Abstract
During S phase in eukaryotes, assembly of chromatin on daughter strands is thought to be coupled to DNA replication. However, conflicting evidence exists concerning the role of the highly conserved core histone tail domains in this process. Here we present a novel in vivo labeling technique that was used to examine the role of the amino-terminal tails of the H2A/H2B dimer in replication-coupled assembly in live cells. Our results show that these domains are dispensable for nuclear import but at least one tail is required for replication-dependent, active assembly of H2A/H2B dimers into chromatin in vivo.
Keywords: Chromatin assembly, histone tail, replication, Physarum
In eukaryotes, assembly of chromatin on daughter strands is believed to be mechanistically coupled to concomitant replication of DNA during S phase (Almouzni and Mechali 1988; Smith and Stillman 1989). The assembly of chromatin is a multistep process in which the H3/H4 tetramer is first deposited onto the DNA, followed shortly by the deposition of H2A/H2B dimers and, in most eukaryotes, association of a linker histone to form the complete nucleosome (Wolffe 1998; Annunziato and Hansen 2000). Crystallographic studies have shown that all four histone proteins are composed of two distinct domains; a central histone-fold domain and a tail domain, both of which have distinct functions in chromatin (Arents and Moudrianakis 1993; Luger et al. 1997). The histone-fold domains are assembled via multiple histone–histone interactions to form a central cylinder around which nucleosomal DNA is wrapped (Luger et al. 1997). In contrast, the histone-tail domains are thought to mediate nucleosome stability and internucleosome interaction and are required for formation of the compact chromatin fiber (Garcia-Ramierez et al. 1992; Carruthers and Hansen 2000). Moreover, the tail domains are key control elements in the regulation of the structural and functional state of chromatin (Wolffe and Hayes 1999). Indeed, many chromatin-associated activities, such as transcription, DNA recombination, and repair have been intimately correlated with posttranslational modifications of the tails (Strahl and Allis 2000).
Early work suggested that, in addition to their role in postreplicative processes, the histone tails are critical for replication-coupled chromatin assembly during S phase (for review, see Annunziato and Hansen 2000). For example, the amino-terminal tail of H4 in newly assembled chromatin was found to have a highly conserved pattern of acetylation on lysines 5 and 12 (Sobel et al. 1995). Consistent with this idea, H3/H4 tails within biochemically fractionated replication complexes exhibit this conserved pattern of acetylation (Kaufmann et al. 1995; Chang et al. 1997; Tyler et al. 1999). Simultaneous deletion of both H3/H4 tails in yeast is lethal and endogenous 2 μm plasmids isolated from theses strains exhibit reduced superhelical density (Morgan et al. 1991; Ling et al. 1996). However, the phenotypes of cells lacking H3/H4 tails suggest that these domains are required for essential functions throughout the cell cycle (Morgan et al. 1991); deposition-related acetylation sites in these proteins can be mutated in yeast with little effect on cell viability or nucleosome assembly (Ma et al. 1998). Moreover, microinjection of mRNAs encoding full-length and truncated H3 and H4 into Xenopus eggs indicates that only the central histone-fold domains of these proteins are required for replication-coupled chromatin assembly (Freeman et al. 1996). In addition, both tails in the H3/H4 tetramer are dispensable for replication-coupled chromatin assembly by purified chromatin assembly factor-1 (CAF-1) in vitro (Shibahara et al. 2000).
The requirement for the amino-terminal tails of the H2A/H2B dimer for chromatin assembly is even less clear. Similar to H3 and H4, deletion of either the H2A or the H2B amino-terminal tail does not affect viability in yeast, but simultaneous deletion of both tails is lethal (Schuster et al. 1986). However, the actual effects of these deletions and the importance of the tails in chromatin assembly remain undefined. Moreover, little is known about the requirement for H2A/H2B-specific chaperones during chromatin assembly in vivo. Indeed, in vitro experiments suggest that these histones may simply spontaneously assemble onto a preformed H3/H4 tetramer–DNA complex (Verreault et al. 1996; Shibahara et al. 2000). However, the delayed kinetics of H2A/H2B deposition after the incorporation of H3/H4 during chromatin assembly suggests that the mere presence of the H3/H4 tetramer–DNA complex is not sufficient for H2A/H2B dimer incorporation (Smith and Stillman 1991; Almouzni and Wolffe 1993).
