Abstract
Inhibitors that reduce viral nucleic acid extraction efficiency and interfere with cDNA synthesis and/or polymerase activity affect the molecular detection of viruses in aquatic environments. To overcome these significant problems, we developed a methodology for assessing nucleic acid yields and DNA amplification efficiencies for environmental water samples. This involved adding particles of adenovirus type 5 and murine norovirus and newly developed primer-sharing controls, which are amplified with the same primer pairs and result in the same amplicon sizes as the targets, to these samples. We found that nucleic acid loss during the extraction process, rather than reverse transcription-PCR (RT-PCR) inhibition, more significantly attributed to underestimation of the presence of viral genomes in the environmental water samples tested in this study. Our success rate for satisfactorily amplifying viral RNAs and DNAs by RT-PCR was higher than that for obtaining adequate nucleic acid preparations. We found that inhibitory properties were greatest when we used larger sample volumes. A magnetic silica bead-based RNA extraction method effectively removed inhibitors that interfere with viral nucleic acid extraction and RT-PCR. To our knowledge, this is the first study to assess the inhibitory properties of environmental water samples by using both control virus particles and primer-sharing controls.
INTRODUCTION
Enteric viruses are one of the most important causative agents of waterborne gastroenteritis because of their high infectivity (13, 43), persistence in water (1, 2), and tolerance to wastewater treatment (9, 33) and chlorination (44). Therefore, monitoring viruses in the environment is important for protecting public health (48). However, the inhibitory effects caused by some substances present in virus concentrates can hinder these tests, as in the case of analysis involving reverse transcription-PCR (RT-PCR), by causing underestimation of virus genomes. Such inhibitory effects also occur in the detection of bacterial genome (7, 45). Humic acid, fulvic acid, a humic acid-like component in beef extract, which can be used as an eluent for virus concentration, and cations such as Ca2+ and Fe3+ are known to inhibit RT-PCR (1, 15, 26, 34, 49). Microfiltration (MF) membrane-based methods using acid rinse procedures followed by alkaline elution were developed to avoid the use of beef extract (16, 21). However, RT-PCR inhibition has been reported to be observed even under these conditions (11, 15), probably because of coconcentration of humic acid, which precipitates in the presence of cations (6) or acid conditions and becomes soluble at alkaline pH (23). Moreover, a silica membrane-based nucleic acid extraction/purification method, which is a commonly used method for molecular detection of viruses in aquatic environments (3, 15, 41), cannot always effectively remove humic acid from samples (3, 34). The principle of this method is based on attachment and detachment of nucleic acids to silica by altering pH and ionic strength (29). Nucleic acids tend to bind silica at acidic pH in the presence of chaotropic salts but do not bind tightly at neutral or mild pH. This accounts for the presence of humic acid in concentrated virus preparations. Factors that interfere with nucleic acid extraction and isolation have not been identified (8, 10).
There have been several techniques reported to reduce inhibitory effects. Addition of T4 gene 32 protein (26), bovine serum albumin (26), or polyvinylpyrrolidone (31) to samples are known to reduce inhibitory effects. Removal of inhibitors by using an antigen-antibody reaction (38), cation exchange resin (1), or gel chromatography (1, 5) has also been reported. However, these techniques cannot reduce the inhibitory effects entirely or may cause loss of viruses during the process (1, 26, 31, 38). Therefore, developing methods to evaluate the magnitude of inhibition is essential (14, 42). Exogenously added control nucleic acids have been used for this purpose (14, 27, 30, 32, 37, 42). Although the inhibition of polymerase activity depends on the target nucleotide sequences (22, 40), most previous studies have not employed sequence-matched controls (27, 30, 32, 37, 42). Gregory et al. (14) used a control that had matched sequences at primer annealing sites with the target and therefore was amplified by the same primer pair as the target, and they successfully predicted the magnitude of RT-PCR inhibition. However, amplicon size must also be considered, because inhibition becomes more pronounced with longer templates (22, 49). Nevertheless, use of a control that considers both primer sequence and amplicon size has not been reported.
To improve these techniques, we developed novel primer-sharing controls (PSCs) to evaluate the magnitude of RT-PCR inhibition. These PSCs were then applied to assess inhibitory effects on viral genome detection in samples acquired from rivers, lakes, and groundwater. Prior to nucleic acid extraction, murine norovirus (MNV) and human adenovirus type 5 (Ad5) were also added to samples as control virus particles in order to evaluate nucleic acid extraction efficiencies. In addition, we compared nucleic acid extraction and purification methods by using PSCs and the virus particles.
MATERIALS AND METHODS
Viruses.
Human Ad5 and MNV (S7-PP3 strain) were kindly provided by M. Ito (Kyoto City Institute of Health and Environmental Sciences, Kyoto, Japan) and Y. Tohya (Nihon University, Kanagawa, Japan) and propagated in HEp-2 (ATCC CCL-23) and RAW 264.7 (ATCC TIB-71) cell lines (American Type Culture Collection, Manassas, VA). Ad40 and a human norovirus (NoV; GII-4; Lordsdale-like strain) were kindly provided by Y. Yoshida (Tokyo Metropolitan Institute of Public Health, Tokyo, Japan) and E. Utagawa (National Institute of Infectious Diseases, Tokyo, Japan), respectively, as supernatants derived from patients' fecal specimens.
Construction of PSC RNA and DNA.
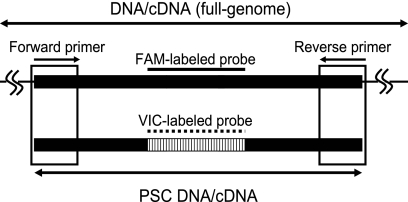
PSC has the same sequence as the amplicon of the target viral nucleic acid except for the TaqMan probe recognition sequence (Fig. 1). Hence, PSC is expected to be reverse transcribed and amplified with the same efficiency as the target nucleic acid even in cases in which RT-PCR inhibition occurs. Therefore, by using PSC as the internal control, the occurrence and magnitude of RT-PCR can be estimated.
Fig. 1.
Schematic diagram of PSC DNA amplified by the same primer set as target genome DNA, which was of the same amplicon size but was recognized by a different TaqMan probe.
