Abstract
Polar auxin transport is crucial for the regulation of auxin action and required for some light-regulated responses during plant development. We have found that two mutants of Arabidopsis—doc1, which displays altered expression of light-regulated genes, and tir3, known for its reduced auxin transport—have similar defects and define mutations in a single gene that we have renamed BIG. BIG is very similar to the Drosophila gene Calossin/Pushover, a member of a gene family also present in Caenorhabditis elegans and human genomes. The protein encoded by BIG is extraordinary in size, 560 kD, and contains several putative Zn-finger domains. Expression-profiling experiments indicate that altered expression of multiple light-regulated genes in doc1 mutants can be suppressed by elevated levels of auxin caused by overexpression of an auxin biosynthetic gene, suggesting that normal auxin distribution is required to maintain low-level expression of these genes in the dark. Double mutants of tir3 with the auxin mutants pin1, pid, and axr1 display severe defects in auxin-dependent growth of the inflorescence. Chemical inhibitors of auxin transport change the intracellular localization of the auxin efflux carrier PIN1 in doc1/tir3 mutants, supporting the idea that BIG is required for normal auxin efflux.
Keywords: Auxin transport, light signaling, Arabidopsis
Regulation of plant growth and development by genetic and environmental signals relies on tightly controlled spatial and temporal distribution of plant hormones. Changes in the level of the auxin, indole-3-acetic acid (IAA), during light-regulated growth responses have been described in several systems (Jones et al. 1991; Behringer and Davies 1992). However, until recently, evidence directly linking light and auxin signal transduction pathways has been lacking. Several gene families encoding primary-response auxin-inducible transcripts have been characterized, including the Aux/IAA genes, the SAUR genes, and the GH3-like genes (Abel and Theologis 1996). In Arabidopsis, gain-of-function mutations in the Aux/IAA genes SHY2/IAA3, AXR2/IAA7, and AXR3/IAA17 promote characteristics of photomorphogenetic development such as short hypocotyls and development of leaves in etiolated seedlings (Tian and Reed 1999; Nagpal et al. 2000). Interestingly, some Aux/IAA proteins may be direct targets of phytochromes because they are phosphorylated by phytochrome A in vitro (Colon-Carmona et al. 2000). The phenotypes caused by mutations in the auxin-responsive GH3-like genes, FIN219 and DFL1, also suggest a role for auxin in light-regulated development. Mutations in FIN219 result in reduced far-red light–mediated inhibition of hypocotyl elongation and induction of light-regulated genes (Hsieh et al. 2000), whereas mutations in the DFL1 gene result in a short hypocotyl phenotype in light-grown seedlings that increases in severity with increasing light fluence (Nakazawa et al. 2001). The NPH4/ARF7 locus encodes a member of the ARF family of proteins, thought to function as transcriptional activators of auxin-regulated genes. Mutations in this gene cause impaired hypocotyl phototropism and other differential growth responses associated with changes in auxin response (Stowe-Evans et al. 1998; Harper et al. 2000).
In aerial parts of the adult plant, IAA is transported basipetally from its site of synthesis at the shoot apex toward the roots in a process referred to as polar auxin transport (Lomax et al. 1995). In roots, auxin moves in the opposite direction in different cell types, acropetally through the root stele and basipetally through epidermal cells (Tsurumi and Ohwaki 1978; Meuwly and Pilet 1991). Disruption of auxin transport affects critical processes such as embryo development, vascular differentiation, stem elongation, flower and root development, apical dominance, and tropic responses (Lomax et al. 1995).
Physiological studies have indicated that the polar auxin transport system requires the activity of specific auxin influx and efflux carriers located on the plasma membrane of transporting cells. These carriers act to move auxin through files of cells by successively transporting auxin into and out of adjacent cells in the file. Net auxin movement is polar because the efflux carriers are asymmetrically localized in transporting cells (Lomax et al. 1995). Chemical inhibitors referred to as phytotropins, such as N-1-naphthylphthalamic acid (NPA), specifically inhibit the efflux component of polar auxin transport (Katekar and Geissler 1977; Hertel et al. 1983), apparently by binding to a plasma membrane–associated protein called the NPA-binding protein (NBP; Muday et al. 1993; Bernasconi et al. 1996). The identity of this protein is not known, but some experiments suggest that it is distinct from the efflux carrier and may act to regulate auxin transport (Morris et al. 1991). A role of intracellular protein trafficking in the regulation of auxin transport has been proposed, because treatments of tobacco cultured cells with the inhibitor of vesicular transport brefeldin A (BFA) are able to block the auxin efflux system (Delbarre et al. 1998).
Much of our current knowledge of the components of polar auxin transport comes from genetic studies in Arabidopsis. The pin1 mutants have reduced auxin transport and a phenotype similar to plants treated with auxin transport inhibitors, including alterations in vascular development and the formation of pin-like structures instead of flower buds (Okada et al. 1991; Galweiler et al. 1998). PIN1 encodes a membrane protein that likely functions as an auxin efflux carrier expressed in vascular tissues (Galweiler et al. 1998). A second related protein, EIR1/AGR1/AtPIN2, performs a similar function in the epidermal and cortical cells in the meristematic and elongation zone of the root (Chen et al. 1998; Luschnig et al. 1998; Muller et al. 1998). Immunolocalization studies have shown that PIN2 is asymmetrically localized at the periclinal side of the carrier cells, and a model has been proposed that explains directional auxin fluxes in the root (Muller et al. 1998). Pid1 mutants are similar to pin1 in appearance and are also deficient in auxin transport (Okada et al. 1991). PID1 encodes a serine-threonine protein kinase, suggesting a signaling or regulatory function (Christensen et al. 2000). The influx carrier, an amino acid permease-like protein, is encoded by the AUX1 gene (Bennett et al. 1996; Marchant et al. 1999). Roots of the aux1 mutant are defective in gravitropic responses and resistant to exogenous auxin (Pickett et al. 1990).
tir3 (transport inhibitor response 3) mutants were isolated in a screen for resistance to the inhibitory effects of NPA on root elongation (Ruegger et al. 1997). The mutants have a pleotropic phenotype, including decreased inflorescence height, decreased petiole and root length, fewer and shorter siliques, and reduced apical dominance. In addition, tir3 plants display reduced lateral root production, a process that depends on polar auxin transport from the shoot (Reed et al. 1998). These defects are associated with decreased polar auxin transport in the inflorescence and a reduction in the number of NPA-binding sites. These results suggest that the NBP has a positive role in polar auxin transport and that TIR3 either encodes the NBP or is required for its expression, localization, or stabilization.
An interesting link has been established between polar auxin transport and light responses in hypocotyl growth in Arabidopsis. Steindler and coworkers (1999) have reported that the light-regulated ATHB-2 gene acts as a negative regulator of shade-induced hypocotyl elongation, and they found this response to be altered in the auxin response mutant axr1-2. A role in this response for auxin transport was supported by experiments in which treatments with NPA caused reduced hypocotyl elongation in response to low red light to far-red light (R:FR) ratios (Steindler et al. 1999). These results are consistent with experiments performed with Arabidopsis seedlings in which auxin transport was blocked with NPA, suggesting that auxin transport is required for phytochrome-regulated hypocotyl elongation in light-grown plants (Jensen et al. 1998). Additional data indicating a strong connection between light and auxin transport include reported changes in auxin transport rate and intensity induced by dim red light treatments in cucumber seedlings (Shinkle et al. 1998) and differential growth mediated by lateral transport of auxin triggered by phototropic responses in pea and tobacco (Li et al. 1991).