Here we present direct evidence that the H2A/H2B histone tail domains have a function in replication-coupled chromatin assembly within a living cell. Our method avoids complications from tail-dependent genetic effects not directly related to chromatin assembly by incorporating trace amounts of fluorescently tagged H2A/H2B dimers into cells of the naturally synchronous macroplasmodial form of the slime mold Physarum polycephalum at the beginning of S phase. Examination of the location and fate of exogenous proteins within the cell reveals that the tails are not required for transport of H2A/H2B into nuclei but are required for efficient assembly into chromatin.
Results and Discussion
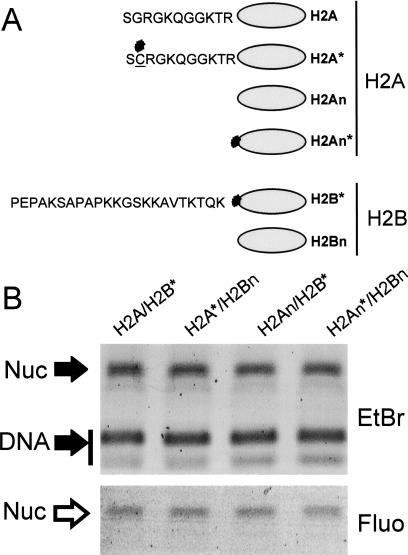
We wished to examine if the H2A/H2B amino-terminal tail domains are required for chromatin assembly in vivo by incorporating trace amounts of fluorescein-tagged exogenous proteins into Physarum macroplasmodia at the beginning of S phase (Thiriet and Hayes 1999, 2001). These H2A/H2B dimers were either full length or lacking one or both amino-terminal tails (Fig. 1A). As a control, we first examined if dimers lacking tail domains compete efficiently with full-length proteins during nucleosome reconstitution in vitro. Standard salt-dialysis reconstitutions were performed with a 215-bp DNA fragment, the appropriate ratio of purified core histones, and trace amounts of the fluorescein-tagged H2A/H2B. Fluorographs of nucleoprotein gels showed that the efficiency of dimer assembly into nucleosomes in vitro was not significantly affected by the absence of the H2A/H2B amino-terminal-tail domains or by the position of the fluorescein tag within the dimer (Fig. 1B and results not shown). This result is in agreement with previous work showing that histone octamers lacking tails can reconstitute nucleosomes in vitro (Ausio et al. 1989; Vitolo et al. 2000).
Figure 1.
Fluorescently tagged H2A/H2B dimers efficiently reconstitute into nucleosomes in vitro. (A) Scheme of the histone proteins used in this study. The stars show the location of the fluorescein tag attached via a unique cysteine substitution. Note that H2A, H2An, and H2B were modified at positions 2, 12, and 26, respectively. (B) Nucleoprotein gel of nucleosomes (Nuc) reconstituted with a 215-bp DNA fragment, native core histones, and trace amounts of fluorescently tagged histone dimers as a competitor. (Top) Ethidium bromide (EtBr)-stained gel. (Bottom) Fluorograph (Fluo) of the same gel.