Enteric AdV (EAdV) PSC DNA (118 bp) was chemically synthesized to have a sequence stretch identical to an AdV41 strain (GenBank accession number X16583) except for the target sequence of the EAdV-specific TaqMan probe (JTVFP) (Table 1), which was replaced with the sequence of the MNV-specific TaqMan probe (MKMNV-TP [Table 1]). Similarly, GII NoV PSC DNA (98 bp) was chemically synthesized to have a sequence stretch identical to Camberwell virus (GenBank accession number AF145896) except for the target sequence of the GII NoV-specific TaqMan probe (RING2-TP [Table 1]), which was replaced with the sequence of MNV-specific TaqMan probe (MKMNV-TP [Table 1]). GII NoV PSC RNA was synthesized by in vitro transcription using GII NoV PSC DNA. Briefly, GII NoV PSC DNA was cloned into a Zero Blunt TOPO pCR2.1 vector (Invitrogen, Carlsbad, CA), which contains T7 promoter sequence recognized by T7 RNA polymerase. The construct was used to transform One Shot TOP10 chemically competent Escherichia coli (Invitrogen). Transformants were incubated at 37°C on a Luria broth agar plate containing kanamycin (50 μg/ml). Colonies were picked and incubated at 37°C on Luria broth agar. Plasmid DNA was extracted using a QIAprep spin miniprep kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Plasmid DNA containing the GII NoV PSC DNA sequence was transcribed using T7 RNA polymerase (Toyobo, Osaka, Japan) according to the manufacturer's instructions. The reaction mixture (10 μl) was then treated with 1 U of RQ1 DNase (Promega, Madison, WI) in DNase I buffer (150 mM Tris-HCl [pH 8.3], 225 mM KCl, and 9 mM MgCl2) at 37°C for 30 min to digest plasmid DNA and then at 75°C for 5 min to inactivate DNase. Constructed PSC DNA and RNA were stored at −80°C until the analysis.
Table 1.
Primers and probes used for RT-qPCR
| Primer | Target | Polarity | Sequence (5′→3′)d | Length | Product size (bp) | Reference |
|---|---|---|---|---|---|---|
| TVFF | EAdVs and EAdV PSCa | + | AACTTTCTCTCTTAATAGACGCC | 23-mer | 118 | 25 |
| JTVFR | EAdVs and EAdV PSC | − | AGGGGGCAGAAAACAAAA | 19-mer | ||
| JTVFP | EAdVs | + | FAM-CTGACACGGGCACTCTTCGC-TAMRA | 20-mer | ||
| AQ2 | Ad5b | + | GCCCCAGTGGTCTTACATGCACATC | 25-mer | 132 | 18 |
| AQ1 | Ad5 | − | GCCACGGTGGGGTTTCTAAACTT | 23-mer | ||
| AP | Ad5 | + | FAM-TGCACCAGACCCGGGCTCAGGTACTCCGA-TAMRA | 29-mer | ||
| COG2F | GII NoVs and GII NoV PSCc | + | CARGARBCNATGTTYAGRTGGATGAG | 26-mer | 98 | 20 |
| COG2R | GII NoVs and GII NoV PSC | − | TCGACGCCATCTTCATTCACA | 21-mer | ||
| RING2-TP | GII NoVs and GII NoV PSC | + | FAM-TGGGAGGGCGATCGCAATCT-TAMRA | 20-mer | ||
| MKMNVF | MNVc | + | CGGTGAAGTGCTTCTGAGGTT | 21-mer | 60 | 24 |
| MKMNVR | MNV | − | GCAGCGTCAGTGCTGTCAA | 19-mer | ||
| MKMNV-TP | MNV, EAdV PSC, and GII NoV PSC | + | VIC–CGAACCTACATGCGTCAG–NFQ-MGB | 18-mer |
EAdV DNA and EAdV PSC DNA were amplified with the following thermal cycling conditions: initial denaturation at 95oC for 10 min and then 50 cycles of amplification with denaturation at 95oC for 15 s and annealing and extension at 55oC for 60 s.
Ad5 DNA was amplified with the following thermal cycling conditions: initial denaturation at 95oC for 10 min and then 50 cycles of amplification with denaturation at 95oC for 15 s and annealing and extension at 55oC for 10 s and 65oC for 60s.
GII NoV DNA, GII NoV PSC DNA, and MNV DNA were amplified with the following thermal cycling conditions: initial denaturation at 95oC for 10 min and then 50 cycles of amplification with denaturation at 95oC for 15 s and annealing and extension at 56oC for 60 s.
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine; VIC, 6-carboxyrhodamine; NFQ-MGB, nonfluorescent quencher-minor groove binder.
Humic acid solution and enrichment.
Thirty-six milligrams of humic acid (Sigma-Aldrich, Tokyo, Japan) was added to 1.0 liter of distilled water and dissolved under alkaline conditions (pH 13) in the presence of NaOH. The humic acid solution was subjected to a virus concentration procedure (21) to obtain humic acid solution that was coconcentrated with viruses and caused inhibition. The humic acid concentration in the eluate was determined as a reference by measuring total dissolved solids in 3 ml of the eluate after evaporation (0.49 mg/ml). Humic acid absorbance at 260 nm (E260) was determined using a Hitachi U-2010 spectrophotometer (Hitachi, Tokyo, Japan).
Environmental water samples. (i) Water sample collection.
Twenty-four environmental water samples were collected from September to October 2009. Thirteen river water samples were collected at seven sites along the Siem Reap River in Cambodia (SR-1 to SR-7) and at six sites along the Chao Phraya River in Thailand (BR-1, BR-2, BC-1, BC-2, BM-1, and BM-2). Five lake water samples were collected from the Tonle Sap Lake in Cambodia (SL-1 to SL-5). Four groundwater samples were also collected from Cambodia (SG-1 to SG-4). Samples were concentrated on-site immediately after collections in polyethylene containers (1.8-liter or 20-liter volume) presterilized by gamma radiation. The concentrated samples were kept cool and transported to the laboratory at The University of Tokyo, Tokyo, Japan, within 7 days according to methods described in a previous study (17). The samples were stored at −80°C immediately after transportation until further concentration.
(ii) Virus concentration.