At the molecular level, one of the best-studied plant responses to light is the transcriptional regulation of nuclear genes encoding photosynthetic proteins, including the chlorophyll a/b-binding proteins of photosystem II (CAB; Terzaghi and Cashmore 1995). CAB genes are expressed at high levels in the light in green plants and at low levels in dark-grown seedlings. The DOC1 gene (dark overexpression of CAB) was identified as a putative component of light signal transduction pathways regulating this response because mutations in the locus lead to overexpression of CAB genes in the dark (Li et al. 1994). Unlike more pleiotropic mutants with increased CAB mRNA levels in the dark, such as shy2-1 and the det/cop/fus class (Kim et al. 1998; Hardtke and Deng 2000), doc1 seedlings have etiolated morphology when germinated in the dark. Thus, the phenotype of dark-grown doc1 plants suggests that the expression of some light-regulated genes could be genetically separated from the morphological changes that occur during the transition from etiolated to photosynthetic growth.
In this report, we show that doc1 mutants are allelic to tir3 mutants. We describe the cloning of the gene identified by these mutants that we have renamed BIG, owing to its large size. The BIG gene is predicted to encode an enormous protein with significant identity to the Drosophila protein Calossin/Pushover. To better understand the connection between the light- and auxin-related phenotypes, we have performed expression-profiling experiments. These results reveal that altered expression of light-regulated genes in etiolated doc1 seedlings can be suppressed by increasing endogenous auxin levels. In addition, we have shown that NPA has an unusual effect on PIN1 localization in the doc1/tir3 mutants. We propose that the reduction in levels of NBP in the doc1/tir3 mutants affects cellular localization or function of auxin efflux carriers.
Results
Isolation of new doc1 alleles
To further characterize the DOC1 locus, we isolated new doc1 alleles using two approaches. In the first approach, a doc1 ap3-1 double mutant in the Columbia background was constructed. The ap3-1 mutant contains a temperature-sensitive allele of the apetala3 homeotic gene, which is male sterile at the nonpermissive temperature (Sablowski and Meyerowitz 1998). This feature was used to facilitate crosses of γ-ray–mutagenized pollen from DOC1 plants of the Landsberg erecta (La-er) accession onto the doc1 ap3-1 mutant. We screened ∼2000 M1 lines for the doc1 visible phenotype and identified doc1-2, a line that contained Landsberg DNA at PCR markers <0.2 cM from docl. The progeny of this line consisted of plants with reduced height and increased numbers of inflorescences. Our second approach to isolate new doc1 alleles was to screen transferred-DNA (T-DNA) mutagenized Arabidopsis lines for the morphological phenotype of doc1. During this screen we identified the doc1-3 mutant and confirmed that it was a new doc1 allele by complementation analysis.
The tir3-1 and doc1-1 mutants define mutations in a single gene and have similar defects in auxin transport and CAB gene expression
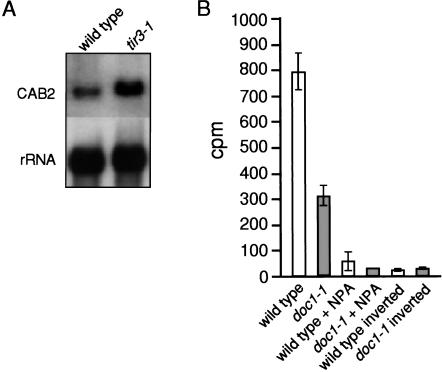
The Arabidopsis tir3 mutations map to the same region of chromosome 3 as the doc1 mutations and cause very similar morphological defects (Li et al. 1994; Ruegger et al. 1997). Although tir3 and doc1 mutants were isolated in very different mutant screens, complementation tests indicated that tir3-1 and doc1-1 were alleles (data not shown). To determine if each mutation conferred a similar set of defects, we performed auxin transport and CAB gene expression studies on the doc1-1 and tir3-1 alleles, respectively (Fig. 1). As for tir3-1, doc1-1 stem segments transported approximately one third the amount of IAA compared with that of wild type during an 18-h period, indicating that doc1-1 has an auxin transport defect (Fig. 1B). Likewise, an analysis of CAB2 gene expression in light- and dark-grown tir3-1 seedlings showed that CAB2 mRNA was more abundant in the dark-grown seedlings compared with wild-type seedlings (Fig. 1A). Thus, the tir3 and doc1 mutations appear to have similar, if not identical, effects on these diverse aspects of plant growth and development.
Figure 1.
(A) CAB expression is increased in dark-grown tir3-1 seedlings. Five micrograms of total RNA from 5-day-old dark-grown seedlings was probed with a CAB2 cDNA. To confirm equal loading, the blot was stripped and re-probed with an 18S rRNA gene. (B) doc1-1 inflorescence stems are deficient in polar auxin transport. Twenty-five millimeters of stem was excised, and the apical end was placed in nutrient solution containing 1 μM 14C-IAA ± NPA for 18 h. The basal end of the inverted segments was placed in nutrient solution where indicated. The amount of radioactive IAA (CPM) transported to the basal end of the stem was assayed by liquid scintillation. Each column represents the mean of 10 stem segments. The average of three stem segments was used for the control columns. Bar, SEM.