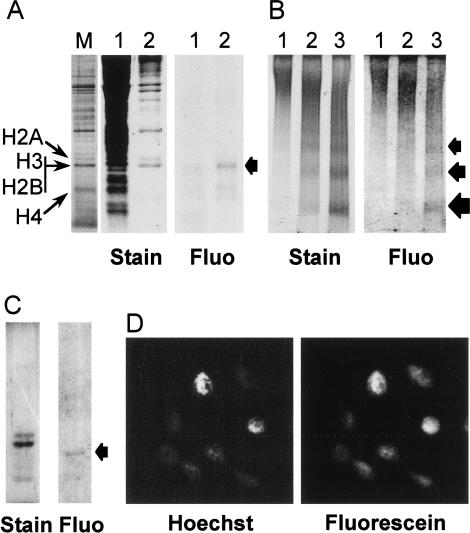
We then determined if exogenous H2A/H2B dimers could be introduced into live Physarum cells and we investigated the fate of the proteins after incorporation (Fig. 2). We and others have previously established that Physarum macroplasmodia are capable of taking up exogenous proteins and using them in cellular metabolism (Bradbury et al. 1974; Thiriet and Hayes 1999, 2001). To exploit this feature, exogenous full-length H2A/H2B* dimer was incorporated into the cell by spreading a solution containing the proteins onto the upper plasmodium surface and the fate of the proteins examined (Fig. 2). After absorption, we found that the majority of the tagged protein remained full length after incorporation and >90% of the exogenous dimer was recovered in the nuclear fraction (Fig. 2A). We then determined if the exogenous H2A/H2B* dimer was assembled into chromatin. Chromatin was purified from Physarum nuclei treated with micrococcal nuclease (MNase) and separated on nucleoprotein gels. The fluorescein-tagged H2A/H2B* comigrated precisely with the bulk nucleosomal ladder visualized by ethidium bromide staining (Fig. 2B). To ensure that the fluorescent signal was due to the full-length peptide, proteins from mononucleosome and dinucleosome bands were prepared and analyzed in SDS-PAGE. The fluorograph of the gel revealed a single band corresponding to full-length H2B (Fig 2C). The subcellular localization was confirmed by direct fluorescence microscopy, which shows an exact colocalization of Hoechst-stained nuclei and fluorescence due to exogenous H2A/H2B* (Fig. 2D). Identical results were obtained with H2A*/H2B (data not shown).
Figure 2.
Exogenous H2A/H2B dimers are internalized into Physarum cells, localized to the nuclei, and incorporated into chromatin. A solution of H2A/H2B* dimers (60 ng/μL) was deposited onto the upper cellular surface of Physarum fragments during S phase. (A) Exogenous H2A/H2B are localized in Physarum nuclei. The cellular localization of the exogenous proteins was analyzed by SDS-PAGE of cytoplasmic and nuclear fractions, shown in lanes 1 and 2, respectively. The gel was stained with Coomassie and the exogenous dimers were imaged by fluorography, as indicated. Physarum nuclear proteins were run as markers, with the histones as indicated (lane M). Fluo, Fluorograph. (B) Exogenous H2A/H2B dimers are incorporated into Physarum chromatin. Physarum nuclei were digested for 2, 5, and 10 min with MNase (lanes 1,2,3, respectively), and the products analyzed by nucleoprotein gel electrophoresis, followed by fluorography and ethidium staining, as indicated. (C) The presence of full-length exogenous proteins in mononucleosome and dinucleosome bands was confirmed by SDS-PAGE. (D) Fluorescently tagged exogenous H2A/H2B colocalizes with Hoechst-stained Physarum nuclei.
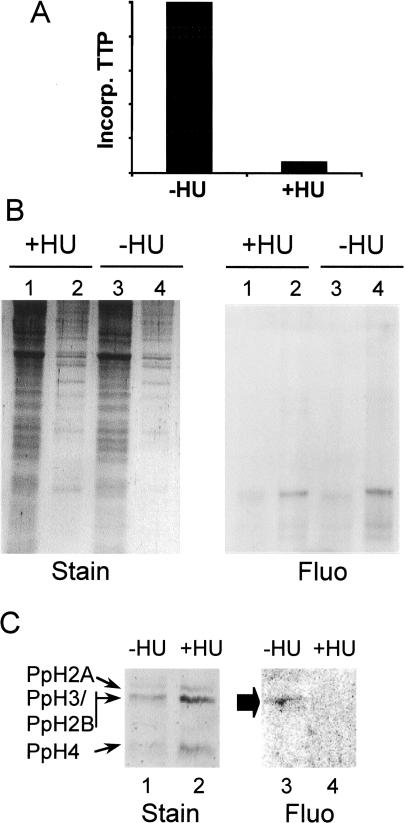
In vivo, the majority of chromatin assembly takes place during S phase and is most likely coupled to DNA replication. To determine if the transport into nuclei or the assembly of the exogenous dimers into nucleosomes depended on DNA replication, we incorporated full-length dimer into macroplasmodial segments deposited onto either normal medium or medium containing the DNA synthesis inhibitor hydroxyurea (Fouquet et al. 1975) (Fig. 3A). Analysis of cytoplasmic and nuclear fractions in SDS-PAGE revealed that the exogenous H2A/H2B* was located in nuclei regardless of the presence or absence of DNA replication, indicating that transport of the exogenous histones into Physarum nuclei is not dependent on this process (Fig. 3B).
