Abstract
Trypanosoma brucei, the causative agent of human African trypanosomiasis, has a complex life cycle that includes multiple life cycle stages and metabolic changes as the parasite switches between insect vector and mammalian host. The parasite's single mitochondrion contains a unique catenated mitochondrial DNA network called kinetoplast DNA (kDNA) that is composed of minicircles and maxicircles. Long-standing uncertainty about the requirement of kDNA in bloodstream form (BF) T. brucei has recently eroded, with reports of posttranscriptional editing and subsequent translation of kDNA-encoded transcripts as essential processes for BF parasites. These studies suggest that kDNA and its faithful replication are indispensable for this life cycle stage. Here we demonstrate that three kDNA replication proteins (mitochondrial DNA polymerases IB, IC, and ID) are required for BF parasite viability. Silencing of each polymerase was lethal, resulting in kDNA loss, persistence of prereplication DNA monomers, and collapse of the mitochondrial membrane potential. These data demonstrate that kDNA replication is indeed crucial for BF T. brucei. The contributions of mitochondrial DNA polymerases IB, IC, and ID to BF parasite viability suggest that these and other kDNA replication proteins warrant further investigation as a new class of targets for the development of antitrypanosomal drugs.
INTRODUCTION
Trypanosoma brucei is the protist parasite responsible for the fatal human disease human African trypanosomiasis (HAT) and a related disease in livestock called nagana. The few current pharmacological options to treat HAT are hampered by high toxicity and the emergence of drug resistant parasites (1). Therefore, there is an urgent need for the development of new drugs. Trypanosomes possess a number of biological features without counterparts in humans that may provide sources of new targets for drug discovery efforts. One of the parasite's most remarkable properties is the unusual mitochondrial DNA network of trypanosomatids called kinetoplast DNA (kDNA). This DNA network is housed within the parasite's single mitochondrion and contains topologically interlocked circular DNA molecules called minicircles and maxicircles (43). Maxicircles are functionally similar to other eukaryotic mitochondrial DNA in that they encode proteins involved in respiratory complexes (13). Nascent maxicircle transcripts require insertion and deletion of uridines in order to create a functional open reading frame (16). This posttranscriptional process, known as RNA editing, is dependent upon minicircle-encoded guide RNAs (16, 45). Therefore, both minicircles and maxicircles are essential for mitochondrial physiology.
The topological complexity of the catenated kDNA network dictates a unique mode of replication in which minicircles are released from the network, replicated as theta structures, and reattached to the network periphery where Okazaki fragment processing occurs (43). A plethora of proteins involved in kDNA replication have been studied in T. brucei, including six helicases (25–27, 40), two DNA ligases (10), two primases (19, 20), a topoisomerase IA (40), a topoisomerase II (50), and five DNA polymerases (Pols) (4, 7, 21, 35). The involvement of multiple DNA polymerases in kDNA replication distinguishes this process from replication of other mitochondrial genomes, which depend solely upon DNA polymerase γ, a family A DNA polymerase. In T. brucei, Pol β and Pol β PAK (two mitochondrial family X DNA polymerases) contribute to Okazaki fragment processing and gap filling in the later stages of minicircle replication (35). The mitochondrial localization of the Pol β enzymes in T. brucei is in contrast to other eukaryotes, where Pol β enzymes participate in nuclear DNA repair. The three other mitochondrial DNA polymerases of T. brucei (POLIB, POLIC, and POLID) are family A proteins that are most related to prokaryotic DNA polymerase I and appear to function in the earlier stages of kDNA replication, each with a specialized function (4, 7, 21). POLIB, POLIC, and POLID lack homologues in mammals, including humans, thus identifying these proteins as potential biological targets for the development of new antitrypanosomal drugs. Analyses of kDNA replication proteins have provided compelling molecular evidence for essential functions in distinct steps of kDNA replication in procyclic form (PF) parasites, a life cycle stage found in its insect vector (4, 7, 20, 26). However, analysis of kDNA replication protein functions in bloodstream form (BF) parasites, the life cycle stage found in the mammalian host and the target for disease intervention (18, 37), is an understudied area of trypanosome biology.
A striking feature of T. brucei is its ability to adapt to diverse environments encountered throughout the stages of its life cycle. Developmental regulation of mitochondrial activity appears to play a central role in these adaptations (18, 30). PF parasites each possess a highly active, branched mitochondrion and generate ATP through oxidative phosphorylation and mitochondrial substrate-level phosphorylation (47). Conversely, BF parasites each have a much-reduced mitochondrion, lack cytochromes, and depend exclusively upon glycolysis for ATP production. A strictly glycolytic metabolism creates a seeming independence of BF parasites from maxicircle-encoded products and contributed to the assumption that kDNA is dispensable in the BF stage, thus diminishing the value of kDNA replication proteins as a source of new drug targets. This notion has been challenged by multiple lines of evidence, beginning with the demonstration that RNA editing is active and essential in BF parasites and that maxicircle-encoded subunit A6 of the ATP synthase complex (complex V) is required for generation of the mitochondrial membrane potential (ΔΨm) (14, 37, 39). More recently, mitochondrial translation was found to be essential for BF T. brucei (9). Further, inhibition of minicircle replication initiation appears to contribute to the trypanosome death elicited by treatment of infected animals with ethidium bromide (34). These findings suggest that kDNA is by no means dispensable in this medically relevant life cycle stage.
Only a single kDNA replication protein, topoisomerase II (TbTopoIImt), has been examined in BF T. brucei thus far. RNA interference (RNAi) resulted in a modest loss of kDNA networks (20 to 30%) accompanied by slowed parasite growth but not cell death (48, 53). The kDNA loss phenotype produced in BF parasites was significantly reduced compared to that produced in PF parasites, where TbTopoIImt RNAi resulted in loss of kDNA in ∼80% of the population (50). Silencing efficiency was not reported in these BF studies. Thus, it remains unclear if the slow-growth phenotype reflected a diminished requirement for this kDNA replication protein in BF parasites or an inefficient knockdown that makes data interpretation difficult. Nonetheless, these data could indicate that TbTopoIImt is crucial for BF survival and strongly suggest that kDNA replication proteins are indeed required for viability of BF parasites.