Expression profiling of doc1 and doc1 yucca1 mutants
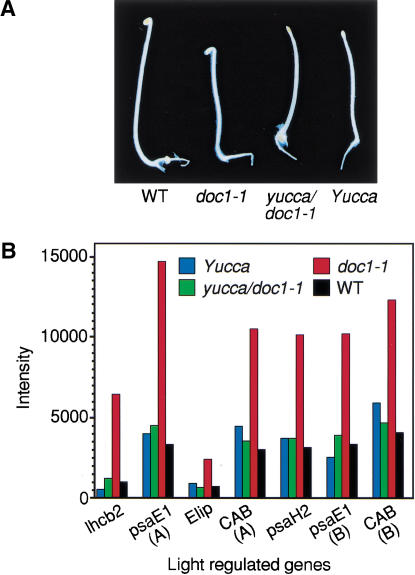
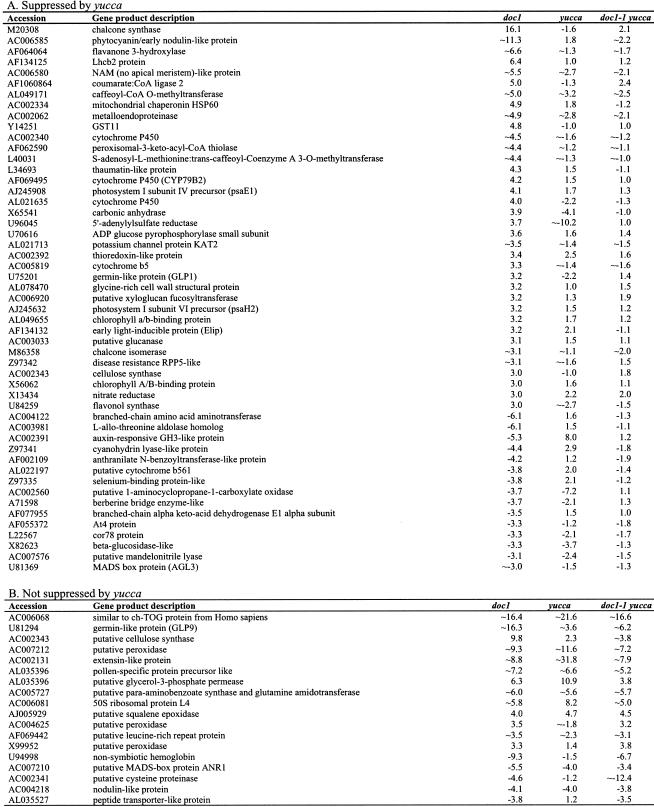
To further examine the effects of the doc1/tir3 mutations on light-regulated gene expression, we performed expression-profiling experiments using the commercially available Arabidopsis Affymetrix GeneChip. These chips monitor the expression of >8200 genes. Of these, >5000 genes gave signals that were significantly above the background in both dark-grown wild-type and doc1-1 seedlings. Any genes that gave a signal below or near the background were assigned as absent on the chip and were not subjected to further analysis. Among 77 genes expressed at least threefold higher in dark-grown doc1-1 than in wild type, we identified many known light-regulated genes, including components of the photosynthetic machinery (Fig. 2B), nitrate reductase, and genes encoding enzymes participating in flavonoid biosynthesis (Table 1). Interestingly, multiple enzymes for the latter pathway were overexpressed, suggesting increased production of anthocyanins and related ultraviolet-protective pigments. When the doc1-1 mutant was grown in the light, transcripts for the CAB genes, light-regulated genes, and flavonoid biosynthetic genes that were overexpressed in dark-grown doc1-1 accumulated to similar levels in the mutant compared with the wild type (Li et al. 1994; data not shown). There were also 22 genes, of no particular classification, that had at least a threefold decrease in expression level in doc1-1. Genes with predicted functions with altered expression in dark-grown doc1-1 are shown in Table 1. Because doc1-1 was originally isolated as a mutant with elevated expression of CAB genes in the dark and was later found to be allelic to tir3, we suspected that the derepression of CAB expression might be the result of altered auxin distribution. To test this hypothesis, we made use of a recently identified auxin overproducing mutant called yucca. The YUCCA gene encodes a flavin monooxygenase-like enzyme, and its overexpression in the yucca mutant has been shown to produce at least 50% more free IAA than wild type (Zhao et al. 2001). Because yucca has a phenotype affecting all parts of the seedling, we hypothesized that yucca would suppress doc1-1 if defects in auxin distribution were responsible for the observed doc1 phenotype. As expected, the dark-grown yucca doc1-1 double mutant looked similar to the yucca mutant (Fig. 2A), and light-regulated genes were not overexpressed (Fig. 2B). Of the 99 genes with altered expression in dark-grown doc1 mutants, 73 genes were restored to wild-type levels by overexpression of YUCCA (Table 1). These results suggest that altered levels of expression of light-regulated genes in dark-grown doc1 are due to lower concentrations of auxin in some cells, resulting from defective polar auxin transport. The hypothesis that auxin needs to be transported to specific target cells to repress the expression of light-regulated genes is consistent with the fact that exogenous auxin failed to suppress the altered CAB gene expression in dark-grown doc1 seedlings (data not shown). In the light, the aerial phenotype of the double-mutant yucca doc1-1 included long hypocotyls and epinastic cotyledons similar to the yucca mutant. However, the double mutant displayed defective lateral root formation like the doc1-1 mutant.
Figure 2.
Suppression of overexpression of light-regulated genes in dark-grown doc1 by auxin overproducing yucca. (A) Four-day-old dark-grown wild type, doc-1, yucca, and doc1-1 yucca double mutant. (B) Expression levels of some light-regulated genes in dark-grown seedlings analysed by microarray. The Affymetrix accession nos. for CAB(A), CAB(B), psaE1(A), psaE1(B), lhcb2, Elip, and psaH2 are 16004-s-at, 13213-s-at, 18088-i-at, 18089-r-at, 15153-at, 16637-s-at, and 18081-at, respectively. The CAB(A) oligos were designed based on the genomic sequence of the CAB gene (GenBank AL049655), and CAB(B) oligos were based on the mRNA sequence of the CAB gene (GenBank X56062). Both sets of oligos for the CAB gene may also pull out other homologous genes such as CAB2 in the experiments shown here. Oligos for both psaE1(A) and psaE1(B) were designed based on the mRNA sequence of the photosystem-I subunit IV precursor (GenBank AJ245908). Both sets of psaE1 oligos were designed for detecting the same gene, but neither of them is perfect based on Affymetrix rules for oligo design. There are <15 oligos in the set for psaE1(A), whereas some rules for oligo design were dropped for the psaE1(B). Microarray experiments were performed using instructions provided by Affymetrix.
Table 1.
mRNAs with altered expression in dark-grown doc1-1
∼ Indicates that the expression level in wild type is very low and the signals are near or below background. The fold changes of those genes are only approximate.
Interactions of tir3 with mutations affecting auxin response and transport
To further investigate the function of DOC1/TIR3 in auxin-regulated processes, we generated double mutants between tir3-1 and mutations affecting either auxin transport or response. The phenotypes of the tir3-1 pin1 and tir3-1 pid1 double mutants were very similar. In each case, the double mutant had smaller rosette leaves and shorter petioles than either single mutant (Fig. 3D,F). In the case of tir3-1 pid1, rosette leaves often had a fused or lobed appearance. In both double mutants, the primary inflorescence failed to elongate. Removal of rosette leaves revealed a very short inflorescence (1 to 2 mm) that terminated in a pin (Fig. 3F). Secondary inflorescences would frequently elongate significantly, although these also terminated in a pin (data not shown).
Figure 3.
The tir3 mutation confers a severe phenotype in combination with auxin transport and response mutants. Plants were photographed after 45 d of growth in potting soil (see Materials and Methods). All bars represent 3 cm. (A) wild type; (B) tir3-1; (C) pin-formed1-1; (D) tir3-1 pin-formed1-1; (E) pinoid1-3; (F) tir3-1 pinoid1-3 (insert: a primary inflorescence dissected from a tir3 pid1 plant; the arrow points to a terminal pin-shaped organ); (G) from left to right: wild type, tir3-1, axr1-12, and tir3-1 axr1-; (H) larger image of tir3-1 axr1-12 plant showing severe reduction in elongation between flowers.
The combination of tir3-1 with the auxin response mutant axr1-12 also results in a severe phenotype. AXR1 encodes a subunit of a heterodimeric RUB-activiting enzyme that functions in the RUB conjugation pathway. The RUB (related to ubiquitin) protein is conjugated to the cullin component of SCF-type ubiquitin protein ligases (E3). RUB conjugation appears to be particularly important for function of SCFTIR1, an E3 implicated in auxin response (Leyser et al. 1993; del Pozo et al. 1998; Gray et al. 1999). Thus, the axr1 mutants are deficient in all aspects of auxin response. The tir3-1 axr1-12 double mutant is an extreme dwarf with a phenotype much more severe than either single mutant (Fig. 3G,H). Like the tir3-1 pin1 plants, growth of the primary inflorescence in these double mutants arrested prematurely, in this case at ∼4 cm of height. Secondary inflorescences continued to elongate, indicative of a loss of apical dominance. In addition, there was very little internode elongation between adjacent flowers, so the flowers had an extremely bunched appearance. The phenotypes of these double mutants suggest that the combination of tir3-1 with either pin1, pid1, or axr1 results in a severe defect in auxin-dependent growth of the inflorescence, presumably associated with a reduction in internode elongation and the cessation of meristem activity shortly after the floral transition.