Figure 3.
Assembly of exogenous H2A/H2B dimers into Physarum chromatin requires DNA replication in S phase. (A) Plot of 32P-dTTP incorporation per unit of DNA in cells treated with or without hydroxyurea (HU) during S phase. (B) Nuclear localization of the exogenous histone dimer is not dependent on DNA replication. SDS-PAGE of cytoplasmic or nuclear fractions (lanes 1 and 3 or 2 and 4, respectively) from cells treated or untreated with HU. (Fluo) Fluorograph. (C) SDS-PAGE of protein content in mononucleosomes and dinucleosomes from cells treated or untreated with HU (−/+HU). A fluorograph and a Coomassie-stained image of the gel are shown.
In contrast, we find that assembly of exogenous H2A/H2B* into chromatin occurs in a DNA-replication dependent manner. We prepared chromatin from cells treated with hydroxyurea and analyzed the protein content of mononucleosome and dinucleosome bands by SDS-PAGE as described earlier. Again, we found the exogenous dimer was present in nucleosomes when cells were grown in normal medium (Fig. 3C, lanes 1,3). However, we failed to detect any fluorescent protein in mononucleosomes and dinucleosomes obtained from cells grown in medium containing hydroxyurea (Fig. 3C, lanes 2,4).
Previous studies have shown that, in Physarum, histones H2A and H2B are also synthesized during G2 phase (Loidl and Grobner 1987). We thus examined exogenous dimer uptake in G2 phase and found that assembly into chromatin occurred but to a lower extent than that observed in S phase (results not shown). The function of histone dimer synthesis and assembly in G2 phase remains unclear but could be due to a general or transcription-related turnover of H2A/H2B.
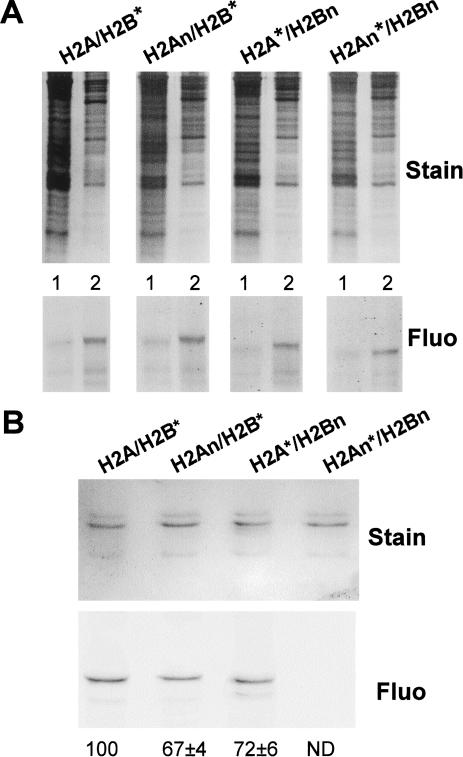
To investigate the role of the amino-terminal tails of H2A/H2B in replication-coupled chromatin assembly in vivo, we examined the incorporation of dimers lacking tail domains. Fluorescein-tagged dimers lacking one or both amino-terminal tails of H2A and H2B were introduced into plasmodium segments in early S phase, as described earlier. Segments treated in parallel with identical concentrations of full-length dimer were used to determine 100% of incorporation. SDS-PAGE analysis of cytoplasmic and nuclear fractions from treated cells showed that all detectable exogenous H2A/H2B dimers were located within nuclei (Fig. 4A). Thus, the amino-terminal tails of H2A and H2B do not play a significant role in the transport of the exogenous dimers into nuclei. Also, because equivalent amounts of labeled intact or tailless proteins were recovered in each nuclear fraction, loss of the tails apparently does not affect protein stability in our experimental system.
