Abstract
The release of vast quantities of DNA sequence data by large-scale genome and expressed sequence tag (EST) projects underlines the necessity for the development of efficient and inexpensive ways to link sequence databases with temporal and spatial expression profiles. Here we demonstrate the power of linking cDNA sequence data (including EST sequences) with transcript profiles revealed by cDNA-AFLP, a highly reproducible differential display method based on restriction enzyme digests and selective amplification under high stringency conditions. We have developed a computer program (GenEST) that predicts the sizes of virtual transcript-derived fragments (TDFs) of in silico-digested cDNA sequences retrieved from databases. The vast majority of the resulting virtual TDFs could be traced back among the thousands of TDFs displayed on cDNA-AFLP gels. Sequencing of the corresponding bands excised from cDNA-AFLP gels revealed no inconsistencies. As a consequence, cDNA sequence databases can be screened very efficiently to identify genes with relevant expression profiles. The other way round, it is possible to switch from cDNA-AFLP gels to sequences in the databases. Using the restriction enzyme recognition sites, the primer extensions and the estimated TDF size as identifiers, the DNA sequence(s) corresponding to a TDF with an interesting expression pattern can be identified. In this paper we show examples in both directions by analyzing the plant parasitic nematode Globodera rostochiensis. Various novel pathogenicity factors were identified by combining ESTs from the infective stage juveniles with expression profiles of ∼4000 genes in five developmental stages produced by cDNA-AFLP.
INTRODUCTION
With the advent of high throughput techniques for DNA sequencing, whole genome sequences from several organisms have become available (1,2) and many others will be available in the near future. At the same time, millions of expressed sequence tags (ESTs), single pass sequences of cDNA clones selected randomly from a library, have been generated and deposited in public and private databases. Searching for homologous sequences in databases is usually the first step towards understanding the functions of newly identified genes. Homology information is useful for orthologous genes, but in the case of paralogs the value of this information may be more limited. Furthermore, it is often found that a significant proportion (40–60%) of newly identified DNA sequences lack homology with genes for which the functions are known (2,3). Additional tools are therefore needed to allow functional analysis of newly identified genes.
Biological responses and developmental processes are precisely controlled at the level of gene expression. Information on the temporal and spatial regulation of gene expression often sheds light on the potential function of a particular gene. Hence, an essential aspect of functional genomics is the transcriptome, i.e. the analysis of expression patterns of genes on a large scale. There are currently three high throughput techniques for large-scale monitoring of gene expression: serial analysis of gene expression (SAGE) (4), hybridization-based methods (5,6), gel-based RNA fingerprinting techniques such as differential display (7) and cDNA-AFLP (8). In principle, SAGE can provide quantitative data concerning gene expression. However, it is expensive and labor intensive when multiple sample points are to be compared. Microarray technology is very powerful in generating a broad view of gene expression. Unlike cDNA arrays, oligonucleotide arrays are able to distinguish between highly homologous sequences. However, the design of oligonucleotide arrays requires comprehensive sequence knowledge at present only available for a small number of organisms. cDNA-AFLP is an inexpensive gel-based method for analysis of gene expression patterns and can be performed in any laboratory.
In the cDNA-AFLP procedure cDNAs synthesized from mRNAs isolated from various sample points are digested by two restriction enzymes. Oligonucleotide adapters are then ligated to the resulting restriction fragments to generate template DNA for PCR. PCR primers complementary to the adapter sequences with additional selective nucleotides at the 3′-ends allow specific amplification of a limited number of cDNA fragments. Unlike differential display methods that make use of small random primers (7), relatively high annealing temperatures can be used and, hence, cDNA-AFLP is more stringent and reproducible. In contrast to most hybridization-based techniques, cDNA-AFLP will distinguish between highly homologous genes from gene families while (contrary to oligonucleotide arrays) no sequence foreknowledge is needed.
Since sequence information is accumulating at an unprecedented rate for a wide variety of organisms, there is an urgent need for efficient and inexpensive ways to screen these databases on genes with interesting expression profiles. Here we report on the advantages of combining ESTs with cDNA-AFLP data. The potential benefits of this combination in gene discovery and functional analysis prompted us to develop a computer program that creates restriction patterns of cDNAs in silico in accordance with the enzyme combinations used in cDNA-AFLP. The resulting virtual cDNA fragments are ordered according to the extensions of the amplifying primers and their sizes. These virtual fragments can then be traced back on cDNA-AFLP gels to identify the corresponding bands, with primer extensions and fragment sizes as a unique identifier. The program can also be used in the opposite direction by using the size and primer extensions of a potentially interesting band identified on a cDNA-AFLP gel as criteria to search the corresponding cDNA. This simplifies the procedure of cloning full-length genes with interesting temporal and spatial expression patterns.