We directly examined this hypothesis by individually silencing the family A mitochondrial DNA polymerases POLIB, POLIC, and POLID in BF parasites. Our previous studies of these polymerases indicated that all three are required for cell growth and revealed nonredundant roles in PF kDNA replication but did not encompass studies in BF parasites (4, 7, 21). Here we report that depletion of these proteins was lethal to BF parasites and resulted in loss of kDNA networks. Network loss appeared to result from inhibition of minicircle replication and was accompanied by depolarization of mitochondrial membrane potential and subsequent parasite death. These findings provide the first direct evidence that BF parasites require kDNA replication for viability. Therefore, kDNA replication proteins warrant further investigation as biological targets for the development of new antitrypanosomal drugs.
MATERIALS AND METHODS
Trypanosome growth.
Bloodstream form T. brucei single marker (SM), a derivative of strain Lister 427 engineered to express T7 RNA polymerase and tetracycline repressor, were maintained at 37°C with 5% CO2 in HMI-9 medium as previously described (52). Cell densities were determined using a Neubauer hemocytometer, and cultures were maintained with between 5 × 104 and 1 × 106 parasites/ml unless otherwise indicated. To avoid generation of revertants, clonal cells were maintained in culture for no longer than 21 days.
RNAi.
Vectors for RNAi were constructed as described previously (2, 42, 51), substituting pT7-stl, a derivative of pLew100, in initial cloning steps. Coding sequences corresponding to 500-bp fragments of POLIB (Tb11.02.2300), POLIC (Tb927.7.3990), and POLID (Tb11.02.0770) were PCR amplified from Lister 427 genomic DNA using gene-specific primers with appropriate linkers. The coding sequences and primers for POLIB and POLID were identical to those previously used for RNAi in PF studies, with no reports of off-target effects (4, 7).
Forward (CGAGAGACAACCGAATCATCC) and reverse (TGCATAGCACCTCACGC) primers were used to amplify the fragment for POLIC. Notably, the gene fragments chosen to target each polymerase lack significant similarity to the other Pol I enzymes and the rest of the parasite's genome (using BLASTN, P value for genes showing the highest similarity to the RNAi target was 1). Following linearization with EcoRV, the stem-loop plasmids were transfected into SM parasites, using the Amaxa Nucleofector system as previously described (5), and stable clonal transfectants were selected using phleomycin (2.5 mg/ml) with limiting dilution. Clonal cell lines used for silencing of POLIB, POLIC, and POLID were termed SMIB, SMIC, and SMID, respectively. RNAi was induced by the addition of 1.0 μg/ml tetracycline in growth medium. Staggered RNAi inductions were performed to minimize variation in sample preparation.
Clonogenic assays.
Parasites that were uninduced or induced for 10 days of RNAi were subjected to limiting dilution cloning in HMI-9 supplemented with appropriate antibiotics but lacking tetracycline using 96-well plates at 1 parasite/ml. Individual wells were examined 5 days later for the presence of motile parasites, and plating efficiencies were determined. Proliferating parasites were diluted in HMI-9 medium and maintained as described above.
Microscopy and fluorescence analyses.
Parasites were pelleted at 800 × g, washed in room temperature trypanosome dilution buffer (TDB; 5 mM KCl, 80 mM NaCl, 1 mM MgSO4, 20 mM Na2HPO4, 2 mM NaH2PO4, 20 mM glucose; pH 7.7), and resuspended in TDB at a concentration of 2 × 107 parasites/ml. Parasites were allowed to settle by gravity onto poly-l-lysine-coated microscopy slides and fixed for 5 min in 1% formaldehyde dissolved in TDB. Following overnight permeabilization in ice-cold methanol, parasites were rehydrated with 3 washes in phosphate-buffered saline (PBS, pH 7.4), stained with 6.7 μg/ml 4′-6′-diamidino-2-phenylindole (DAPI), and mounted in Vectashield (Vector Laboratories). Slides were viewed using a Nikon Eclipse E600 microscope, and images were acquired using a Spot digital camera obtained from Diagnostic Instruments. Quantification of kDNA network morphology was performed as previously described (4, 7). To eliminate any potential for bias, the identities of samples were withheld from the individual performing the quantification.
Southern blot analysis.
Total DNA (isolated using Purescript genomic DNA isolation kit by Gentra Systems) from 1 × 106 parasites was digested overnight with HindIII and XbaI. Digested DNA was fractionated on a 300-ml (26-cm-long) 1% agarose gel containing 1.0 μg/ml ethidium bromide (EtBr) for 20 h at 2.4 V/cm. Ethidium bromide (1.0 μg/ml) was included in Tris-borate-EDTA running buffer, which was continuously recirculated. The gel was processed using standard depurination, denaturation, and neutralization treatments, and DNA was transferred to a GeneScreen Plus membrane as described previously (50). The resulting membrane was separated into three portions based upon migration of molecular mass markers and anticipated molecular masses of bands detected with random primed radiolabeled probes for minicircles (∼1 kb), maxicircles (∼1.4 kb), and the loading control α-tubulin (∼3.9 kb). The resulting three membranes were detected with approximately equal specific activities of the appropriate radiolabeled probes. Quantification and normalization were performed as described previously (4).
Neutral/alkaline two-dimensional electrophoresis.
Two-dimensional fractionation of total DNA was performed as previously described (4, 24). Briefly, total DNA isolated from parental or SMIB RNAi cells was separated in the first dimension for 18 h in the presence of 1.0 μg/ml ethidium bromide and then equilibrated and electrophoresed in the second dimension for 20 h in the presence of 50 mM NaOH. Following standard depurination, denaturation, and neutralization treatments, DNA was transferred to a GeneScreen Plus membrane. Leading- and lagging-strand minicircle replication intermediates were detected using strand-specific T4 polynucleotide kinase 5′-end-labeled oligonucleotide probes.
Analysis of mitochondrial membrane potential.