Figure 4.
Transport of histone H2A/H2B dimers into nuclei does not require the amino-terminal tails, but assembly into chromatin requires at least one tail domain. (Fluo) Fluorograph. (A) Cellular localization of exogenous histone H2A/H2B dimers. Cytoplasmic and nuclear fractions (lanes 1 and 2, respectively in each gel) were analyzed for each of the dimers shown at the top. The gels were stained and fluorographed, as indicated. (B) Assembly of exogenous H2A/H2B chromatin requires the amino-terminal-tail domains. Soluble chromatin was prepared from cells treated with each of the dimers listed at the top and analyzed as in Fig. 3C. The quantitation is the average of two independent experiments. ND, not determined.
To determine if dimers lacking tail domains were actually incorporated into nucleosomes, we analyzed the protein composition of mononucleosomes and dinucleosomes by SDS-PAGE, as described earlier (Fig. 4B). Interestingly, the fluorograph of the gel revealed that exogenous H2A/H2B was present in nucleosomes when either the H2A or H2B tail was deleted (Fig. 4B, lanes 2,3). Quantitation of the gels showed that the efficiency of incorporation into chromatin was moderately (∼30%) lower than the incorporation of full-length dimer containing both tails. In striking contrast, mononucleosomes and dinucleosomes from nuclei containing completely tailless H2A/H2B dimer did not reveal any detectable fluorescence in the protein gel (Fig. 4B, lane 4) or in the nucleoprotein gel (data not shown) despite the presence of this dimer in the nucleus (Fig. 4A). We thus conclude that H2A/H2B dimers lacking both amino-terminal tails are not assembled into chromatin, whereas the presence of either tail is sufficient to allow replication-coupled assembly but with reduced efficiency. Furthermore, the absence of signal with completely tailless dimers indicates that the exogenous dimers are not disrupted on incorporation and do not exchange with endogenous proteins. These results also indicate that in Physarum, histone H2A and H2B amino-terminal tails have redundant functions in chromatin assembly.
Genetic experiments in yeast showed that deletion of both amino-terminal tails of the H2A/H2B dimer is lethal (Schuster et al. 1986). Interestingly, when we incorporated much higher amounts of completely tailless dimer (>1 mg/mL) into plasmodia, we observed growth defects and a detectable loss in the ability to produce nucleosome ladders on nuclease digestion, similar to observations with H3/H4 tail-deletion mutants in yeast (Ling et al. 1996). However, similar incorporation of full-length proteins did not result in such phenotypes (data not shown). Moreover, under conditions of high input of exogenous proteins, we detected a reduction in the amount of endogenous H2A/H2B from isolated nuclei. These results further suggest that the exogenous proteins compete for the same chromatin assembly pathways used by endogenous H2A/H2B dimer.
Yeast genetic experiments have also been used to demonstrate that the H2A/H2B amino-terminal tail domains have redundant functions essential for viability of the cell (Schuster et al. 1986). Although it is possible that the tails are required for S-phase-dependent chromatin assembly in yeast, the exact growth-dependent function(s) of these domains were not determined from these genetic studies. Besides these studies, there is little evidence concerning the role of the H2A/H2B amino-terminal tails or dimer tail posttranslational modifications for chromatin assembly in vivo (Annunziato and Hansen 2000). Moreover, in vitro experiments suggest that assembly of the H2A/H2B dimer into nascent chromatin may occur passively, with the dimer spontaneously binding the H3/H4 tetramer–DNA complex without the assistance of specialized assembly factors (Verreault et al. 1996). CAF-1 has been shown to specifically associate with the H3/H4 tetramer (Verreault et al 1996; Chang et al. 1997), and CAF-1 alone can mediate assembly of nucleosomes in vitro with core histones and replicating DNA (Verreault et al. 1996). Thus nucleosome assembly could depend only on CAF-1-dependent loading of the H3/H4 tetramer onto DNA. However, our results demonstrate that the H2A/H2B tail domains play an essential role in the assembly of these proteins into chromatin in vivo and, moreover, that dimer assembly likely occurs through an active mechanism.