In this paper we demonstrate the utility of the program by linking EST sequence data and expression profiles of ∼4000 genes from the potato cyst nematode Globodera rostochiensis, which causes extensive damage to solanaceous crops. Genes potentially related to the nematode’s ability to parasitize plants were identified within a pool of hundreds of ESTs. We show that this program could be useful in any system where stage- or tissue-specific genes are to be selected from pools of (uncharacterized) cDNAs.
MATERIALS AND METHODS
The nucleotide sequences of the ESTs described in this study are available in the GenBank EST division (dbEST) under accession nos BE607308 (GE1867), AW506364 (GE1782), AW506154 (GE1483), AW506045 (GE1349), AW505895 (GE1133), AW506065 (GE1373), AW506280 (GE1659), AW506299 (GE1699), AW505736 (GE99), AW506094 (GE1409), AW505716 (GE54), AW505855 (GE1084), AW506406 (GE1816), BE607310 (GE2075) and BE607309 (GE1156).
Generation of ESTs
The ESTs described by Popeijus et al. (9) were used in this study. Briefly, total RNA was extracted from infective second stage juveniles (J2) of the potato cyst nematode G.rostochiensis pathotype Ro1 Mierenbos freshly hatched in potato root diffusate (PRD). cDNA primed with an oligo(dT) primer was directionally cloned in the pcDNA II vector (Invitrogen, Leek, The Netherlands). The resulting library contained at least 2.5 × 106 recombinant plasmids. ESTs were obtained by random sequencing of the library inserts from the 5′-end.
cDNA-AFLP profile
cDNA-AFLP profiles were generated as described by Qin et al. (10). Briefly, total RNA was extracted from five developmental stages of G.rostochiensis: D, dehydrated unhatched J2 in cysts (in diapause); S, rehydrated unhatched J2 in 1-year-old cysts after exposure to sterile tap water for 2 days; H, pre-parasitic J2 (dry cysts incubated in sterile tap water for 1 week, then PRD for a second week); U, developing nematodes (mostly J1) in gravid females 2 months post-inoculation; P, developing nematodes (J2) in gravid females 3 months post-inoculation. cDNA was synthesized with oligo(dT)12–18 as primer. The resulting cDNAs were then digested with the restriction enzymes EcoRI and TaqI and ligated to corresponding adapters. The ligated cDNA fragments were subsequently amplified by EcoRI and TaqI primers that annealed to the adapters in PCR reactions and displayed on polyacrylamide gels.
GenEST program
GenEST was developed on Linux using the GNU C++ toolset and the Standard Template Library. It is command line based and can be run on Unix/Linux and from the MS-DOS prompt under Microsoft Windows. GenEST (the program, source code and a detailed manual) can be downloaded from http://www.spg.wau.nl/nema /nm_res-e.htm (GenEST hyperlink in Section I. Plant–nematode interactions).
A command file can be created, with a text editor, which contains restriction enzyme recognition sites to be used as the begin and end tags and the marker length modifier. Multiple combinations can be defined in a command file. GenEST uses the begin tag to search for the tag sequence in the cDNA data, which are contained in the input files in FASTA format. If such a tag is found, it will continue its search for a matching end tag. This search action is executed in both directions for all begin/end tag combinations. The marker length modifier is designed to compensate for the additional adapter sequences present in the transcript-derived fragments (TDFs) as they appear on a cDNA-AFLP gel.
Furthermore, the identifiers of a band on a gel (restriction enzyme recognition sequences, primer extensions and band size) can be used as a search query to quickly identify the corresponding EST(s) in an automated procedure.
RESULTS
ESTs and cDNA-AFLP-based expression profiles
A cDNA library from second stage juveniles in the H stage of the potato cyst nematode G.rostochiensis was used to sequence 985 cDNA clones. Starting from the 5′-end, the average read was ∼600 bp (9). In parallel, cDNA-AFLP-based gene expression profiles were generated from five distinct developmental stages, D, S, H, U and P, of this nematode species. The expression profiles were highly reproducible and no significant differences were observed between independent replicates. An average of 32 bands per lane were displayed using EcoRI and TaqI primers with two selective nucleotides (E+NN and T+NN, respectively) extending beyond the adapters into the cDNA. Approximately 8200 TDFs were displayed using the whole set of 256 (16 × 16) primer combinations. In a previous study (10) it was shown that genes involved in plant parasitism are usually up-regulated in developmental stages S and H or in stage H only. Bands showing such expression patterns were excised from gels, cloned and sequenced. Sequencing of >100 TDFs revealed that the marker-based size estimations corresponded well to the actual sizes of these TDFs (with an accuracy of ±1 nt for bands <300 nt and ±3 nt for bands >300 bp).