Identification of the DOC1/TIR3 gene
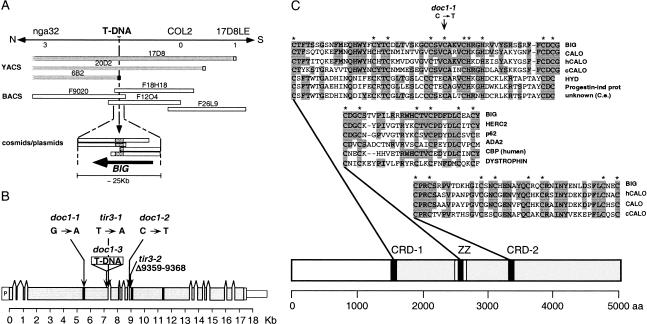
To investigate the molecular mechanism of DOC1/TIR3 action, we cloned the gene by a combination of chromosome walking and T-DNA tagging. First, we performed an extensive fine-mapping of the doc1 locus by identifying recombinant chromosomes from a mapping cross between doc1-1 (Col-0) and La-er using cleaved amplified polymorphic sequence (CAPS) and single sequence length polymorphism (SSLP) markers (Konieczny and Ausubel 1993; Bell and Ecker 1994). DOC1 was mapped between the SSLP marker nga32 and the CAPS marker 17D8LE on the top of chromosome 3. A set of overlapping yeast artificial chromosomes (YACs) spanning the region between both markers had been identified previously (Bartel and Fink 1995). YAC ends from yUP6B and yUP20D2, and the COL2 gene (Ledger et al. 1996) that maps between them, were isolated and subsequently used as probes to screen cosmid and bacterial artificial chromosome (BAC) libraries. Multiple cosmids and BACs overlapping the region between markers nga32 and 17D8LE were identified (Fig. 4A). Fragments from BACs F9O20, F18H18, and F26L9 were mobilized into a binary vector and together with cosmid clones were used to generate a complete contig.
Figure 4.
(A) Physical mapping and cloning of the BIG gene. The BIG gene was mapped in a ∼260-kb region on the top arm of chromosome 3 by identifying three recombinant chromosomes at marker nga32 and one recombinant chromosome at marker 17D8LE. This region was covered by YACs and BACs, cosmids, and large-insert plasmids. A T-DNA insertion in the doc1-3 allele identified by RFLP analysis in the ∼2-kb right-end fragment from the YAC yUP6B2 served to clone the BIG gene. (B) Structure of the BIG gene. Scale diagram of the BIG gene consisting of 14 exons (shaded boxes) and 13 introns confirmed by sequencing overlapping cDNA clones and RT–PCR-generated DNA fragments. Positions of the putative Zn-finger domains (solid black boxes) are shown. Molecular analysis of mutations in three doc1 and two tir3 alleles and their relative positions is shown. (C) BIG protein. The similarity between BIG, Calossin/Pushover (CalO), a large fragment of human calossin (hCALO; KIA0462 protein; accession no. AB007931) and a Caenorhabditis elegans predicted calossin (cCALO, AF003140) extends over >3000 amino acids at the C-terminal part of the protein (shaded gray). BIG is >60% identical to CALO at two cysteine-rich regions (shaded in black). The first cysteine-rich domain (CRD-1) has similarity to a domain present in ubiquitin ligases. HYD indicates hyperplastic discs L14644; Progestin-ind prot, human Progestin-induced protein AF006010; and unknown (C.e.) C. elegans putative protein Z81077. The second cysteine-rich domain (CRD-2) conserved among the pushover family of proteins, shows some similarity to Zn-finger domains in eukaryotic transcription factors. A third domain not conserved in the Calossin-like proteins is the ZZ domain, a Zinc-binding domain seen in both chromatin and cytoskeletal proteins. HERC2 indicates mouse HERC2 protein (AAD08658); p62, human phosphotyrosine-independent ligand p62B for the Lck SH2 domain B-cell isoform (U46752); ADA2, Saccharomyces cerevisiae Transcriptional Adaptor 2, (NP_010736); CBP (human), human CREB-binding protein, (AAC51331); DYSTROPHIN, human dystrophin (AAA53189).
Southern blot analyses using BAC F18H18 as a probe indicated that the doc1-3 allele contains a T-DNA insertion of ∼4 kb. DNA probes derived from this BAC were used in Southern blot analyses to further delimit the T-DNA insertion site to a 2-kb genomic region defined by the 6B2RE YAC end (Fig. 4A). Sequencing of T-DNA flanking sequences revealed that the insertion had disrupted an open reading frame (ORF) spanning 17,331 bp of genomic DNA represented as incomplete fragments in several clones of our contig. Given the remarkably large size of this gene (for Arabidopsis), we have renamed it BIG. The putative start codon (Net Start score, 0.770) was located immediately downstream of a DNA region of 257 bp with 60% identity to the 5′ region of an Arabidopsis L-isoaspartylmethyltransferase gene (Mudgett and Clarke 1996), including a putative TATA box (TATTAAT) at position −208 with respect to the putative start codon. BAC sequences from the putative gene were used to screen a cDNA library and to perform BLAST searches of the dbEST database. Several expressed sequence tags (ESTs) from Arabidopsis identical to regions of the putative transcript were present in GenBank. EST accession Z47688 included a poly(A) tail that indicated that mRNAs for this gene include a 3′ untranslated region (3′UTR) of 165 bases. Overlapping cDNA clones and RT–PCR products were assembled to reconstruct a transcript 15.4 kb long consisting of 14 exons (Fig. 4B).
To confirm that the BIG gene was DOC1/TIR3, we identified the specific mutations in five doc1 and tir3 alleles (Fig. 4B). The T-DNA insertion in doc1-3 was localized before position +7228 with respect to the putative start codon. The mutation in doc1-1 was a base pair change from G to A at position +5513 that causes an amino acid change from Cys to Tyr. The doc1-2 and tir3-1 mutations were also found to be single base pair changes (from C at position +8872 to T and from T at position +7331 to A, respectively) likely to affect pre-mRNA splicing as they disrupt the acceptor site of intron 7 and the donor site of intron 4, respectively. The tir3-2 mutation was a small deletion from position +8874 to +8883 (CTTCAGGTTC) that causes a frame-shift of the ORF, causing a stop codon after seven amino acids. Southern analysis using fragments of the gene as probes suggested that BIG is a single copy gene (data not shown). This was later confirmed as BLAST searches to the complete Arabidopsis genome failed to identify any DNA or protein sequence similar to the BIG gene.
BIG encodes a putative member of the Calossin/Pushover family of proteins
The predicted BIG protein is 5077 amino acids long (MW 566 kD, pI 5.7) and very similar in size and features to the Drosophila protein Calossin/Pushover (CalO; GenBank no. AF096897; Xu et al. 1998). CalO was first identified because it binds to calmodulin in vitro (Xu et al. 1998). Mutations in the pushover (push) locus cause defects in behavior, altered synaptic transmission at the neuromuscular junctions, male sterility, and defective achiasmatic segregation of chromosomes in female meiosis (Richards et al. 1996; Sekelsky et al. 1999). There are very similar predicted proteins of unknown function present in the human and Caenorhabditis elegans genomes (GenBank accession nos. AB007931 and AF003140, respectively). In addition, numerous ESTs from several animals and plants encoding putative proteins highly similar to these proteins have been identified, suggesting that BIG belongs to a class of proteins ubiquitously present in the plant and animal kingdoms.