Several “molecular chaperones” such as nucleoplasmin and NAP-1 have been shown to be specifically associated with a maternal pool of H2A/H2B dimers in Xenopus and Drosophila, respectively, and to facilitate nonspecific nucleosome assembly in vitro (Dilworth et al. 1987; Ito et al. 1996). Our findings suggest that H2A/H2B chaperones are required in vivo and are likely to interact with the amino-terminal tail domains. A recent in vitro study showed that histone acetylation by p300 facilitates the transfer of H2A/H2B dimers from nucleosomes to the Drosophila molecular chaperone NAP-1 in ATP-dependent remodeling processes related to transcription (Ito et al. 2000). Opposite mechanisms might take place during chromatin assembly. However, the actual identity of such putative molecular chaperones and the mechanism by which H2A/H2B is assembled onto the tetramer–DNA complex in vivo remains to be determined.
In addition, our results also demonstrate that the H2A/H2B tail domains are dispensable for nuclear import. Nuclear import of H2A/H2B has recently been shown to be mediated by several members of the karyopherin/importin family (Mosammaparast et al. 2001). Thus, facilitated nuclear import of these histones requires elements of structure other than the tail domains, in agreement with previous work (Mosammaparast et al. 2001). Further, our results show that chromatin assembly and nuclear import can be uncoupled, suggesting that they occur by distinct molecular mechanisms. Perhaps histones are shuttled between different chaperones or the histone chaperones responsible for nuclear import require the addition of signals or cofactors to assemble the proteins into nascent chromatin.
We have also described a powerful strategy for following the assembly of mutant proteins into a living cell in a cell-cycle dependent manner. This novel strategy presents important advantages over genetic methods or microinjection of RNAs (Freeman et al. 1996; Schuster et al. 1986). The incorporation of trace amounts of chemically tagged proteins avoids deleterious genetic effects and allows the investigation of preassembled exogenous complexes in live cells. Coupled with in vitro mutagenesis, this approach provides insights into the roles of protein domains when genetic analyses would lead to cell lethality in which the underlying cause is often not readily apparent (Schuster et al. 1986). The natural synchrony of millions of nuclei in a single Physarum plasmodium makes it possible to incorporate at specific cell cycle stages any exogenous protein or complex, given sufficient similarity to the endogenous cellular homolog(s). Given the high degree of phylogenic conservation in histone proteins, it is likely that the incorporation of trace amounts of exogenous histones we report depends on interactions with endogenous histone chaperones involved in nuclear import and chromatin assembly. Indeed, incorporation of large amounts of exogenous H2A/H2B appears to co-opt the assembly pathway used by endogenous H2A/H2B. An important advantage of this system is that if a mutant protein or complex is incompatible with any interaction or process required for a cellular function, introduction of trace amounts into the cell is unlikely to cause cell lethality, because the plasmodium will simply use endogenous homologs.
Materials and methods
Physarum culture
P. polycephalum, strain TU291, was cultured and cell cycle advancement within macroplasmodia was followed by observation of mitosis by phase contrast microscopy, as described (Thiriet and Hayes 1999). Specimens for direct fluorescence microscopy were fixed with ethanol, mounted in glycerol/ethanol (1 : 1), and illuminated with the appropriate wavelengths to visualize either Hoescht-stained DNA or the fluorescein-tagged proteins. For experiments involving inhibition of DNA replication, the macroplasmodium fragments were cultured normally until the desired cell cycle stage, 80 mM hydroxyurea was added to the media, and protein solution was immediately deposited onto the cell surface as described in the following sections.