Detection of mitochondrial membrane potential was performed essentially as described previously (3). Uninduced and induced parasites were sedimented, resuspended in HMI-9 at 2.5 × 106 cells/ml, and incubated for 30 min at 37°C with 5% CO2 in HMI-9 containing MitoTracker Red CM-H2XRos (Invitrogen) provided at 1 μM for microscopy analyses or at 2.5 μM for flow cytometry analyses. Cells were then washed 3 times in PBS and fixed for microscopy as described above or resuspended in 1 ml of PBS for flow cytometry. Control cells were pretreated for 60 min with 5 μM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) or the FCCP carrier (100% ethanol), washed, and then resuspended in PBS. Changes in mitochondrial fluorescence intensity were analyzed using a Becton Dickinson LSR II flow cytometer. FlowJo software (version 7.6.1) was used to analyze and graph experimental results.
RESULTS
POLIB, POLIC, and POLID are required for viability of BF T. brucei.
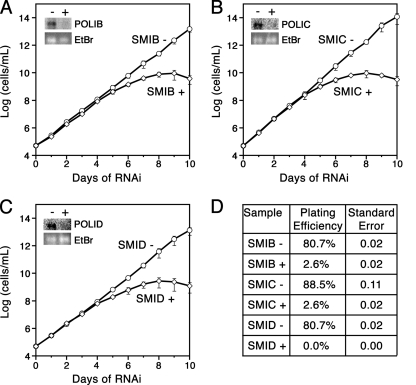
To determine if POLIB, POLIC, or POLID is required for T. brucei BF growth, inducible stem-loop RNAi constructs for each of the polymerases were stably integrated and selected for in SM parasites, which express T7 RNA polymerase and tetracycline repressor protein. The individual clonal cell lines, referred to as SMIB, SMIC, and SMID, grew with average doubling times of approximately 8.5 h, which was slightly slower than that of the parental line (∼8.2 h per doubling). Induction for silencing of POLIB, POLIC, and POLID all resulted in slowed growth after 4 days of RNAi, with subsequent parasite cell death (Fig. 1A to C). Northern blot analysis of parasites induced for 48 h of RNAi revealed knockdown efficiencies ranging from 90 to 95% for each target transcript (Fig. 1A to C, insets). Notably, parasites nonresponsive to RNAi, commonly referred to as “revertants,” did not emerge after 10 days of the SMIB, SMIC, or SMID RNAi induction, as previously reported for silencing other essential proteins in BF parasites (8, 49).
Fig. 1.
Effect of DNA polymerase RNAi on cell viability. (A to C) Clonal cell lines were grown in the absence (open circles) or presence (open diamonds) of tetracycline (1 μg/ml) to induce RNAi. Inset, Northern blot of RNA isolated from 5 × 107 parasites induced for 0 (−) or 48 (+) hours of RNAi of the gene indicated. Membranes were hybridized with 32P-labeled probes as described previously (4). Ethidium bromide (EtBr)-stained rRNA is included to show similar loading. Graph, cell density was plotted as the product of cell number and total dilution. Means and standard errors from three separate RNAi inductions are presented for clonal cell lines SMIB A24 (A), SMIC A15 (B), and SMID A13 (C). (D) Results from clonogenic assays performed with parasites that were uninduced (−) or induced (+) for 10 days of RNAi prior to plating. Viability (plating efficiency) and standard errors from two separate inductions are presented for each clonal cell line.
The low cell culture density required to cultivate BF parasites makes it challenging to observe cells that have potentially recovered or following prolonged periods of time in culture. Therefore, we performed clonogenic assays by limiting dilution to further assess the contributions of these kDNA replication proteins to BF parasite viability. Parasite cultures, which were uninduced or induced for 10 days of RNAi, were diluted to a single parasite per ml and plated in 96-well plates. Five days later, wells were examined for the presence of parasites. Parasites induced for RNAi prior to plating exhibited dramatically reduced plating efficiencies compared to those of the uninduced controls, which were 80 to 90% viable. Parasites induced for silencing of POLIB, POLIC, and POLID proliferated, with efficiencies of 2.6%, 2.6%, and 0%, respectively (Fig. 1D). These experiments demonstrate, for the first time, a lethal phenotype upon silencing of kDNA replication proteins in BF T. brucei.
POLIB, POLIC, and POLID perform essential kDNA maintenance roles in BF parasites.