Small volumes of environmental water samples (50 to 2,000 ml) were concentrated as previously described (21), with minor modifications. Briefly, 2.5 M MgCl2 was added to the samples to obtain a final concentration of 25 mM and was filtered through an electro-negative filter (45-mm diameter, 0.45-μm pore size; Millipore). Next, 200 ml H2SO4 (pH 3.0) was passed through the filter, and virus was eluted with 5 ml NaOH (pH 10.8). The eluate was recovered in a tube containing 25 μl H2SO4 (pH 1.0) and 50 μl 100× Tris-EDTA buffer. Samples were further concentrated using a Centriprep YM-50 apparatus (Millipore) to approximately 650 μl after the transportation. Large volumes of environmental water samples (8 to 200 liters) were concentrated using a cartridge with a Durapore polyvinylidene difluoride filter (Opticap XL2 disposable capsule filters; 0.45-μm pore size, 0.1-m2 filtration area; Millipore). Briefly, water samples were suction filtered using a sterilized tube and an aspirator (AS-01; AS ONE, Osaka, Japan). The sample was then mixed with 2.5 M MgCl2 by injecting 1 meter upstream of the filter to obtain a final concentration of 25 mM. Next, the filter was rinsed with 4 liters of H2SO4 (pH 3) and 2 liters of distilled water. Viruses were eluted with 200 ml NaOH (pH 10.8) and collected in a tube containing 1 ml H2SO4 (pH 1.0) and 2 ml 100× Tris-EDTA buffer. Sample (60 ml) from the 200-ml concentrate was reduced to approximately 250 μl by using a Centricon plus-70 filter (Millipore) after the transportation. The further-concentrated samples were stored at −80°C until purification or nucleic acid extraction.
(iii) Gel chromatography.
Gel chromatography was performed using a MicroSpin S-300 HR apparatus (Amersham Biosciences, Tokyo, Japan) to remove inhibitors. Briefly, 350 μl distilled water was added to the column and centrifuged seven times for 1 min at 735 × g. Two hundred microliters of concentrated sample was then applied to the column and centrifuged for 2 min at 735 × g before DNA extraction.
(iv) Nucleic acid extraction.
Silica membrane-based nucleic acid extraction was performed using a QIAamp DNA minikit (Qiagen) and a QIAamp viral RNA minikit (Qiagen) to extract DNA and RNA, respectively, according to the manufacturer's instructions. Briefly, 200 μl of a sample was subjected to DNA extraction to obtain a final volume of 200 μl, while 140 μl of the sample was subjected to RNA extraction, to obtain a final volume of 60 μl. Magnetic silica bead-based RNA extraction was performed using a Mag Extractor viral RNA apparatus (Toyobo) according to the manufacturer's instructions. Briefly, 60 μl of the sample was subjected to RNA extraction and purification to obtain a final volume of 60 μl. The extracted nucleic acid was stored at −80°C until analysis.
(v) Water sample processing.
To examine the effects of the presence of inhibitory substances on extraction of EAdV DNA and GII NoV RNA from concentrated environmental water samples, Ad5 and MNV were added to the respective samples. Nucleic acid yields of the added Ad5 and MNV were evaluated by RT-quantitative PCR (RT-qPCR). If the yields of the added Ad5 or MNV were lower than the positive controls by <10%, the extracted DNA or RNA was diluted 100 or 20 times to overcome RT-PCR inhibition. The genomes were then reamplified and requantified. If these yields were still lower than the positive control by <10% after dilution, we concluded that the sample caused loss of nucleic acid during extractions. On the contrary, if the yields were satisfactorily improved after the dilution, we concluded that the original sample did not cause loss of nucleic acid during extraction but caused RT-PCR inhibition. Furthermore, samples exhibiting lower-than-expected Ad5 DNA values were subjected to gel chromatography before DNA extraction, and those exhibiting lower-than-expected MNV RNA values were subjected to magnetic silica bead-based RNA extraction (see Fig. S1 in the supplemental material).
RT-qPCR. (i) RT.
RT was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Tokyo, Japan). Five microliters of RNA was added to the 5 μl of reaction mixture containing 1 μl of 10× reverse transcription buffer, 25 units of MultiScribe reverse transcriptase, 0.4 μl of 25× deoxynucleoside triphosphates, 10 units of RNase inhibitor, and 0.5 μl each of 10 μM antisense primers. Reverse primer COG2R was used for indigenous GII NoV RNA and GII NoV PSC RNA, and MKMNVR was used for MNV RNA (Table 1). The RT reaction mixture was sequentially incubated for 10 min at 25°C, 120 min at 37°C, and 5 min at 85°C in a GeneAmp PCR system 9600 (Applied Biosystems) to synthesize cDNA.
(ii) qPCR.
Twenty-five microliters of reaction mixture contained 5 μl sample, 12.5 μl TaqMan gene expression master mix (Applied Biosystems), 2 μl each of 100 μM primers, and 0.5 μl each of 5 μM TaqMan probes. Primer and TaqMan probe sequences, product sizes, and thermal cycling conditions for each target are summarized in Table 1. PCR mixtures were amplified using an ABI sequence detection system 7500 (Applied Biosystems). Amplification data were collected and analyzed using Sequence Detector software version 1.3 (Applied Biosystems). Tenfold serial dilutions of the DNA standard whose concentrations ranged from 1.0 × 104 to 1.0 × 101 copies per tube were amplified to quantify viral genomes. The copy numbers of each undiluted DNA standard was determined by using a NanoDrop ND 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions.
RESULTS
Quantification of viral DNA and PSC DNA by monoplex qPCR.
Standard curves for each genome were generated by monoplex qPCR of serial 10-fold-diluted Ad40 DNA, EAdV PSC DNA, GII NoV cDNA, and GII NoV PSC DNA. DNA concentrations ranged from 1.0 × 101 to 1.0 × 104 copies per tube in triplicate. In each qPCR assay, 1.0 × 101 copies per tube of DNA were sufficient to generate positive signals that reproducibly gave standard deviations of cycle threshold values that were lower than 0.40 (see Fig. S2 in the supplemental material). The standard curves obtained from Ad40 DNA and EAdV PSC DNA had similar PCR efficiencies, i.e., 98% and 96%, which were equivalent to slope values of −3.40 and −3.45 (see Fig. S2A), respectively. Similarly, PCR efficiencies for GII NoV cDNA and GII NoV PSC DNA were 100% and 103%, which were equivalent to slope values of −3.31 and −3.23, respectively (see Fig. S2B). Cross-reactions between target DNA and TaqMan probe for PSC (MKMNV-TP) or between PSC DNA and TaqMan probe for target DNA (JTVFP or NV-G2P) were not observed in the case of each DNA concentration at ranges from 0.0 to 1.0 × 104 copies per tube.