Gapped alignment indicated that BIG and CalO are 28% identical at the amino acid level over >3000 amino acids at their C terminus, with a BLAST score of 885 and an E-value of 0.00 (Fig. 4C; Altschul et al. 1997). However, the similarity between these proteins is not uniform. Rather, it is particularly high (>60% identity) in several stretches 30 to 50 amino acids long, including two cysteine-rich domains that resemble Zinc fingers. The first cysteine-rich domain (CRD-1) at amino acid position +1564 has similarity to a putative Zinc finger domain present in some ubiquitin ligases (Fig. 4C; Pfam accession PF02207; E value = 2.3e-13). This domain seems to be important for BIG function because the doc1-1 mutation is a substitution of one of the conserved Cys to Tyr (Fig. 4C). The second conserved cysteine-rich domain (CRD-2) at position +3442 is highly conserved among the Calossin family of proteins (Fig. 4C) and is similar to Zn-finger domains in eukaryotic transcription factors. A third cysteine-rich domain at position +2597 belongs to the ZZ domain family but is not conserved in the Calossin proteins (Fig. 4C; Pfam accession no. PF00569; E = 1.6e-07). Proteins containing this domain include proteins that function in the ubiquitin pathway such as HERC2 and p62 (GenBank T14346; Vadlamudi and Shin 1998), transcriptional coactivators such as the CREB-binding protein (CBP) and ADA2 (Chrivia et al. 1993; Marcus et al. 1994), and Dystrophyn-related proteins (Roberts and Bobrow 1998).
Expression of BIG
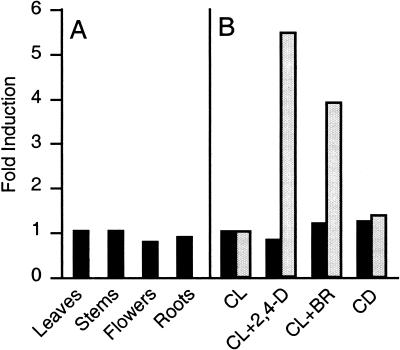
Doc1/tir3 mutations cause reduced growth in most plant organs, suggesting that BIG is expressed ubiquitously during development (Li et al. 1994; Ruegger et al. 1997). However, several putative components of the polar auxin transport machinery are expressed preferentially in particular organs or cell types. To test if the BIG gene was expressed in specific tissues, we performed quantitative RT–PCR analyses to monitor changes in mRNA accumulation. We compared BIG mRNA levels in roots, rosette leaves, inflorescence stems, and flowers. As shown in Figure 5A, BIG mRNA accumulated to similar levels in different organs of wild-type Arabidopsis. Because doc1/tir3 mutants display increased accumulation of CAB mRNA in the dark, BIG mRNA levels were compared between light-grown and dark-grown seedlings. Similarly, because of the effects of the doc1/tir3 mutations on auxin transport, we monitored BIG mRNA levels after seedlings were treated with 50 μM of the synthetic auxin 2,4-D. In addition, we also tested induction of BIG with 1 μM brassinolide, a steroid hormone known to act synergistically in several responses controlled by auxin (Kim et al. 2000). In all cases, BIG mRNA levels remained unchanged (Fig. 5B). As a control for the auxin treatments, we checked and detected elevated mRNA levels of the auxin-inducible gene SAUR-AC1 (Fig. 5B; Gil et al. 1994). Interestingly, this auxin-induced transcript served as a double control, because it was also induced by brassinolide treatment. The results of these experiments are consistent with BIG being ubiquitously and constitutively expressed throughout the plant.
Figure 5.
(A) BIG expression in Arabidopsis organs. RT–PCR analyses of tissue samples of 4-week-old plants grown in soil (leaves, stems, and flowers) or vertical plates (roots). (B) RT–PCR analyses of 7-day-old seedlings grown under continuous light (CL) or continuous dark (CD) or treated with 50 μM 2,4-D (CL+2,4-D) or 1 μM Brassinolide (CL+BR) as described (Gil et al. 1994). Radioactive signals were quantified and normalized using elongation factor EIF-4A expression as an internal standard. Relative BIG transcript levels are shown in black and levels of the auxin-inducible gene SAUR-AC1 in gray.
Localization of PIN1 in tir3 and doc1 seedlings
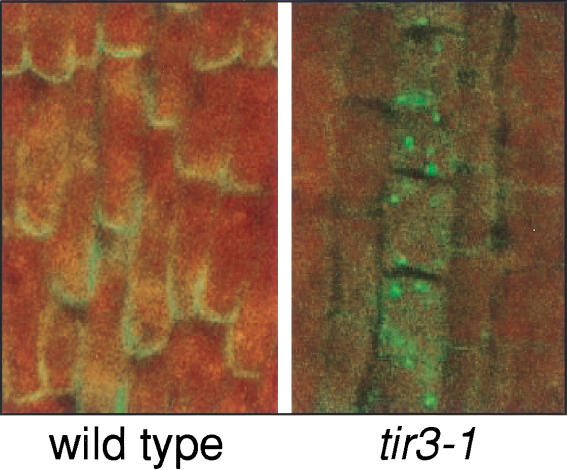
Because the mutant phenotype suggests a key role for BIG in polar auxin transport (Ruegger et al. 1997), the cellular localization of the putative auxin efflux carrier PIN1 (Galweiler et al. 1998) in tir3-1 and doc1-3 was determined. Using affinity-purified anti-PIN1 antibody, whole mount immunolocalization experiments on Arabidopsis seedlings were performed. In wild-type roots, PIN1 is localized at the bottom (acropetal) side of stele cells, in some cases accompanied by a weaker signal in the endodermis (Friml et al., in prep.). The polarity of PIN1 localization was partially disrupted by treatment of seedlings with polar auxin transport inhibitors like NPA (Geldner et al., in prep.). In untreated tir3 or doc1 seedlings, no difference in PIN1 localization was detected compared with that of wild type (data not shown). However, in mutants treated with NPA, >50% of roots (doc1: 52%, n = 83; tir3: 61%, n = 165), showed mislocalization of PIN1 protein to an unidentified intracellular compartment (Fig. 6). This phenomenon was never observed in wild-type roots (n = 167) treated with NPA.
Figure 6.
Localization of PIN1 in the stele region of 4-day-old Arabidopsis wild-type and tir3-1 seedling root tips after treatment with 150 μM NPA for 3 h. Immunocytochemical analysis was performed using primary antibodies raised against PIN1 and a FITC-conjugated secondary antibody. Fluorescent staining was imaged by laser-confocal microscopy. Indirectly visualized signals of the FITC-conjugated antibody are indicated by green color. The tissue autofluorescence indicated by red color of stele tissue is overlaid with the green immunofluorescence channel, facilitating the visualization of PIN1.
Discussion
Mutations in the BIG gene were identified in two independent screens and originally characterized as mutants affecting photomorphogenesis and auxin transport. In this report, we present evidence indicating that the defect in light-regulated gene expression first identified in the doc1 mutant is caused by an alteration in auxin distribution in dark-grown seedlings. BIG was identified as a putative component of the polar auxin transport machinery because tir3 mutations promote resistance to inhibition of root growth by phytotropins, decrease IAA movement through inflorescence stems, and result in a reduction in the number of NPA-binding sites in microsomal fractions. Moreover, many of the morphological defects observed in the mutants are consistent with a defect in some aspect of auxin physiology. In this study, we provide several additional lines of evidence indicating the involvement of BIG in polar auxin transport. These include synergistic effects of the tir3 mutation with mutations that alter auxin signaling or transport, suppression of some doc1 defects by elevated levels of endogenous auxin, and effects of doc1/tir3 mutations on the intracellular localization of the auxin efflux carrier PIN1.
