2001;20:5022–5031. doi: 10.1093/emboj/20.17.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 35.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yogev O, Goldberg R, Anzi S, Yogev O, Shaulian E. Jun proteins are starvation-regulated inhibitors of autophagy. Cancer Res. 2010;70:2318–2327. doi: 10.1158/0008-5472.CAN-09-3408. [DOI] [PubMed] [Google Scholar]
- 38.Motohashi H, Kimura M, Fujita R, Inoue A, Pan X, Takayama M, et al. NF-E2 domination over Nrf2 promotes ROS accumulation and megakaryocytic maturation. Blood. 2010;115:677–686. doi: 10.1182/blood-2009-05-223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuroha T, Takahashi S, Komeno T, Itoh K, Nagasawa T, Yamamoto M. Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption. J Biochem. 1998;123:376–379. doi: 10.1093/oxfordjournals.jbchem.a021947. [DOI] [PubMed] [Google Scholar]
- 40.Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Jiang T, Li J, Proctor G, McManaman JL, Lucia S, et al. Regulation of renal lipid metabolism, lipid accumulation and glomerulosclerosis in FVBdb/db mice with type 2 diabetes. Diabetes. 2005;54:2328–2335. doi: 10.2337/diabetes.54.8.2328. [DOI] [PubMed] [Google Scholar]
- 44.Yokoyama K, Hayashi H, Hinoshita F, Yamada A, Suzuki Y, Ogura Y, et al. Renal lesion of type Ia glycogen storage disease: the glomerular size and renal localization of apolipoprotein. Nephron. 1995;70:348–352. doi: 10.1159/000188616. [DOI] [PubMed] [Google Scholar]
- 45.Man N, Chen Y, Zheng F, Zhou W, Wen LP. Induction of genuine autophagy by cationic lipids in mammalian cells. Autophagy. 2010;6:449–454. doi: 10.4161/auto.6.4.11612. [DOI] [PubMed] [Google Scholar]
- 46.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovsan J, Bashan N, Greenberg AS, Rudich A. Potential role of autophagy in modulation of lipid metabolism. Am J Physiol Endocrinol Metab. 2010;298:1–7. doi: 10.1152/ajpendo.00562.2009. [DOI] [PubMed] [Google Scholar]
- 48.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weidberg H, Shvets E, Elazar Z. Lipophagy: selective catabolism designed for lipids. Dev Cell. 2009;16:628–630. doi: 10.1016/j.devcel.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hill BG, Haberzettl P, Ahmed Y, Srivastava S, Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem J. 2008;410:525–534. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 53.Sessa A, Meroni M, Battini G, Righetti M, Maglio A, Tosoni A, et al. Renal involvement in Anderson-Fabry disease. J Nephrol. 2003;16:310–313. [PubMed] [Google Scholar]
- 54.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 55.Han D, Yang B, Olson LK, Greenstein A, Baek SH, Claycombe KJ, et al. Activation of autophagy through modulation of 5′-AMP-activated protein kinase protects pancreatic beta-cells from high glucose. Biochem J. 2010;425:541–551. doi: 10.1042/BJ20090429. [DOI] [PubMed] [Google Scholar]
- 56.Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci USA. 2000;97:11371–11376. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Satoh JI. MicroRNAs and Their Therapeutic Potential for Human Diseases: Aberrant MicroRNA Expression in Alzheimer's Disease Brains. J Pharmacol Sci. 2010;114:269–275. doi: 10.1254/jphs.10r11fm. [DOI] [PubMed] [Google Scholar]
- 58.Sonntag KC. MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res. 2010;1338:48–57. doi: 10.1016/j.brainres.2010.03.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18:233–240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe E, Muenzer JT, Hawkins WG, Davis CG, Dixon DJ, McDunn JE, et al. Sepsis induces extensive autophagic vacuolization in hepatocytes: a clinical and laboratory-based study. Lab Invest. 2009;89:549–561. doi: 10.1038/labinvest.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.