Quantification of viral DNA and PSC DNA by duplex qPCR.
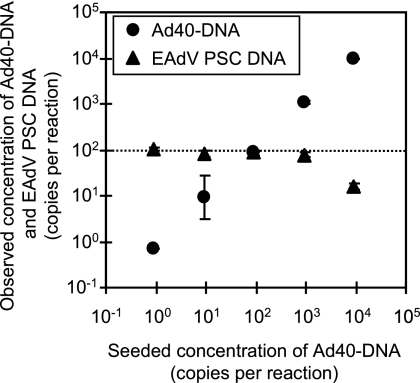
To determine whether PSC might be affected by primer competition, we performed duplex qPCR targeting Ad40 DNA and EAdV PSC DNA. Serial 10-fold-diluted Ad40 DNA and 1.0 × 102 copies each of EAdV PSC DNA in distilled water were quantified by duplex qPCR (Fig. 2). Primer competition was not evident at Ad40 DNA levels below or equal to 1.0 × 103 copies per reaction mixture. However, EAdV PSC DNA was underestimated to be 1.6 × 101 copies in the presence of Ad40 DNA at 1.0 × 104 copies per reaction mixture.
Fig. 2.
Input levels and detection of serially diluted Ad40 DNA and the indicated amount (1.0 × 102 copies per reaction) of EAdV PSC DNA by duplex qPCR. Horizontal and vertical axes indicate the added and observed concentrations of Ad40 DNA and EAdV PSC DNA, respectively. The dotted line indicates the added concentration of EAdV PSC DNA. Error bars indicate the standard deviations (n = 3).
Effects of humic acid.
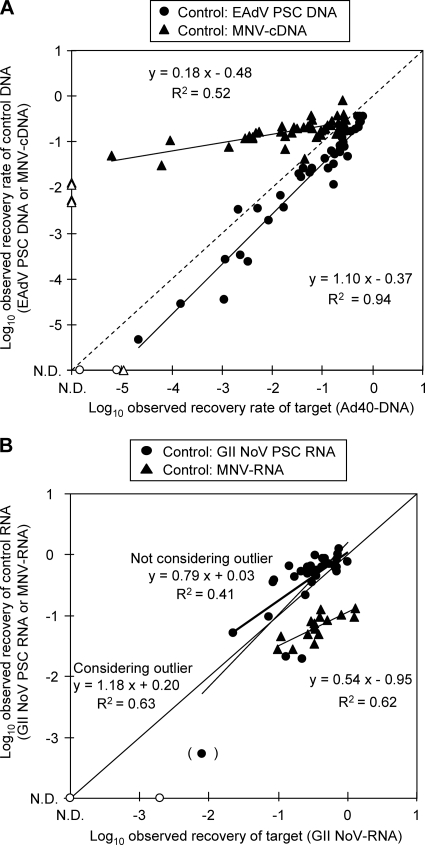
To assess the ability of PSCs to indicate inhibition of target genome amplification, Ad40 DNA or GII NoV RNA was simultaneously quantified by duplex RT-qPCR in the presence of control nucleic acids and humic acid (Fig. 3). Ad40 DNA was inoculated to between 1.0 × 102 and 1.0 × 103 copies per tube, and EAdV PSC DNA or MNV cDNA was inoculated to 1.0 × 102 copies per tube. Humic acid was included in the reaction mixtures at concentrations between 0.049 and 0.12 mg/ml. The relationship between the log-transformed underestimation rates of Ad40 DNA and EAdV PSC DNA were linearly approximated using the equation y = 1.10x − 0.37 (R2 = 0.94), whereas that between log-transformed underestimation rates of Ad40 DNA and MNV cDNA was linearly approximated using the equation y = 0.18x − 0.48 (R2 = 0.52) (Fig. 3A). Comparing EAdV PSC DNA to MNV cDNA as an internal control, the slopes of the plots for EAdV PSC DNA approximated y = x, which indicated that the magnitude of inhibitions was the same between target and control nucleic acids and more closely than those for MNV cDNA. This result indicates inhibition of only EAdV PSC DNA can closely approximate that of Ad40 DNA.
Fig. 3.
Inhibition of target viral genome and control amplification in the presence of humic acid. (A) Underestimation of Ad40 DNA and EAdV PSC DNA and the underestimation of Ad40 DNA and MNV cDNA. The vertical axis indicates the observed recovery of EAdV PSC DNA or MNV cDNA. The horizontal axis indicates the observed recovery of Ad40 DNA. (B) Underestimation of GII NoV RNA and GII NoV PSC RNA. A plot bracket on (x, y) of (−2.1, −3.3) indicates the possible outlier. The underestimation of GII NoVRNA and MNV-RNA is indicated. The vertical axis indicates the observed recoveries of GII NoV PSC RNA or MNV RNA. The horizontal axis indicates the observed recovery of GII NoV RNA. In both panels, ND on the vertical and horizontal axes indicates not detected. Open circles and triangles indicate the lower limits of detection.