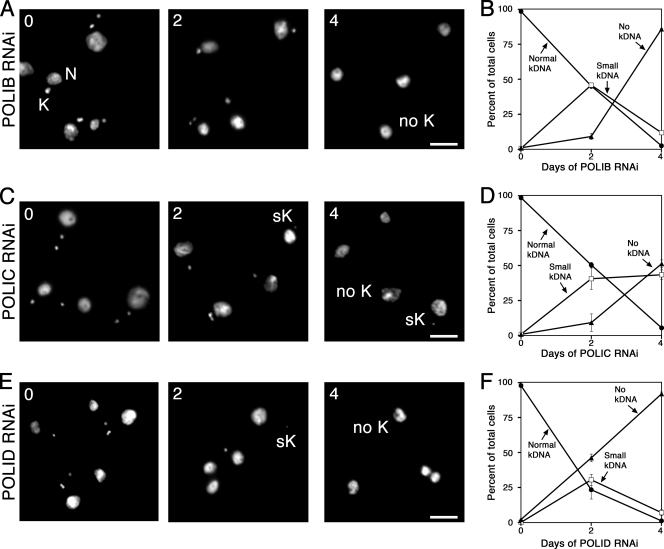
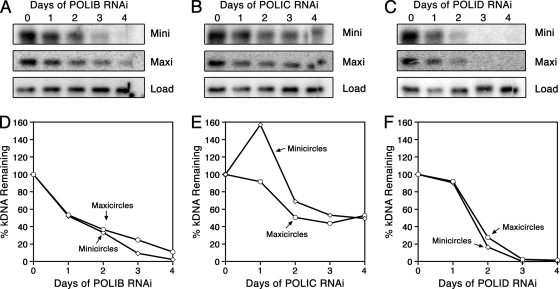
Functional studies have implicated numerous essential proteins for kDNA replication in PF parasites, yet only a single kDNA replication protein has been examined in disease-causing BF parasites. Silencing of this topoisomerase II (TbTopoIImt) in BF parasites resulted in a modest loss of kDNA compared to the extent of network loss observed when silencing this gene in PF parasites (48, 53). Therefore, we sought to assess the role of three mitochondrial DNA polymerases in BF kDNA maintenance. Parasites were stained with DAPI, which intercalates into both nuclear and mitochondrial DNA, and viewed using fluorescence microscopy. While normal-sized kDNA networks were clearly distinguishable in uninduced cultures, parasites induced for 4 days of polymerase silencing exhibited obvious network shrinkage and loss (Fig. 2A, C, and E). To quantify these striking observations, parasites induced for silencing of POLIB, POLIC, or POLID were observed and scored according to network size. More than 300 parasites per time point were classified as possessing normal-size networks, small kDNA (networks unambiguously less than one-half the size of normal-sized networks seen in uninduced cells), or no kDNA if no extranuclear DAPI staining was observed despite viewing multiple focal planes. Loss of kDNA resulted when each polymerase was silenced (Fig. 2B, D, and F). For example, after 4 days of POLIB silencing, the percentage of parasites possessing normal-sized kDNA fell to less than 3%, while parasites with no kDNA represented more than 85% of the cells at this time point. Kinetics of kDNA loss during silencing of POLIC and POLID were also rapid, with the majority of parasites completely lacking kDNA after 4 days of silencing. Interestingly, the kinetics of network loss seen during POLID silencing were almost indistinguishable from those produced during POLIB silencing, with less than 3% of the cells viewed possessing intact networks following 4 days of RNAi. To confirm loss of both minicircles and maxicircles, Southern blot analyses were performed during silencing of each polymerase (Fig. 3). Southern blot data revealed that silencing of POLIB and POLID produced robust loss of both kDNA species, while silencing of POLIC resulted in modest loss of minicircles and maxicircles, as reported in our previous studies of PF parasites (4, 7, 21).
Fig. 2.
Kinetics of kDNA loss during DNA polymerase silencing. (A, C, E) Representative images of parasites induced for indicated day of RNAi for POLIB (A), POLIC (C), or POLID (E). (B, D, F) Kinetics of kDNA loss were determined by classifying cells as possessing normal-sized kDNA (closed circles), small kDNA (open squares), or no kDNA (closed triangles). The means and standard errors from two inductions are presented for parasites depleted of POLIB (B), POLIC (D), and POLID (F). Abbreviations: N, nucleus; K, normal-sized kDNA; sK, small kDNA; no K, no kDNA. Scale bar, 5 μM.
Fig. 3.
Loss of minicircles and maxicircles during RNAi. (A to C) Membranes from Southern blot analysis of the abundance of minicircles and maxicircles during RNAi of POLIB (A), POLIC (B), and POLID (C). (D to F). Phosphorimaging quantification of total minicircle and maxicircle abundances from the membranes presented in panels A to C. Values were normalized against the α-tubulin loading control. Open diamonds, minicircles; open circles, maxicircles.
Dyskinetoplastid BF parasites produced during RNAi are not viable.
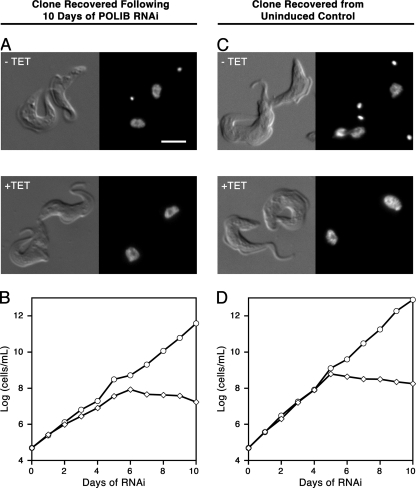
Previously, viable T. brucei that lacked portions of kDNA (dyskinetoplastids) were reported following extended treatment with the highly mutagenic DNA-binding compounds acriflavine and ethidium bromide (38, 44). A small percentage of parasites surviving 10 days of polymerase RNAi were viable in the clonogenic assays (Fig. 1D). To address the possibility that the viable cells following silencing had become nonresponsive to RNAi or were in fact dyskinetoplastid, individual clones recovered from clonogenic assays were expanded and further analyzed. Recovered parasites from POLIB RNAi-induced and -uninduced control cultures were stained with DAPI. All recovered cells examined possessed kDNA networks, as evident from DAPI staining (Fig. 4A and C), and remained sensitive to RNAi induction, with growth inhibition patterns similar to those of the parental cells (Fig. 4B and D). Similar results were seen for cells recovered following POLIC RNAi clonogenic assays as well (see Fig. S1 in the supplemental material). These data suggest incomplete knockdown rather than the development of insensitivity to RNAi or survival of parasites in the absence of kDNA.
Fig. 4.
Analysis of parasites recovered from POLIB clonogenic assays. (A, B) Parasites viable in clonogenic assays of cultures induced for POLIB RNAi were recovered and examined for the presence of kDNA and sensitivity to RNAi. (A) Differential interference contrast (DIC) and fluorescence microscopy images of DAPI-stained parasites that were grown in the presence or absence of tetracycline. (B) Growth curves of parasites recovered from clonogenic assays of POLIB RNAi-induced cultures. (C, D) Parasites viable in clonogenic assays of uninduced control cultures were recovered and assessed for RNAi sensitivity, as described in the legends to panels A and B. Scale bar, 5 μM.
Disruption of network-free minicircle replication precedes parasite death.