Preparation of histone dimers
Genes coding for full-length and various tailless Xenopus laevis H2As and H2Bs were modified by PCR to contain single cysteine substitutions as outlined in Figure 1, and the proteins were overexpressed in bacteria and purified as described (Hayes and Lee 1997). The purified proteins were modified with fluorescein-5-maleimide (∼threefold excess, Pierce) for 30 min at room temperature. The reaction was stopped by addition of 10 mM dithiothreitol (DTT) and excess reagent removed by ion exchange chromatography on a Biorex 70 column. Following exhaustive washes with 0.6M NaCl, the modified dimers were then eluted with 1M NaCl in TE and dialyzed against 20 mM Tris-HCl (pH 7.2; Thiriet and Hayes 2001).
In vitro reconstitution of nucleosomes
A 215-bp DNA fragment containing a Xenopus borealis 5SrRNA gene was obtained by EcoRI and DdeI digestion of plasmid pXP-10 (Vitolo et al. 2000). Five micrograms of purified fragment was mixed with ∼3 μg purified core histones and ∼100 ng fluorescently tagged H2A/H2B dimers and nucleosomes reconstituted via a salt dialysis method as described (Hayes and Lee 1997). Nucleosomes were analyzed by electrophoresis on 0.7% agarose, 0.5× TBE gels. The fluorescent proteins within the gel were imaged by fluorometry before staining the gel with ethidium bromide (Thiriet and Hayes 2001).
Incorporation of dimers into Physarum
Macroplasmodia were cut into halves and 400 μL H2A/H2B dimers at the concentrations indicated in the figure legends were deposited into the upper surface as described (Thiriet and Hayes 1999) with minor modifications. The incorporation was performed in the S phase of the cell cycle unless indicated in the text and treated plasmodia were kept in growth medium. Unless otherwise stated, the amounts of exogenous histones used in this study did not lead to detectable effects on growth of the cell or progression of the cell cycle (cf. Thiriet and Hayes 1999, 2001). Controls were treated in the same way but with buffer only. Note that plasmodia cut into separated pieces remain synchronous for several cell cycles when cultured in normal conditions (Thiriet and Hayes 1999).
Isolation of nuclei and preparation of chromatin
Macroplasmodia segments on filter paper supports were washed in 1 mM ethylenediamine tetraacetic acid (EDTA). The cells were then harvested and disrupted by Dounce homogenization in nuclei isolation buffer (15 mM MgCl2, 15 mM Tris-HCl at pH 8.0, 5 mM EDTA, 0.25 M hexylene glycol, 0.6% surfynol, 3 mM DTT). The nuclei were then pelleted by centrifugation at 400g for 5 min. The nuclear pellet was resuspended in 2 mL isolation buffer and carefully loaded onto a 1 M sucrose cushion in nuclei isolation buffer after centrifugation at 8000g for 20 min, and the nuclei were recovered from the bottom of the tubes. Nuclei were then washed with MNase buffer (60 mM KCl, 15 mM NaCl, 10 mM Tris-HCl at pH 8.0, 1 mM CaCl2), pelleted by centrifugation, and resuspended in the same buffer. Chromatin was prepared from freshly isolated nuclei by addition of one-twenthieth volume MNase (0.3 U/μL) for the times indicated in text. The digestion was stopped by addition of 5 mM EDTA and kept on ice for 10 min. Microfuge tubes were centrifuged for 5 min at maximal speed. The supernatant containing soluble chromatin was then dialyzed for ∼2 h against TE to remove salts.
Protein and chromatin analyses
Proteins in aliquots of cytoplasmic and nuclear fractions were analyzed in SDS-PAGE. The fluorescent proteins were detected by fluorometry of the gels before staining. Chromatin was analyzed by native 5% PAGE (acrylamide/bisacrylamide: 40 : 1) in 0.5× TBE. Mononucleosome and dinucleosome species were isolated from preparative-scale 1% agarose gels containing 0.5× TBE. One analytical lane was loaded next to the preparative, stained with ethidium bromide, and used for localizing the nucleosomal species to avoid destabilization of nucleoprotein complexes by ethidium bromide. Mononucleosomes and dinucleosomes were electroeluted from the gel and samples concentrated by evaporation in a speed-vac. Samples were normalized according to DNA content (500 ng per lane) and protein composition analyzed by SDS-PAGE.