A number of additional mutations have been isolated that affect both auxin physiology and photomorphogenesis. Mutations in the shy2, axr2, and axr3 loci cause photomorphogenesis in dark-grown seedlings, including reduced hypocotyl length and formation of leaves (Tian and Reed 1999; Nagpal et al. 2000). However, unlike these signaling mutants, the doc1/tir3 defects in auxin transport have little effect on seedling morphology and hypocotyl elongation in the dark (Li et al. 1994) but result in decreased hypocotyl elongation in the light. This is consistent with results from Jensen et al. (1998) that indicate that in Arabidopsis, polar auxin transport is important for hypocotyl elongation in the light but not in the dark. On the other hand, our results indicate that overexpression of the auxin biosynthetic gene YUCCA in the doc1-1 mutant suppresses the aberrant expression of light-regulated genes in the dark. This result indicates a novel and unexpected role for auxin in the repression of light-regulated genes in dark-grown seedlings and suggests that polar auxin transport is important in etiolated seedlings.
The tir3-1 mutation shows a strong synergistic interaction with mutations that affect either auxin response or transport, consistent with the proposed role for BIG in auxin transport. The phenotype of the tir3 pin1 double mutant includes a drastic reduction in inflorescence growth and premature arrest of this growth, suggesting that BIG and PIN1 have overlapping functions in the inflorescence. Because BIG is expressed throughout the plant, it will be interesting to determine if combination of tir3 with mutations in other members of the PIN family will result in additional synergistic effects. Multiple growth responses are also compromised in the auxin-signaling mutant axr1-12. The tir3-1 axr1-12 double mutant also had a very severe phenotype as expected for plants with defects in both auxin transport and response. One of the most interesting aspects of all three double-mutant phenotypes was the early arrest of inflorescence elongation. This observation suggests that auxin levels and/or response are limiting for inflorescence elongation and organ formation.
Any model for BIG function must explain the contradictory responses of the doc1/tir3 mutants to NPA. On the one hand, the tir3 mutants were isolated because they are resistant to the growth-inhibiting properties of NPA. In contrast, the doc1/tir3 mutants appear to be hypersensitive to NPA with respect to effects on PIN1 localization. However, this paradox can be resolved by considering the effects of NPA on root elongation and the basis for tir3 resistance to the compound. Evidence from a number of studies now indicates that NPA inhibits root elongation by promoting accumulation of IAA to inhibitor levels in the root tip (Ruegger et al. 1997; Sabatini et al. 1999; Casimiro et al. 2001). As the tir3 mutant is deficient in auxin transport in the shoot, it follows that less auxin is transported down into the mutant root system. As a consequence, NPA-mediated accumulation of IAA in the root tip is reduced, leading to less inhibition of root elongation. This view is also consistent with the decrease in lateral root formation in tir3 seedlings, a process that depends on IAA from the shoot system. The key element of this model is that decreased inhibition of root elongation in tir3 is not primarily due to a decrease in the number of NPA-binding sites but rather to a decrease in auxin transport into the root system.
In contrast, the effects of NPA on PIN1 localization are likely to be related to a decrease in NPA-binding sites. A reduction of NPA-binding sites is also evident in the tir3-1 mutant (Ruegger et al. 1997), suggesting that BIG encodes or is required for expression, localization, or stabilization of the NBP. Because none of the doc1/tir3 mutations are proven null alleles, the additive effects of doc1/tir3 mutations and NPA-treatments on PIN1 localization are not conclusive in defining a role for BIG in normal NBP microsomal localization. Recent results have suggested that NPA does not have a specific role in auxin transport. Rather, NPA appears to affect trafficking of a number of unrelated proteins to the plasma membrane (Geldner et al., in prep.). The fact that auxin transport is particularly sensitive to NPA and related compounds may be a reflection of the importance of correct polar localization of the efflux carriers to the auxin transport process. Consistent with this, intracellular vesicle transport is clearly required for polar auxin transport (Delbarre et al. 1998; Steinmann et al. 1999). In Drosophila, CalO affects synaptic transmission at the neuromuscular junctions, with specific defects in neurotransmitter release evoked by nerve stimulation. This process is dependent on the synaptic vesicle cycle (Sudhof 1995). Thus, it is possible that BIG and CalO participate in intracellular vesicle transport. There is also a well-established role for changes of intracellular Ca2+ levels in this process.
Calossins are among the biggest proteins in eukaryotic cells. The Drosophila CalO protein was first identified by screening a retinal expression library for calmodulin-binding proteins (Xu et al. 1998). At this point it is not known if BIG also binds calmodulin, but it is possible that this protein participates in Ca2+/calmodulin regulation of auxin transport because Ca2+ and calmodulin have been implicated in auxin transport responses (Lomax 1995). Although the precise biochemical role of BIG and other calossin-like proteins in cellular metabolism awaits further study, the presence of a number of protein domains suggests a few general possibilities. The three Zn-finger domains are likely to participate in protein-protein interactions, suggesting that BIG is likely to interact with a number of other proteins. Among the Zn-finger domains, the CDR-1 motif is very well conserved in calossins and several ubiquitin ligases represented by the Drosophila protein hyperplastic discs (Hyd; Mansfield et al. 1994). This domain is critical for BIG function because the doc1-1 mutation replaces one of the putative Zn-binding cysteines. The function and binding partners of the CDR-1 motif are not known. Interestingly, among the proteins similar to BIG at another Zn-binding domain, the ZZ domain, there is a family of very large proteins that includes dystrophin. This protein is associated with the plasma membrane and the cytoskeleton in postsynaptic neuronal complexes and plays a role in the maintenance of membrane-associated complexes at points of intercellular contact (Roberts and Bobrow 1998). Moreover, the ZZ domain has been found to interact with calmodulin (Anderson et al. 1996). In the future, it will be important to determine the cellular localization of BIG and the identity of interacting proteins. Clearly, BIG and other calossins play a critical role in some aspect of cellular function in both plants and animals.
Materials and methods
Plant materials and growth conditions
For all experiments described, unless specified otherwise, the Arabidopsis wild type is ecotype Columbia. For mutant screens to identify new doc1 alleles, plants were grown on 0.5× MS media plates and transferred to soil under conditions described previously (Li et al. 1994). To generate male sterile doc1-1 ap3-1 mutants, plants were grown initially in the greenhouse and then transferred to 28°C before bolting. For microarray experiments, wild-type and mutant plants (doc1-1, doc1-1 yucca, and yucca) were grown in 0.5× MS media without sugar in the dark at 22°C for 4 d before they were harvested for RNA preparation. For immunolocalization experiments, seeds were surface sterilized using 5% calcium hypochlorite and plated on 0.5× MS/1% sucrose medium. After 2-d vernalization at 4°C, seedlings were grown in vertical position in 16-h light/8-h dark cycle at 22°C. Three- to five-day-old seedlings were transferred into 0.5× MS/1% sucrose liquid medium supplemented with 50 to 150 μM NPA (dissolved in DMF) and incubated for 1 to 3 h. After incubation, they were immediately fixed for immunolocalization experiments.