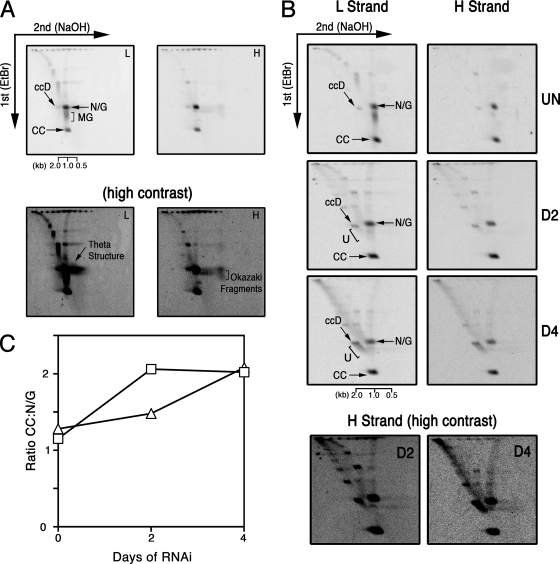
During kDNA replication, minicircles are released as covalently closed (CC) monomers and replicated as theta structures to produce nicked-and-gapped (N/G) nascent minicircles (11). Discontinuities in the DNA backbones of nascent minicircles decreases susceptibility to ethidium bromide-induced supertwisting; thus, newly replicated N/G species exhibit decreased electrophoretic mobility compared to that of unreplicated CC minicircles. The pattern of minicircle replication intermediates is well established in PF parasites, with CC and N/G minicircles present in approximately equimolar amounts (4, 24, 40). To ensure that the network-free mode of minicircle replication utilized by PF parasites is conserved in BF parasites, we performed two-dimensional analysis of free minicircles from single-marker cells, the parental line of our RNAi clones. Hybridization with radiolabeled oligonucleotides that detect leading- and lagging-strand replication intermediates revealed that the pattern of free minicircles in BF parasites was virtually indistinguishable from that of PF parasites (Fig. 5A, top). Higher-contrast images of detected membranes revealed theta structures as well as leading-strand (multiply gapped [MG])- and lagging-strand (Okazaki fragments)-specific intermediates (Fig. 5A, bottom).
Fig. 5.
Analysis of minicircle replication intermediates in parental and POLIB-depleted parasites. Neutral/alkaline two-dimensional gel electrophoresis of free minicircles. (A, top) Total DNA from parental parasites was separated in the presence of ethidium bromide and then under denaturing conditions (NaOH) prior to transfer to the membrane. Minicircle replication intermediates were detected with oligomers that specifically hybridize to leading (L)- or lagging (H)-strand intermediates. (Bottom) Higher-contrast images of membranes presented above. Contrast was adjusted equally in images of membranes to visualize the abundance of theta structures and Okazaki fragments. (B, top) Two-dimensional analysis of parasites induced for the indicated number of days of POLIB RNAi. (Bottom) Higher-contrast images of H-strand membranes presented above. (C) Phosphorimager quantification of changes in the relative abundance of unreplicated CC and newly replicated N/G intermediates in blots presented in panel B. Open squares, L-strand detection; open triangles, H-strand detection. Abbreviations: CC, covalently closed; ccD, covalently closed dimer; MG, multiply gapped; N/G, nicked/gapped; U, fraction U.
To provide further evidence that parasite cell death during RNAi resulted from inhibition of kDNA replication and to determine if functions of kDNA replication proteins appear consistent between PF and BF stages, we used two-dimensional electrophoresis. Previously reported analysis of minicircle replication disruption during POLIB RNAi indicated that this enzyme functions at the core of minicircle replication machinery (4). Silencing of POLIB in PF parasites resulted in the persistence of unreplicated CC minicircles accompanied by the accumulation of a multicatenane dimeric minicircle species known as fraction U (4). Therefore, we chose to focus our BF studies on POLIB. Analysis of minicircle replication intermediates produced during BF POLIB silencing revealed a decline in the detection of Okazaki fragments (Fig. 5B) and an increase in the abundance of unreplicated minicircles relative to newly replicated progeny, beginning 4 days postinduction for silencing (Fig. 5B and C). As in PF parasites, the persistence of unreplicated minicircles was accompanied by the accumulation of fraction U (Fig. 5B). These data indicate that POLIB performs a conserved role in minicircle replication in both PF and BF parasites and that disruption of kDNA replication leads to parasite cell death.
Disruption of mitochondrial membrane potential accompanies loss of kDNA.
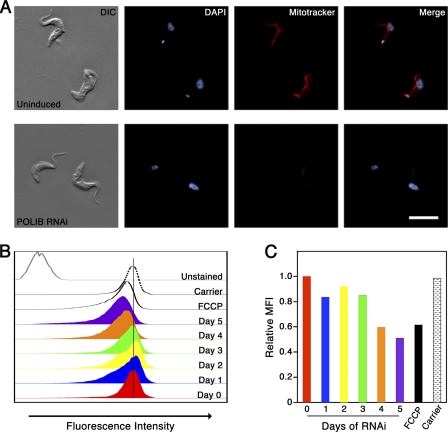
Viability of BF trypanosomes requires an intact ΔΨm (3). The ATP synthase complex is responsible for ΔΨm generation in BF trypanosomes, and Schnaufer and colleagues demonstrated ΔΨm depolarization and lethality upon RNAi silencing of the α subunit (37). Loss of kDNA networks during RNAi of mitochondrial DNA polymerases resulted in depletion of maxicircles (4, 7). We anticipated that the same loss of maxicircles in BF parasites (including the maxicircle-encoded subunit A6 of the ATP synthase complex) would subsequently lead to the collapse of ΔΨm and cell death. To determine if a collapse in ΔΨm could be contributing to lethality in parasites depleted of POLIB RNAi, we used the fluorescent dye MitoTracker Red CM-H2XRos. This cell-permeable dye is provided in a reduced form that fluoresces when oxidized within a polarized mitochondrion. Fluorescence microscopy analysis of uninduced parasites revealed staining of the tubular mitochondrion, indicating an intact ΔΨm (Fig. 6A). Parasites induced for RNAi exhibited depolarization of ΔΨm, with the MitoTracker fluorescence signal dramatically declining within 4 days of POLIB depletion (Fig. 6A). To provide a more quantitative analysis of ΔΨm collapse, flow cytometry analysis of MitoTracker Red-stained cells was performed. The fluorescence intensity of control cells pretreated with the protonophore FCCP to uncouple ΔΨm was significantly decreased compared to that of cells that were uninduced or pretreated with ethanol or dimethyl sulfoxide (DMSO), the solvents used for FCCP and MitoTracker Red, respectively. The fluorescence intensity of POLIB-depleted cells decreased over the time course of RNAi. The mean fluorescence intensity (MFI) of parasites induced for more than 3 days was similar to that of FCCP-treated negative-control parasites. Together, our findings demonstrate that loss of kDNA during DNA polymerase silencing results in depolarization of ΔΨm, which contributes to cell death.