Concentrations of inoculated GII NoV RNA were quantified together with GII NoV PSC RNA or MNV RNA by duplex RT-qPCR. GII NoV RNA ranged between 1.0 × 102 and 1.0 × 103 copies per reaction mixture, and GII NoV PSC RNA or MNV RNA was inoculated to be 1.0 × 102 copies per reaction mixture. Humic acid solutions with E260 values between 0.25 and 0.98 (which were equivalent, with values of 0.012 and 0.049 mg/ml of humic acid solutions before extraction) were inoculated into the samples. Figure 3B shows the relationship between the log-transformed underestimation rates of GII NoV RNA and GII NoV PSC RNA. One plot located on (x, y) of (−2.1, −3.3) was likely to be an outlier. If the plot was included in the analysis, the relationship between the log-transformed underestimation rates of GII NoV RNA and GII NoV PSC RNA were linearly approximated using the following equation: y = 1.18x + 0.20 (R2 = 0.63). If the plot was not included, the relationship was linearly approximated using the following equation: y = 0.79x + 0.03 (R2 = 0.41). On the other hand, the relationship between the log-transformed underestimation rates of GII NoV RNA and MNV RNA were approximated by the equation y = 0.54x − 0.95 (R2 = 0.62) (Fig. 3B). Comparing GII NoV PSC RNA to MNV RNA as an internal control, the slopes of the plots for GII NoV PSC RNA approximated y = x more closely than those for MNV RNA, even if the possible outlier was considered or not. This indicates that inhibition only of GII NoV PSC RNA can approximate that of GII NoV RNA.
Loss of viral genomes concentrated from environmental samples during extractions.
Table 2 displays the results showing that the Ad5 and MNV genomes were underestimated. Of the 16 samples obtained from large volumes of environmental water samples, the Ad5 DNA yields from five samples were lower than expected by <10%. Of the nine samples obtained from small-volume water samples, the Ad5 DNA yield of only one sample was lower than expected by <10% (Table 2).
Table 2.
Observed recovery of exogenously added Ad5 and MNV in concentrated environmental samples
| Sample | Concn vola | % recovery |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Ad5 |
MNV |
||||||||
| Not purifiedb | Purifiedc | Membraned | Beadse | ||||||
| Not diluted | Diluted | Not diluted | Diluted | Not diluted | Diluted | Not diluted | Diluted | ||
| SR-1 | L | NDf | 38 | 0.03 | 60 | ND | 5.30 | 41 | |
| SR-2 | S | 22 | 48 | ||||||
| SR-3 | S | 23 | 60 | ||||||
| SR-4 | S | 74 | 83 | ||||||
| SR-5 | S | 73 | 74 | ||||||
| SR-6 | S | 64 | 76 | ||||||
| SR-7 | S | 63 | 79 | ||||||
| SL-1 | S | 100 | 100 | ||||||
| SL-2 | L | 20 | 3.40 | 1.90 | 120 | ||||
| SL-3 | L | 9.50 | 14 | 26 | 29 | 3.60 | 16 | 86 | |
| SL-4 | L | 43 | 18 | ||||||
| SL-5 | L | 31 | 15 | ||||||
| SG-1 | L | 0.63 | 0.90 | 0.66 | 0.75 | 0.41 | ND | 6.90 | 4.20 |
| SG-2 | L | 97 | 87 | ||||||
| SG-3 | L | 71 | 39 | ||||||
| SG-4 | S | 9.90 | 12 | 32 | 30 | 110 | |||
| SG-4 | L | 69 | 130 | ||||||
| BR-1 | S | 61 | 83 | ||||||
| BR-1 | L | 7.30 | 45 | 35 | 32 | 4.00 | 32 | 230 | |
| BR-2 | L | 44 | 5.60 | 48 | 220 | ||||
| BC-1 | L | 28 | 20 | ||||||
| BC-2 | L | 7.90 | 10 | 10 | 15 | 8.90 | 0.84 | 20 | |
| BM-1 | L | 60 | 82 | ||||||
| BM-2 | L | 78 | 90 | ||||||
L, concentrated from a large volume of water; S, concentrated from a small volume of water.
Recoveries without gel chromatography.
Recoveries with gel chromatography.
Recoveries with silica membrane-based method.
Recoveries with magnetic silica bead-based method.
ND, not detected.
Hundredfold dilution of DNA improved Ad5 DNA yields in two samples (SR-1 and BR-1) by >10% (38% and 45%, respectively), but the yields from rest of the four samples were lower (Table 2). Samples with yields of inoculated Ad5 DNA and/or EAdV PSC DNA lower than the positive controls by <10% were gel purified. Gel chromatography recovered 55% of stock Ad5, suggesting that it caused no significant loss of virions. Ad5 levels increased in four samples (SR-1, BR-1, SL-3, and SG-4) after two purifications. Hundredfold dilutions of DNA after gel chromatography and DNA extraction improved the Ad5 DNA yield (SR-1) from 0.032% to 60%, but yields from the other five samples were not significantly changed (Table 2).
The MNV RNA yields from 7 out of 16 samples were lower than expected (Table 2). Three of seven samples (BR-1, BR-2, and SL-3) showed acceptable yields (32%, 48%, and 16%, respectively) after 20-fold dilution of extracted RNA. Although the yield was still not acceptable, 20-fold dilution of SR-1 also improved its yield (Table 2). MNV RNA yields extracted by the magnetic silica bead-based method were 2.6 times higher than those extracted by the silica membrane-based method. For all samples, the MNV RNA yields of the magnetic silica bead-based method were 2.3 to 4,000 times higher than those of the silica membrane-based method. Despite the 16 times improvement in MNV RNA yields, 20-fold dilution of SG-1 did not result in an acceptable value.
Inhibition of RT-qPCR by concentrated environmental water samples.
EAdV PSC DNA, inoculated into environmental samples to estimate the magnitude of PCR inhibition occurring during quantifications of EAdV DNA, showed that the yields of EAdV PSC DNA (SR-1 and BR-1) were lower than expected by <10% (Table 3). Even after gel chromatography, the inoculated EAdV PSC DNA was not detected in SR-1. The underestimation of control EAdV PSC DNA in BR-1 improved from 0.29% to 130% following gel chromatography (Table 3). In only one sample (SR-1), the yield of inoculated GII NoV PSC RNA was not detected by the silica membrane-based method. However, magnetic silica bead-based extraction resulted in an acceptable yield of GII NoV PSC RNA in SR-1 (77%) and the other samples (Table 3).
Table 3.