IAA transport assays
IAA transport assays were performed according to the procedure of Okada et al. (1991). Stem segments (2.5 cm) of primary inflorescences were incubated in 30 μL of nutrient solution (5 mM KNO3, 2.5 mM KPO4 adjusted to pH 5.5, 2 mM MgSO4, 2 mM Ca(NO3)2, 50 μM Fe-EDTA, 70 μM H3BO3, 14 μM MnCl2, 0.5 μM CuSO4, 1 μM ZnSO4, 0.2 μM Na2MoO4, 10 μM NaCl, and 10 nM CoCl2) in a 1.5-mL Eppendorf plastic tube. The solution contained 1 μM 14C-IAA (specific activity, 57mCi/mmole; American Radiolabeled Biochemicals) with or without 15 μM NPA. The segments were incubated in the apical to basal orientation in time increments of 4 h, from 2 to 18 h. After the incubation, a 5-mm section was removed from the basal end of the segment and added to 3 mL of liquid scintillation cocktail (Bio-Safe II). After shaking the samples at 100 rpm for at least 2 h and incubating overnight at room temperature, the amount of radioactive IAA in the stem segments was determined by scintillation counting. The plants were grown in 24-h light (105 μE/m2sec). Results were expressed in counts per minute ±SEM.
Identification of new alleles of doc1 and genetic analysis
For the generation of the doc1-2 mutant, La-er pollen was γ-ray mutagenized by exposing inflorescence sections to 17 to 20 Krads in a 60Co irradiator. Irradiated flowers were used for manual fertilization of doc1-1 ap3-1 mutants. The doc1-3 allele was identified by its visible phenotype among a collection of transgenic plants containing T-DNA insertions (by-products of several projects) compiled by Dr. Jianming Li in Dr. Joanne Chory's laboratory at the Salk Institute. To determine allelism between tir3-1 and doc1-1, these mutants were crossed, and F1 seeds were plated on nutrient media containing 5 μM of the auxin transport inhibitor 2-carboxyphenyl-3-phenylpropane-1,2-dione (CPD) to assay resistance to inhibition of root growth. F1 seedlings (25/25) were resistant to CPD. The seedlings were transferred to soil for later confirmation of a mutant aerial phenotype.
Mapping of doc1
A homozygous doc1-1 line was crossed to La-er, and the resulting F1 plants were self-pollinated to generate F2 plants segregating the doc1-1 mutation. DNA for SSLP and CAPS analysis from doc1 F2 plants was prepared as described previously (Dellaporta et al. 1983). To place the doc1 locus in the physical map between markers nga32 and 17D8LE, 1091 chromosomes were scored for recombination events. Standard methods for YAC manipulation and generation of end probes were as described (Gibson and Somerville 1992). Cosmid library (Olszewski et al. 1988) and BAC filters (provided by Dr. Joe Ecker, University of Pennsylvania) were hybridized using YAC ends as described (Ausubel et al. 1994; http://http.tamu.edu:8000/∼creel/BACVEC.html). The dCAPS marker (Neff et al. 1998) COL2 required oligonucleotides 5′-CGCCATAGGAGTTTAGAAGG-3′ and 5′-CAT CACAAGTATGCTTCGCTAGCAATTGATTC-3′ as primers for PCR amplification followed by digestion with the restriction enzyme BcgI.
DNA and RNA analysis
DNA from yeast and plant tissues was isolated according to standard protocols (Ausubel et al. 1994). Total RNA was purified with TRIzol (GIBCO BRL) as indicated by the manufacturer. cDNA synthesis for RT–PCR analyses was performed using Superscript II (GIBCO BRL) with 3 μg of total RNA as indicated by the manufacturer. PCR reactions of 12, 13, and 14 cycles (in the exponential range of amplification), were performed by denaturing the template cDNAs at 95°C for 5 min followed by cycles of 90°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min. A DOC1 fragment was amplified using oligonucleotides 5′- CCTGTCAAGGGTCCCTCAGTTCCC-3′ from exon 13 and 5′-CCAGTTGATGATAGCTGGGATCCAGGAG-3′ from exon 14 as primers. As an internal control, we included in the PCR reactions oligonucleotides 5′-CTCTGGTGTCTTG CAGCAGCTGGACA-3′ and 5′-GATCGGCCACGTTCGAAG GCAGCTC-3′ as primers to amplify a fragment of the EIF-4α cDNA (Taylor and Green 1995). PCR products were size fractionated by electrophoresis in 1.5% (w/v) agarose gel, blotted to a membrane and hybridized with radiolabeled DOC1 and EIF-4α probes. Signal intensities were determined with a PhosphorImager (Molecular Dynamics).
Microarray experiments
Total RNA extraction, cDNA synthesis, in vitro labeling, hybridization, wash, scanning, and data analysis were performed according to Affymetrix's instructions.
Antibodies and immunolocalization
The whole-mount immunolocalization experiments on Arabidopsis seedlings were performed as described (Muller et al. 1998). Seedlings were fixed for 1 h under vacuum in 4% formaldehyde, permeabilized, and incubated with primary and subsequently secondary antibody. Affinity-purified primary anti-PIN1 antibody (Galweiler et al. 1998) was used in dilution 1:150. The secondary antibody, DATF-conjugated anti-rabbit antibody (Dianova), was diluted 1:200. The samples were inspected by laser scanning confocal microscope Leica DMIRBE, TCS 4D (Leica). The images were further processed in Photoshop 4 (Adobe) and CorelDraw 7 (Corel Corporation).
Acknowledgments
We thank Kim Hanson and Melissa McCarthy for technical support, and Adan Colon-Carmona, Jianming Li, and Karin Schumacher for their help in generating and identifying the doc1-3 T-DNA line. Seeds of ap3-1 and a cosmid library were supplied by the ABRC stock center. Jennifer Nemhauser made useful comments concerning this manuscript. This work was supported by grants from the Department of Energy (DE-FG03-89ER13993) and the National Science Foundation (MCB96-31390) to J.C., by grants from the Department of Energy (DE-FG02-98ER20313) and the National Institutes of Health (GM43644) to M.E., by a grant from DAAD to J.F., by a grant from DFG to K.P., and by a Marsden grant of New Zealand to J.P. and K.S. J.C. is an Associate Investigator of the Howard Hughes Medical Institute (HHMI), and Y.Z. is a HHMI fellow of the Life Sciences Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL chory@salk.edu; FAX (858) 558-6379.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.905201.