Fig. 6.
Disruption of mitochondrial membrane potential during DNA polymerase silencing. (A) Representative images of MitoTracker Red-stained parasites that were either uninduced or induced for 4 days of POLIB RNAi. (B, C) Flow cytometry analysis of MitoTracker Red-stained parasites. Unstained control, parasites treated with DMSO (used as a solvent for MitoTracker solutions); FCCP, protonophore used as a negative control; carrier, parasites treated with ethanol (used as a carrier for FCCP). (B) Histogram showing fluorescence intensities of indicated samples. (C) Relative mean fluorescence intensities (MFI) of parasites presented in panel B. The unstained background was subtracted from raw MFI values prior to graphing the adjusted MFI relative to that of uninduced cells.
DISCUSSION
Individual silencing experiments for three mitochondrial DNA polymerases, POLIB, POLIC, and POLID, have previously revealed essential kDNA replication roles in the PF insect stage of the parasite. This stage relies on maxicircle-encoded proteins for its oxidative phosphorylation metabolism. Alternatively, the metabolism of the disease-causing BF stage of the parasite is exclusively glycolytic (12, 47). Therefore, the loss of kDNA would be lethal to BF parasites only if the kDNA-encoded proteins function in cellular processes besides oxidative phosphorylation. Recent studies indicate that RNA editing proteins, the A6 subunit of ATP synthase, and mitochondrial translation are essential in BF trypanosomes (9, 37). However, silencing of TbTopoIImt, the enzyme involved in reattaching newly synthesized minicircles to the network, in BF parasites resulted in only mild growth and kDNA loss defects. The goal of this study was to determine whether kDNA replication proteins were essential for BF parasite viability. Here we report the rapid loss of kDNA networks upon silencing of POLIB, POLIC, and POLID, and for each polymerase, loss of kDNA was followed by cell death. This marks the first time that ablation of kDNA replication proteins results in the lethality of BF T. brucei.
Knockdown of each polymerase gene resulted in cell death 5 to 6 days post-RNAi induction (Fig. 1). This likely indicates that loss of essential proteins encoded by kDNA (such as ATP synthase subunit A6), rather than depletion of enzymes required for its replication, is the primary cause of cell death. Proper mitochondrial function is required for numerous processes critical to cell physiology, including energy metabolism, calcium homeostasis and signaling, and generation of membrane potential (3). This creates an attractive paradigm in which inhibition of a single kDNA replication enzyme could lead to the disruption of multiple essential cellular pathways, effectively creating a multipotent effect from inhibiting a single target. Maintenance of the mitochondrial membrane potential is clearly among these pathways, as we demonstrate here with membrane potential collapse beginning when POLIB-depleted parasites have lost more than 80% of minicircle and maxicircle mass (Fig. 6). Loss of kDNA produced in response to ablation of other kDNA replication proteins in BF parasites would likely lead to a similar depolarization of membrane potential and subsequent parasite death. Yet unknown functions for kDNA-encoded proteins likely exist and may also prove to be essential in BF parasites. Alternative editing of maxicircle transcripts is hypothesized to increase mitochondrial protein diversity (33). Indeed, AEP1, a product of alternative editing of cytochrome oxidase III, was identified as a kDNA maintenance factor in BF T. brucei (32). Additionally, a maxicircle coding sequence contains three “maxicircle unidentified reading frames” (MURFs) and a series of GC-rich regions predicted to encode a series of highly hydrophobic proteins of unknown function (38). A more complete understanding of these components and the repertoire of proteins produced by alternative editing of maxicircle transcripts may reveal additional indispensable functions of kDNA-encoded components for BF parasites.
Our current functional analyses of the kDNA replication proteins POLIB, POLIC, and POLID indicate that the essential roles of these proteins in kDNA replication appear consistent in both life cycle stages examined. Silencing each of the polymerases resulted in loss of kDNA networks and was accompanied by changes in the repertoire of free minicircle species. For example, when silencing POLIB, unreplicated CC monomers persisted and fraction U accumulated, with the BF results indistinguishable from those obtained when silencing POLIB in PF parasites (Fig. 5). Additionally, the kinetics of kDNA loss for POLIB and POLID were nearly identical, again similar to the results obtained from the PF silencing experiments. Interestingly, when comparing the rate of kDNA loss, however, the BF parasites appear to lose their kDNA with faster kinetics. While it took nearly 20 doublings for cells to lose their kDNA in PF POLIB silencing (52% with no kDNA, 40% with small kDNA), it took only 12 generations for BF parasites to lose their kDNA (90% with no kDNA). Currently we do not understand why kDNA loss occurs more rapidly in BF parasites, but life stage-specific cell cycle checkpoints have been identified and may contribute to these differences (17).
This study is the first in-depth analysis of kDNA replication protein function in BF parasites. Previous functional studies of kDNA replication proteins have been performed in PF parasites, including those from our laboratory, yet the relevance of these analyses to drug development mandates essential function in disease-causing BF parasites. Focus on the PF stage is largely the result of highly efficient stable transfection methods for this form of the parasite. While standard transfection methodologies yield efficiencies of 10−3 to 10−6 in PF T. brucei, the technique is remarkably less successful in BF parasites (10−7 to 10−8) (6, 23, 29, 46). However, the recent application of nucleofection to BF parasites increased stable transformation efficiency nearly 1,000-fold, providing greater opportunity to examine kDNA replication protein function in this disease-causing life cycle stage (5). When silencing the three DNA polymerases, we found that parasites that survived RNAi (proliferated in clonogenic assays) still contained intact kDNA networks and remained responsive to induction for RNAi, as evidenced from growth kinetics and loss of kDNA during RNAi (Fig. 4; see also Fig. S1 in the supplemental material). Sustained sensitivity to induction for RNAi is noteworthy here, since RNAi-resistant “revertant” parasites have been widely reported in both PF and BF T. brucei (8, 15, 22, 30, 31, 49). The reasons why revertants were not produced when silencing POLIB, POLIC, or POLID is beyond the scope of our analyses. Sustained RNAi sensitivity, however, was critical in determining the essential contribution that each of these mitochondrial DNA polymerases makes to the viability of BF T. brucei.