Observed recovery of EAdV PSC DNA and GII NoV PSC RNA in concentrated environmental water samples
| Sample | Concn vola | % recovery |
|||
|---|---|---|---|---|---|
| E-AdV PSC DNA |
GII NoV PSC RNA |
||||
| Not purifiedb | Purifiedc | Membraned | Beadse | ||
| SR-1 | L | ND | NDf | ND | 77 |
| SR-2 | S | 82 | 35 | ||
| SR-3 | S | 79 | 50 | ||
| SR-4 | S | 85 | 49 | ||
| SR-5 | S | 120 | 58 | ||
| SR-6 | S | 110 | 44 | ||
| SR-7 | S | 98 | 61 | ||
| SL-1 | S | 71 | 48 | ||
| SL-2 | L | 53 | 67 | 120 | |
| SL-3 | L | 87 | 120 | 54 | 89 |
| SL-4 | L | 93 | 55 | ||
| SL-5 | L | 110 | 47 | ||
| SG-1 | L | 90 | 100 | 35 | 140 |
| SG-2 | L | 87 | 49 | ||
| SG-3 | L | 110 | 72 | ||
| SG-4 | S | 90 | 120 | 57 | |
| SG-4 | L | 93 | 60 | ||
| BR-1 | S | 110 | 61 | ||
| BR-1 | L | 0.29 | 130 | 70 | 110 |
| BR-2 | L | 110 | 51 | 140 | |
| BC-1 | L | 110 | 63 | ||
| BC-2 | L | 110 | 120 | 51 | 96 |
| BM-1 | L | 91 | 62 | ||
| BM-2 | L | 98 | 65 | ||
L, concentrated from a large volume of water; S, concentrated from a small volume of water.
Recoveries without gel chromatography.
Recoveries with gel chromatography.
Recoveries with the silica membrane-based method.
Recoveries with the magnetic silica bead-based method.
ND, not detected.
Detection of indigenous enteric viruses in environmental water samples.
Twenty-five samples were tested for indigenous EAdVs and GII NoVs together with EAdV PSC DNA and GII NoV PSC RNA, respectively, by duplex RT-qPCR. Indigenous EAdV DNA was not detected in any tested sample even after gel chromatography. In contrast, indigenous GII NoV RNA was detected in 3 of 24 samples (13%) (Table 4). These samples were collected from different sites along the same river. BR-1 concentrated from small volumes of water was positive for GII NoV-RNA, whereas the same sample from large volumes of water was negative. Using the magnetic silica bead method, which showed higher yields of exogenously added MNV and GII NoV PSC RNA, higher yields of GII NoV RNA were observed in both BR-2 and BC-2.
Table 4.
Detection of indigenous GII NoVs and inhibition of quantification of controls based on two different RNA extraction and purification methods
| Sample | Original water vol | No. of copies detected/ liter when using: |
|
|---|---|---|---|
| Silica membranea | Silica beadsb | ||
| BR-1 | Small (50 ml) | 7.9 × 102 | Not applied |
| BR-2 | Large (20 liters) | NDc | 2.2 × 100 |
| BC-2 | Large (150 liters) | 1.1 × 10−1 | 2.8 × 10−1 |
Samples extracted and purified by the silica membrane-based method.
Samples extracted and purified by the magnetic silica bead-based method.
ND, not detected.
DISCUSSION
In this study, we examined the inhibitory effects of concentrated water samples on nucleic acid extraction and RT-PCR by using virus particles and PSCs as controls. We also took advantage of these controls to determine the abilities of different purification methods to remove inhibitory factors.
Although the mechanism of RT-PCR inhibition has not been elucidated, inhibition of annealing and/or extension by polymerase may be involved in this mechanism (49). To estimate the magnitude of RT-PCR inhibition, we constructed PSC DNA and RNA molecules that could be amplified using the same primer set and could produce the same amplicon size as that of the target viral genome. RT-qPCR assays for EAdV and GII NoV utilized in this study were developed to detect each target gene specifically in previous studies (20, 25). Our assays for EAdV PSC DNA and GII NoV PSC DNA, which need the same primer pairs as the targets, did not cause cross-reactions with EAdV DNA or GII NoV cDNA, respectively, ensuring the assays' specificities. When added together, 1.0 × 102 and ≤1.0 × 103 copies of EAdV PSC DNA and Ad40 DNA, respectively, did not affect each other's amplification in the absence of inhibitors (Fig. 2). However, the presence of >1.0 × 103 copies of Ad40 DNA led to the underestimation of PSC DNA, which was possibly due to primer competition between target and PSC DNA as described previously (14). If the yield of target virus nucleic acid is greater than 1.0 × 103 copies per tube, dilution of the sample is recommended to reduce primer competition with PSCs as well as RT-PCR inhibition. In the duplex RT-qPCR test with humic acid, the extent of underestimation of target and PSC nucleic acids was almost the same (Fig. 3), demonstrating that PSC reliably predicts inhibition of target amplification at least for the assays shown in this study. However, tests using MNV nucleic acid, which needs other primer sets producing different amplicon sizes from EAdV DNA and GII NoV RNA, could not predict RT-qPCR inhibition accurately. Hence, to evaluate the magnitude of RT-PCR inhibition, the primer set and the amplicon size of the control nucleic acid may need to be considered.
The range of inhibition for GII NoV RNA and GII NoV PSC RNA was 2 log10, which is considerably narrower than that for EAdV (Fig. 3B). This difference seemed to be the result of RT. Under the experimental conditions used in this study, which adopted a two-step RT-qPCR, the RT reaction mixture contained higher concentrations of humic acid than the subsequent PCR mixture. Hence, the inhibitory effect may be more pronounced during RT than during PCR. Furthermore, RT inhibition, which underestimates the product yield by <1%, reduces template availability for PCR. In contrast, an observed yield of <1 copy per tube can readily occur during qPCR for EAdV DNA without RT because the inhibition appears as a delay in amplification.
Costafreda et al. (8) showed that morphologically similar viruses have similar sensitivities in terms of nucleic acid extraction inhibition. In this study, Ad5 and MNV were used as internal controls to evaluate the inhibitory effects of samples on DNA extraction of EAdVs and RNA extraction of GII NoVs, respectively. Because both Ad5 and MNV do not seem to be abundant in aquatic environments (11), the presence of indigenous Ad5 and MNV may be negligible, and the detected DNA and cDNA very possibly belong to the control viruses. Such effects on viral nucleic acid extraction have been reported using internal controls (8, 10). However, in these studies, RNA extraction inhibition was not distinguished from RT-PCR inhibition, and the reason for the underestimation of the internal control could not be determined. In this study, we diluted the extracted samples to minimize RT-PCR inhibition, thereby allowing successful differentiation of RT-PCR inhibition from nucleic acid extraction inhibition. If the reaction yields from the added virus improve after dilution, we can conclude that RT-PCR inhibition has occurred. In contrast, if dilution has no effect, or dilution has an effect but still underestimation occurs, which was not observed in this study, we can conclude that nucleic acid extraction inhibition has occurred.