Acknowledgments
We thank Dr. Marty Gorovsky for a careful reading the manuscript and for helpful discussions. This work was supported by NIH grant RO1GM52426. We dedicate this paper to the memory of our friend and colleague Dr. Alan P. Wolffe, whose manifold scientific contributions will continue to stimulate countless minds.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-mail jjhs@uhura.cc.rochester.edu; FAX (716) 271-2683.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.910201.
References
- Almouzni G, Mechali M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988;7:665–672. doi: 10.1002/j.1460-2075.1988.tb02861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, Wolffe AP. Nuclear assembly, structure, and function: The use of Xenopus in vitro systems. Exp Cell Res. 1993;205:1–15. doi: 10.1006/excr.1993.1051. [DOI] [PubMed] [Google Scholar]
- Annunziato AA, Hansen JC. Role of istone acetylation in the assembly and modulation of chromatin structure. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. Topography of the histone octamer surface: Repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausio J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Bradbury EM, Inglis RJ, Matthews HR, Langan TA. Molecular basis of control of mitotic cell division in eukaryotes. Nature. 1974;249:553–556. doi: 10.1038/249553a0. [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Hansen JC. The core histone N termini function independently of linker histones during chromatin condensation. J Biol Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- Chang L, Loranger SS, Mizzen C, Ernst SG, Allis CD, Annunziato AT. Histones in transit: Cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- Dilworth SM, Black SJ, Laskey RA. Two complexes that contain histones are required for nucleosome assembly in vitro: Role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Fouquet H, Bohme R, Wick R, Sauer HW, Scheller K. Some evidence for replication-transcription coupling in Physarum polycephalum. J Cell Sci. 1975;18:27–39. doi: 10.1242/jcs.18.1.27. [DOI] [PubMed] [Google Scholar]
- Freeman L, Kurumizaka H, Wolffe AP. Functional domains for assembly of histones H3 and H4 into the chromatin of Xenopus embryos. Proc Natl Acad Sci. 1996;93:12780–12785. doi: 10.1073/pnas.93.23.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ramierez M, Dong F, Ausio J. Role of the histone ‘tails’ in the folding of oligonucleosomes depleted of histone H1. J Biol Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- Hayes JJ, Lee K-M. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods. 1997;12:2–9. doi: 10.1006/meth.1997.0441. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. p300-Mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes & Dev. 2000;14:1899–1907. [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: A molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Ling X, Harkness TA, Schultz MC, Fisher-Adams G, Grunstein M. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: Redundant and position-independent functions in assembly but not in gene regulation. Genes & Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- Loidl P, Grobner P. Histone synthesis during the cell cycle of Physarum polycephalum. Synthesis of different histone species. J Biol Chem. 1987;262:10195–10199. [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Jackson KR, Guo Y, Brame C J, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J Cell Biol. 2001;153:251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster T, Han M, Grunstein M. Yeast histone H2A and H2B amino termini have interchangeable functions. Cell. 1986;45:445–451. doi: 10.1016/0092-8674(86)90330-2. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Verreault A, Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1–mediated nucleosome assembly onto replicated DNA in vitro. Proc Natl Acad Sci. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- ————— Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ. Histone proteins in vivo: Cell-cycle-dependent physiological effects of exogenous linker histones incorporated into Physarum polycephalum. Methods. 1999;17:140–150. doi: 10.1006/meth.1998.0725. [DOI] [PubMed] [Google Scholar]
- ————— Assembly into chromatin and subtype-specific transcriptional effects of exogenous linker histones directly introduced into a living Physarum cell. J Cell Sci. 2001;114:965–973. doi: 10.1242/jcs.114.5.965. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Vitolo JM, Thiriet C, Hayes JJ. The H3-H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Mol Cell Biol. 2000;20:2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Chromatin structure and function. San Diego: Academic; 1998. [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]