The demonstration that BF T. brucei cannot survive without kDNA is fundamental in evaluating kDNA replication proteins as drug targets. Yet, trypanosomes lacking functional portions of their kDNA genome exist in nature and have been generated through prolonged culture in the presence of mutagenic conditions (38, 44). Isolates of naturally occurring dyskinetoplastid trypanosomes (T. evansi and T. equiperdum) vary in regards to the abundance and identity of residual minicircles and maxicircles (38). Although permissive in these BF parasites, partial loss of kDNA locks the parasite into a monomorphic life cycle that is unable to survive in the tsetse fly vector and, therefore, spread from an infected host (28, 36). Naturally dyskinetoplastid T. equiperdum and T. evansi, as well as an acriflavine-induced dyskinetoplastid strain of T. brucei, were recently found to possess mutations in the nuclear encoded γ subunit of the ATP synthase complex. These mutations are proposed to compensate for the loss of maxicircle-encoded subunit A6 of this complex (28, 36, 37). The compound ethidium bromide inhibits kDNA replication in BF parasites, yet viable dyskinetoplastid T. brucei has not emerged despite decades of the compound's use in treating infected animals, suggesting that compensatory nuclear mutations occur at a low frequency (34). Additionally, RNAi of other proteins required for kDNA function (particularly RNA editing) fails to produce viable dyskinetoplastid parasites (39). Chemical inhibitors of kDNA replication proteins would likely inactivate target proteins even more rapidly than RNAi, thus decreasing the window of time for selection of these low-frequency mutations.
Our current study adds to a rapidly growing body of literature indicating that kDNA is required for BF mitochondrial physiology and, thus, viability. Indeed, two available treatments for sleeping sickness, pentamidine (in humans) and ethidium bromide (in livestock), appear to target kDNA (34, 41). The historical success of drugs targeting kDNA and our finding that mitochondrial DNA polymerases IB, IC, and ID are essential in BF parasites indicate that targeting kDNA replication proteins remains a promising approach for the discovery of new antitrypanosomal drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bibo Li (Cleveland State University) for performing transfections and providing the cell lines used in this study and Christina Arieta (University of Massachusetts Amherst) for assistance with flow cytometry. We are additionally grateful to Arthur Günzl (University of Connecticut Health Center) for his gift of the pT7-stl RNAi vector and the laboratory of Nagendra Yadava (Pioneer Valley Life Sciences Institute, Springfield, MA) for providing the FCCP membrane potential uncoupler. We appreciate useful comments and discussion on the manuscript from James Morris, Arthur Günzl, and Derrick Robinson.
This research was supported by NIH grant AI066279 to M.M.K.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Baral T. N. 2010. Immunobiology of African trypanosomes: need of alternative interventions. J. Biomed. Biotechnol. 2010:389153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brandenburg J., et al. 2007. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 26:4856–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown S. V., Hosking P., Li J., Williams N. 2006. ATP synthase is responsible for maintaining mitochondrial membrane potential in bloodstream form Trypanosoma brucei. Eukaryot. Cell 5:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruhn D. F., Mozeleski B., Falkin L., Klingbeil M. M. 2010. Mitochondrial DNA polymerase POLIB is essential for minicircle DNA replication in African trypanosomes. Mol. Microbiol. 75:1414–1425 [DOI] [PubMed] [Google Scholar]
- 5. Burkard G., Fragoso C. M., Roditi I. 2007. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 153:220–223 [DOI] [PubMed] [Google Scholar]
- 6. Carruthers V. B., H. van der Ploeg L., Cross G. A. 1993. DNA-mediated transformation of bloodstream-form Trypanosoma brucei. Nucleic Acids Res. 21:2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandler J., Vandoros A. V., Mozeleski B., Klingbeil M. M. 2008. Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes. Eukaryot. Cell 7:2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y., Hung C. H., Burderer T., Lee G. S. 2003. Development of RNA interference revertants in Trypanosoma brucei cell lines generated with a double stranded RNA expression construct driven by two opposing promoters. Mol. Biochem. Parasitol. 126:275–279 [DOI] [PubMed] [Google Scholar]
- 9. Cristodero M., Seebeck T., Schneider A. 2010. Mitochondrial translation is essential in bloodstream forms of Trypanosoma brucei. Mol. Microbiol. 78:757–769 [DOI] [PubMed] [Google Scholar]
- 10. Downey N., Hines J. C., Sinha K. M., Ray D. S. 2005. Mitochondrial DNA ligases of Trypanosoma brucei. Eukaryot. Cell 4:765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drew M. E., Englund P. T. 2001. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell Biol. 153:735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durieux P. O., Schutz P., Brun R., Kohler P. 1991. Alterations in Krebs cycle enzyme activities and carbohydrate catabolism in two strains of Trypanosoma brucei during in vitro differentiation of their bloodstream to procyclic stages. Mol. Biochem. Parasitol. 45:19–27 [DOI] [PubMed] [Google Scholar]
- 13. Feagin J. E. 2000. Mitochondrial genome diversity in parasites. Int. J. Parasitol. 30:371–390 [DOI] [PubMed] [Google Scholar]
- 14. Fisk J. C., Ammerman M. L., Presnyak V., Read L. K. 2008. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 283:23016–23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galland N., et al. 2007. Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei. Biochim. Biophys. Acta 1773:521–535 [DOI] [PubMed] [Google Scholar]
- 16. Hajduk S., Ochsenreiter T. 2010. RNA editing in kinetoplastids. RNA Biol. 7:229–236 [DOI] [PubMed] [Google Scholar]