In our evaluation of inhibitory effects of samples on the detection of EAdVs and GII NoVs, six samples showed nucleic acid extraction inhibition during the silica membrane-based extractions and two showed RT-PCR inhibition. Samples processed from large volumes tended to cause inhibitory effects more frequently than those from small volumes. This result indicates that substances coconcentrated with viruses caused inhibition. Samples that adversely affected nucleic acid extraction stained the silica membranes with a brownish color and clogged them (data not shown). Humic acids may contribute to these phenomena because of their color and presence in virus samples. Furthermore, membrane clogging resulted in poor nucleic acid yields. Magnetic silica beads may provide a suitable alternative to silica membranes because they are free from clogging. Several previous reports utilized the magnetic or nonmagnetic silica bead-based method (12, 19, 36), and some of those studies showed that RNA recovery using the magnetic silica bead-based method was lower than that when using the silica membrane-based method (12, 36). In this study, the use of magnetic silica beads resulted in higher RNA yields, less-inhibitory RNA preparations, and enhanced PCR amplification. It is known that ionic strength influences nucleic acid binding to silica (28). Hence, the ionic conditions in the virus concentrate might be suitable for the Mag Extractor viral RNA magnetic silica bead-based RNA extraction kit used in this study. The structures of magnetic silica beads, which do not result in clogging and are less likely to trap organic inhibitors than silica membrane, can explain the less-inhibitory RNA preparations and enhanced RT-PCR results.
Gel chromatography has been reported to be effective for reducing RT-PCR inhibition (1, 5, 39, 45). In this study, gel chromatography provided mixed results wherein one sample (BR-1) showed improved amplification of EAdV PSC DNA but it was not effective for the other samples, which were false negative for EAdV PSC DNA.
Samples that inhibited both nucleic acid extraction and RT-PCR were not encountered. Furthermore, gel chromatography, which made some beneficial contributions to overcome PCR inhibition, was not effective in reducing nucleic acid extraction inhibition. These results indicate that different substances are associated with inhibition of nucleic acid extraction and RT-PCR. Humic acid may be responsible for both of these problems because of its ubiquity, diversity in molecular structure, and presence in water samples.
Enteric adenoviruses have been frequently detected in aquatic samples (4, 46, 50). In this study, we did not detect EAdV DNA in any sample, even after gel chromatography, which was shown to improve sample quality. Thus, our methods should help in djudging environmental samples to be truly negative for EAdVs.
GII NoVs are the leading cause of viral gastroenteritis and are frequently detected in environmental samples (35, 47). We successfully detected GII NoV RNA in three river water samples collected from the same site. Higher yields were observed using magnetic silica beads, which improved RNA preparation quality and enhanced RT-PCR, than using silica membranes. Thus, we conclude that use of silica membranes may contribute to the underestimation of the presence of viruses in environmental samples. Magnetic silica beads similarly improved the ratio of RNA extraction efficiency of exogenously added MNV and indigenous GII NoV RNA (2.5 and 2.3 times, respectively), thereby demonstrating that loss of GII NoV RNA can be evaluated using MNV as a control.
In conclusion, PSC DNA and RNA can be used to assess RT-PCR inhibition with higher accuracy than other nucleic acids (internal controls) amplified by other primers that produce different amplicon sizes. Thus, our results highlight the importance of evaluating inhibitory effects using control virus particles and PSCs.
Supplementary Material
ACKNOWLEDGMENT
This work was partly supported by a Grant-in-Aid for Scientific Research (A, 101200000049) from the Japan Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Abbaszadegan M., Huber M. S., Gerba C. P., Pepper I. L. 1993. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allwood P. B., Malik Y. S., Hedberg C. W., Goyal S. M. 2003. Survival of F-specific RNA coliphage, feline calicivirus, and Escherichia coli in water: a comparative study. Appl. Environ. Microbiol. 69:5707–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aw T. G., Gin K. Y.-H., Ean Oon L. L., Chen E. X., Woo C. H. 2009. Prevalence and genotypes of human noroviruses in tropical urban surface waters and clinical samples in Singapore. Appl. Environ. Microbiol. 75:4984–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bofill-Mas S., Pina S., Girones R. 2000. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 66:238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borchardt M. A., Bertz P. D., Spencer S. K., Battigelli D. A. 2003. Incidence of enteric viruses in groundwater from household wells in Wisconsin. Appl. Environ. Microbiol. 69:1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brigante M., Zanini G., Avena M. 2007. On the dissolution kinetics of humic acid particles: effects of pH, temperature and Ca2+ concentration. Colloids Surf. A Physicochem. Eng. Asp. 294:64–70 [Google Scholar]
- 7. Chandler D. P., Wagnon C. A., Bolton H., Jr 1998. Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl. Environ. Microbiol. 64:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costafreda M. I., Bosch A., Pintó R. M. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72:3846–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costán-Longares A., et al. 2008. Microbial indicators and pathogens: removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 42:4439–4448 [DOI] [PubMed] [Google Scholar]
- 10. da Silva A. K., et al. 2007. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: different behaviors of genogroups I and II. Appl. Environ. Microbiol. 73:7891–7897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fong T.-T., Phanikumar M. S., Xagoraraki I., Rose J. B. 2010. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 76:715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gentry J. J., Vinjé, Lipp E. K. 2009. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J. Virol. Methods 156:59–65 [DOI] [PubMed] [Google Scholar]
- 13. Glass R. I., et al. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254–S261 [DOI] [PubMed] [Google Scholar]