References
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, Rogers RP, Jarrett HW. Ca2+-calmodulin binds to the carboxyl-terminal domain of dystrophin. J Biol Chem. 1996;271:6605–6610. doi: 10.1074/jbc.271.12.6605. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Bartel B, Fink GR. ILR1: An amidohydrolase that releases active indole-3-acetic acid from conjugates. Science. 1995;268:1745–1748. doi: 10.1126/science.7792599. [DOI] [PubMed] [Google Scholar]
- Behringer FJ, Davies PJ. Indole-3-acetic acid levels after phytochrome-mediated changes in the stem elongation rate of dark- and light-grown Pisum seedlings. Planta. 1992;188:85–92. doi: 10.1007/BF00198943. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1gene: A permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Bernasconi P, Patel B, Reagan CJ, Subamanian MV. The N-1-naphtylptalamic acid–binding protein is an integral membrane protein. Plant Physiol. 1996;111:427–432. doi: 10.1104/pp.111.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, et al. Auxin transport promotes Arabidopsislateral root initiation. Plant Cell. 2001;13:843–852. doi: 10.1105/tpc.13.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. The Arabidopsis thalianaAGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci. 1998;95:15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100:469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124:1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre A, Muller P, Guern J. Short-lived and phosphorylated proteins contribute to carrier-mediated efflux, but not to influx, of auxin in suspension-cultured tobacco cells. Plant Physiol. 1998;116:833–844. doi: 10.1104/pp.116.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S, Wood J, Hicks J. A plant DNA minipreparation: Version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- Galweiler L, Guan C, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsisvascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Gibson SI, Somerville C. Chromosome walking in Arabidopsis thalianausing yeast artificial chromosomes. In: Koncz C, et al., editors. Methods in Arabidopsis research. Singapore: World Scientific; 1992. pp. 119–143. [Google Scholar]
- Gil P, Liu Y, Orbovic V, Verkamp E, Poff KL, Green PJ. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 1994;104:777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes & Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng XW. The cell biology of the COP/DET/FUS proteins: Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7: A conditional regulator of differential growth in aerial Arabidopsistissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel R, Lomax TL, Briggs WR. Auxin transport in membrane vesicles from Cucurbita pepoL. Planta. 1983;157:193–201. doi: 10.1007/BF00405182. [DOI] [PubMed] [Google Scholar]
- Hsieh HL, Okamoto H, Wang M, Ang LH, Matsui M, Goodman H, Deng XW. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsisdevelopment. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998;116:455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Cochran DS, Lamerson PM, Evans ML, Cohen JD. Red light–regulated growth: I. Changes in the abundance of indoleacetic acid and a 22-kilodalton auxin-binding protein in the maize mesocotyl. Plant Physiol. 1991;97:352–358. doi: 10.1104/pp.97.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katekar GF, Geissler AE. Auxin transport inhibitors: III. Chemical requirements of a class of auxin transport inhibitors phytotropins. Plant Physiol. 1977;60:826–829. doi: 10.1104/pp.60.6.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. Photomorphogenic development of the Arabidopsisshy2-1D mutation and its interaction with phytochromes in darkness. Plant J. 1998;15:61–68. doi: 10.1046/j.1365-313x.1998.00179.x. [DOI] [PubMed] [Google Scholar]
- Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol. 2000;123:997–1004. doi: 10.1104/pp.123.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsismutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Ledger S, Dare A, Putterill J. COL2 (accession nos. L81119 and L81120) is a homologue of the Arabidopsis flowering time gene CONSTANS (PGR 96-081) Plant Physiol. 1996;112:862. [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsisauxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Li HM, Altschmied L, Chory J. Arabidopsismutants define downstream branches in the phototransduction pathway. Genes & Dev. 1994;8:339–349. doi: 10.1101/gad.8.3.339. [DOI] [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ. An auxin-responsive promoter is differentially induced by auxin gradients during tropisms. Plant Cell. 1991;3:1167–1175. doi: 10.1105/tpc.3.11.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax TL, Muday GK, Rubery PH. Auxin transport. In: Davies PJ, editor. Plant hormones: Physiology, biochemistry and molecular biology. Dordrech, Netherlands: Kluwer Academic Publishers; 1995. pp. 509–530. [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes & Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield E, Hersperger E, Biggs J, Shearn A. Genetic and molecular analysis of hyperplastic discs, a gene whose product is required for regulation of cell proliferation in Drosophila melanogasterimaginal discs and germ cells. Dev Biol. 1994;165:507–526. doi: 10.1006/dbio.1994.1271. [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsisby facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: Putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwly P, Pilet PE. Local treatment with indole-3-acetic acid influences differential growth responses in Zea maysL. roots. Planta. 1991;185:58–64. doi: 10.1007/BF00194515. [DOI] [PubMed] [Google Scholar]
- Morris DA, Rubery PH, Jarman J, Sabater M. Effects of inhibitors of protein synthesis on transmembrane auxin transport in Cucurbita pep. J Exp Bot. 1991;29:147–157. [Google Scholar]
- Muday GK, Brunn SA, Haworth P, Subramanian M. Evidence for a single naphthylphthalamic acid–binding site on the zucchini plasma membrane. Plant Physiol. 1993;103:449–456. doi: 10.1104/pp.103.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB, Clarke S. A distinctly regulated protein repair L-isoaspartylmethyltransferase from Arabidopsis thaliana. Plant Mol Biol. 1996;30:723–737. doi: 10.1007/BF00019007. [DOI] [PubMed] [Google Scholar]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. AtPIN2 defines a locus of Arabidopsisfor root gravitropism control. EMBO J. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25:213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thalianagenetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport-system in early stages of Arabidopsisfloral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, Martin FB, Ausubel FM. Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 1988;16:10765–10782. doi: 10.1093/nar/16.22.10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M. The aux1 mutation of Arabidopsisconfers both auxin and ethylene resistance. Plant Physiol. 1990;94:1462–1466. doi: 10.1104/pp.94.3.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol. 2000;122:481–490. doi: 10.1104/pp.122.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 1998;118:1369–1378. doi: 10.1104/pp.118.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Hillman T, Stern M. Mutations in the Drosophilapushover gene confer increased neuronal excitability and spontaneous synaptic vesicle fusion. Genetics. 1996;142:1215–1223. doi: 10.1093/genetics/142.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RG, Bobrow M. Dystrophins in vertebrates and invertebrates. Hum Mol Genet. 1998;7:589–595. doi: 10.1093/hmg/7.4.589. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M. Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsisis associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell. 1997;9:745–757. doi: 10.1105/tpc.9.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsisroot. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sablowski RW, Meyerowitz EM. Temperature-sensitive splicing in the floral homeotic mutant apetala3-1. Plant Cell. 1998;10:1453–1463. doi: 10.1105/tpc.10.9.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, McKim KS, Messina L, French RL, Hurley WD, Arbel T, Chin GM, Deneen B, Force SJ, Hari KL, et al. Identification of novel Drosophilameiotic genes recovered in a P- element screen. Genetics. 1999;152:529–542. doi: 10.1093/genetics/152.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkle JR, Kadakia R, Jones AM. Dim-red-light-induced increase in polar auxin transport in cucumber seedlings: I. Development of altered capacity, velocity, and response to inhibitors. Plant Physiol. 1998;116:1505–1513. doi: 10.1104/pp.116.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Stowe-Evans EL, Harper RM, Motchoulski AV, Liscum E. NPH4: A conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 1998;118:1265–1275. doi: 10.1104/pp.118.4.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle: A cascade of protein–protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Taylor CB, Green PJ. Identification and characterization of genes with unstable transcripts (GUTs) in tobacco. Plant Mol Biol. 1995;28:27–38. doi: 10.1007/BF00042035. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thalianaSHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- Tsurumi S, Ohwaki Y. Transport of 14C-labeled indoleacetic acid in Viciaroot segments. Plant Cell Physiol. 1978;19:1195–1206. [Google Scholar]
- Vadlamudi RK, Shin J. Genomic structure and promoter analysis of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Lett. 1998;435:138–142. doi: 10.1016/s0014-5793(98)01021-7. [DOI] [PubMed] [Google Scholar]
- Xu XZ, Wes PD, Chen H, Li HS, Yu M, Morgan S, Liu Y, Montell C. Retinal targets for calmodulin include proteins implicated in synaptic transmission. J Biol Chem. 1998;273:31297–31307. doi: 10.1074/jbc.273.47.31297. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]