- 17. Hammarton T. C. 2007. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 153:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hannaert V., Bringaud F., Opperdoes F. R., Michels P. A. 2003. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol. Dis. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hines J. C., Ray D. S. 2010. A mitochondrial DNA primase is essential for cell growth and kinetoplast DNA replication in Trypanosoma brucei. Mol. Cell. Biol. 30:1319–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hines J. C., Ray D. S. 2011. A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei. Eukaryot. Cell 10:445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klingbeil M. M., Motyka S. A., Englund P. T. 2002. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell 10:175–186 [DOI] [PubMed] [Google Scholar]
- 22. Krazy H., Michels P. A. 2006. Identification and characterization of three peroxins—PEX6, PEX10 and PEX12—involved in glycosome biogenesis in Trypanosoma brucei. Biochim. Biophys. Acta 1763:6–17 [DOI] [PubMed] [Google Scholar]
- 23. Li F., Gottesdiener K. M. 1996. An efficient method for stable transfection of bloodstream-form Trypanosoma brucei. Nucleic Acids Res. 24:534–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu B., et al. 2006. Role of p38 in replication of Trypanosoma brucei kinetoplast DNA. Mol. Cell. Biol. 26:5382–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu B., et al. 2009. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol. Cell 35:490–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu B., Wang J., Yildirir G., Englund P. T. 2009. TbPIF5 is a Trypanosoma brucei mitochondrial DNA helicase involved in processing of minicircle Okazaki fragments. PLoS Pathog. 5:e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu B., et al. 2010. TbPIF1, a Trypanosoma brucei mitochondrial DNA helicase, is essential for kinetoplast minicircle replication. J. Biol. Chem. 285:7056–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lun Z. R., Lai D. H., Li F. J., Lukes J., Ayala F. J. 2010. Trypanosoma brucei: two steps to spread out from Africa. Trends Parasitol. 26:424–427 [DOI] [PubMed] [Google Scholar]
- 29. McCulloch R., Vassella E., Burton P., Boshart M., Barry J. D. 2004. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262:53–86 [DOI] [PubMed] [Google Scholar]
- 30. Milman N., Motyka S. A., Englund P. T., Robinson D., Shlomai J. 2007. Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proc. Natl. Acad. Sci. U. S. A. 104:19250–19255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moyersoen J., et al. 2003. Characterization of Trypanosoma brucei PEX14 and its role in the import of glycosomal matrix proteins. Eur. J. Biochem. 270:2059–2067 [DOI] [PubMed] [Google Scholar]
- 32. Ochsenreiter T., Anderson S., Wood Z. A., Hajduk S. L. 2008. Alternative RNA editing produces a novel protein involved in mitochondrial DNA maintenance in trypanosomes. Mol. Cell. Biol. 28:5595–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ochsenreiter T., Cipriano M., Hajduk S. L. 2008. Alternative mRNA editing in trypanosomes is extensive and may contribute to mitochondrial protein diversity. PLoS One 3:e1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roy Chowdhury A., et al. 2010. The killing of African trypanosomes by ethidium bromide. PLoS Pathog. 6:e1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saxowsky T. T., Choudhary G., Klingbeil M. M., Englund P. T. 2003. Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J. Biol. Chem. 278:49095–49101 [DOI] [PubMed] [Google Scholar]
- 36. Schnaufer A. 2010. Evolution of dyskinetoplastic trypanosomes: how, and how often? Trends Parasitol. 26:557–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schnaufer A., Clark-Walker G. D., Steinberg A. G., Stuart K. 2005. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 24:4029–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schnaufer A., Domingo G. J., Stuart K. 2002. Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int. J. Parasitol. 32:1071–1084 [DOI] [PubMed] [Google Scholar]
- 39. Schnaufer A., et al. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159–2162 [DOI] [PubMed] [Google Scholar]
- 40. Scocca J. R., Shapiro T. A. 2008. A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes. Mol. Microbiol. 67:820–829 [DOI] [PubMed] [Google Scholar]
- 41. Shapiro T. A., Englund P. T. 1990. Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc. Natl. Acad. Sci. U. S. A. 87:950–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi H., et al. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shlomai J. 2004. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 4:623–647 [DOI] [PubMed] [Google Scholar]
- 44. Stuart K. D. 1971. Evidence for the retention of kinetoplast DNA in an acriflavine-induced dyskinetoplastic strain of Trypanosoma brucei which replicates the altered central element of the kinetoplast. J. Cell Biol. 49:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stuart K. D., Schnaufer A., Ernst N. L., Panigrahi A. K. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30:97–105 [DOI] [PubMed] [Google Scholar]
- 46. ten Asbroek A. L., Ouellette M., Borst P. 1990. Targeted insertion of the neomycin phosphotransferase gene into the tubulin gene cluster of Trypanosoma brucei. Nature 348:174–175 [DOI] [PubMed] [Google Scholar]
- 47. Tielens A. G., van Hellemond J. J. 2009. Surprising variety in energy metabolism within Trypanosomatidae. Trends Parasitol. 25:482–490 [DOI] [PubMed] [Google Scholar]
- 48. Timms M. W., van Deursen F. J., Hendriks E. F., Matthews K. R. 2002. Mitochondrial development during life cycle differentiation of African trypanosomes: evidence for a kinetoplast-dependent differentiation control point. Mol. Biol. Cell 13:3747–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Urbaniak M. D. 2009. Casein kinase 1 isoform 2 is essential for bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 166:183–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Z., Englund P. T. 2001. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 20:4674–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Z., Morris J. C., Drew M. E., Englund P. T. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174–40179 [DOI] [PubMed] [Google Scholar]
- 52. Wirtz E., Leal S., Ochatt C., Cross G. A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 53. Worthen C., Jensen B. C., Parsons M. 2010. Diverse effects on mitochondrial and nuclear functions elicited by drugs and genetic knockdowns in bloodstream stage Trypanosoma brucei. PLoS Negl. Trop. Dis. 4:e678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.