- 14. Gregory J. B., Litaker R. W., Noble R. T. 2006. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal control for detection of enteroviruses in environmental samples. Appl. Environ. Microbiol. 72:3960–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamza I. A., et al. 2009. Detection of human viruses in rivers of a densly-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 43:2657–2668 [DOI] [PubMed] [Google Scholar]
- 16. Haramoto E., Katayama H., Ohgaki S. 2004. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 70:2154–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haramoto E., Katayama H., Utagawa E., Ohgaki S. 2008. Development of sample storage methods for detecting enteric viruses in environmental water. J. Virol. Methods 151:1–6 [DOI] [PubMed] [Google Scholar]
- 18. Heim A., Ebnet C., Harste G., Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228–239 [DOI] [PubMed] [Google Scholar]
- 19. Jiang S., Noble R., Chu W. 2001. Human adenoviruses and coliphages in urban runoff-impact coastal waters of southern California. Appl. Environ. Microbiol. 67:179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kageyama T., et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katayama H., Shimasaki A., Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kermekchiev M. B., Kirilova L. I., Vail E. E., Barnes W. M. 2009. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 37:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kipton H., Powell J., Town R. M. 1992. Solubility and fractionation of humic acid; effect of pH and ionic medium. Anal. Chim. Acta 267:47–54 [Google Scholar]
- 24. Kitajima M., et al. 2008. Use of murine norovirus as a novel surrogate to evaluate resistance of human norovirus to free chlorine disinfection in drinking water supply system. Environ. Eng. Res. 45:361–370(In Japanese.) [Google Scholar]
- 25. Ko G., Jothikumar N., Hill V. R., Sobsey M. D. 2005. Rapid detection of infectious adenovirus by mRNA real-time RT-PCR. J. Virol. Methods 127:148–153 [DOI] [PubMed] [Google Scholar]
- 26. Kreader C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malorny B., Hoorfar J., Bunge C., Helmuth R. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melzak K. A., Sherwood C. S., Turner R. F. B., Haynes C. A. 1996. Driving forces for DNA adsorption to silica in perchlorate solutions. J. Colloid. Interface Sci. 181:635–644 [Google Scholar]
- 29. Mio K., Kirkham J., Bonass W. A. 2006. Tips for extracting total RNA from chondrocytes cultured in agarose using silica-based membrane kit. Anal. Biochem. 351:314–316 [DOI] [PubMed] [Google Scholar]
- 30. Molenkamp R., van der Ham A., Schinkel J., Beld M. 2007. Simultaneous detection of five different DNA targets by real-time Taqman PCR using the Roche LightCycler480: application in viral molecular diagnostics. J. Virol. Methods 141:205–211 [DOI] [PubMed] [Google Scholar]
- 31. Monpoeho S., et al. 2000. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. Biotechniques 29:88–93 [DOI] [PubMed] [Google Scholar]
- 32. Mormann S., Dabisch M., Becker B. 2010. Effects of technological processes on the tenacity and inactivation of norovirus genogroup II in experimentally contaminated foods. Appl. Environ. Microbiol. 76:536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ottoson J., Hansen A., Björlenius B., Norder H., Stenström T. A. 2006. Removal of viruses, parasitic protozoa and microbial indicators in conventional and membrane processes in a wastewater pilot plant. Water Res. 40:1449–1457 [DOI] [PubMed] [Google Scholar]
- 34. Rock C., Alum A., Abbaszadegan M. 2010. PCR inhibitor levels in concentrates of biosolid samples predicted by a new method based on excitation-emission matrix spectroscopy. Appl. Environ. Microbiol. 76:8102–8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodríguez-Díaz J., et al. 2009. Detection and characterization of waterborne gastroenteritis viruses in urban sewage and sewage-polluted river waters in Caracas, Venezuela. Appl. Environ. Microbiol. 75:387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rutjes S. A., Italiaander R., van den Berg H. H., Lodder W. J., de Roda Husman A. M. 2005. Isolation and detection of enterovirus RNA from large-volume water samples by using the NucliSens miniMAG system and real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 71:3734–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schönenbrücher H., Abdulmawjood A., Failing K., Bülte M. 2008. New triplex real-time PCR assay for detection of Mycobacterium avium subsp. paratuherculosis in bovine feces. Appl. Environ. Microbiol. 74:2751–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwab K. J., De Leon R., Sobsey M. D. 1996. Immunoaffinity concentration and purification of waterborne enteric viruses for detection by reverse transcriptase PCR. Appl. Environ. Microbiol. 62:2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shieh Y.-S. C., Wait D., Tai L., Sobsey M. D. 1995. Methods to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the polymerase chain reaction. J. Virol. Methods 54:51–66 [DOI] [PubMed] [Google Scholar]
- 40. Sidhu M. K., Liao M. J., Rashidbaigi A. 1996. Dimethyl sulfoxide improves RNA amplification. Biotechniques 21:44–47 [DOI] [PubMed] [Google Scholar]
- 41. Skraber S., et al. 2009. Occurrence and persistence of enteroviruses, noroviruses and F-specific RNA phages in natural wastewater biofilms. Water Res. 43:4780–4789 [DOI] [PubMed] [Google Scholar]
- 42. Stals A., et al. 2009. Multiplex real-time RT-PCR for simultaneous detection of GI/GII noroviruses and murine norovirus 1. J. Virol. Methods 161:247–253 [DOI] [PubMed] [Google Scholar]
- 43. Teunis P. F., et al. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 [DOI] [PubMed] [Google Scholar]
- 44. Tree J. A., Adams M. R., Lees D. N. 2003. Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl. Environ. Microbiol. 69:2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsai Y.-L., Olson B. H. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Verheyen J., et al. 2009. Detection of adenoviruses and rotaviruses in drinking water sources used in rural areas of Benin, West Africa. Appl. Environ. Microbiol. 75:2798–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Westrell T., et al. 2006. Short- and long-term variations of norovirus concentrations in the Meuse river during a 2-year study period. Water Res. 40:2613–2620 [DOI] [PubMed] [Google Scholar]
- 48. WHO 2008. Microbial aspects, p. 121–144 Guidelines for drinking water quality, 3rd ed. World Health Organization, Geneva, Switzerland [Google Scholar]
- 49. Wilson I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wong M., et al. 2009. Evaluation of public health risks at recreational beaches in Lake Michigan via detection of enteric viruses and a human-specific bacteriological marker. Water Res. 43:1137–1149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.