Generation of virtual TDFs from ESTs using GenEST
We used GenEST to generate virtual TDFs from 985 ESTs. EcoRI and TaqI recognition sites were used as begin and end tags with a length modifier of 22 nt to account for the additional adapter sequences. The following lines were included in the command file:
GAATTC TCGA 22
TCGA GAATTC 22
A total of 228 virtual TDFs derived from 159 ESTs were predicted by GenEST (Table 1). Of these 159 ESTs, 100 were predicted to produce a single virtual TDF, 51 were predicted to give rise to two virtual TDFs each (thereby generating 102 TDFs), six ESTs were predicted to result in three TDFs each (generating 18 TDFs) and two ESTs were predicted to generate four TDFs each (eight TDFs in total).
Table 1. Virtual TDFs generated after in silico restriction with EcoRI and TaqI of 985 ESTs randomly picked from a cDNA library from infective juveniles of the potato cyst nematode G.rostochiensis using GenEST.
| |
E+AN |
E+CN |
E+GN |
E+TN |
Total |
| N = A | 20 | 17 | 32a | 2 | |
| C | 6 | 11 | 15 | 11 | |
| G | 12 | 14 | 19 | 14 | |
| T | 14 | 33 | 22 | 18 | |
| Total | 52 | 75 | 56a | 45 | 228a |
E+AN/CN/GN/TN are the extensions of the EcoRI primer (E, core primer). Each EcoRI primer was combined with all TaqI primers (T+NN). Note: E+GA will constitute both a TaqI and an EcoRI recognition sequence (GAATTCGA). In this case TDFs will not be amplified and cannot be traced back on a cDNA-AFLP gel. Therefore, these TDFs were not included in the total counta.
To estimate how many genes are represented by the 8200 TDFs displayed in our study we have randomly extracted 1000 full-length cDNAs of Caenorhabditis elegans from GenBank (both the size and average GC content of the G.rostochiensis genome are similar to C.elegans; 11). These sequences were processed by GenEST and 336 cDNAs (≈34%, the remaining cDNAs not containing both restriction sites) generated 693 virtual EcoRI/TaqI TDFs. The percentage of genes which produced TDFs is ∼48% (336/693 = 48%) of the total TDF number. Assuming that the average mRNA size and the number of genes of the potato cyst nematode do not differ substantially from C.elegans, the 8200 TDFs displayed on cDNA-AFLP gels in this study would represent ∼4000 expressed genes.
From ESTs to the corresponding TDFs on cDNA-AFLP gels
The vast majority of the virtual TDFs predicted could be located at the expected position on the cDNA-AFLP gel. The cases where no matches were found between virtual and real TDFs could usually be explained by the system used. Here we describe detailed analyses of 52 virtual TDFs that were generated in silico using the primers E+AN in combination with all TaqI primers (T+NN) (Table 1). Multiple TDFs that originated from a single EST sequence were all checked. Matching bands could be found on gels for 41 TDFs. Eight virtual TDFs were smaller than the exclusion limit of 50 nt used in this study. As expected, these TDFs were not displayed. Lowering the exclusion limit would allow the display of bands down to 10 nt. Among these eight ESTs, six would produce a second virtual TDF. All these TDFs were identified on gels. Within the size range analyzed only three virtual TDFs could not be traced back on the cDNA-AFLP gels (see below).
The TDF computed from EST GE1867 could not be detected. This EST aligned almost completely with the cloned GR-eng-2 gene from G.rostochiensis (12). Careful examination of the sequence suggested that a 10 bp fragment at the 5′-end of the EST, in which a TaqI recognition site was located, may have originated from another gene. We therefore assumed that a rare recombination event occurred during construction of the cDNA library. A second band predicted for GE1867, 399 nt in length with extensions E+TT/T+TG, was readily identified on the gel.
For one particular EST, GE1782, a TaqI recognition sequence (bold) was found to be partially nested inside the EcoRI recognition site (underlined) (GAATTCGA). Contrary to the E+GA group mentioned in Table 1, the TCGA sequence was located at the outside of the TDF. Following the cDNA-AFLP protocol the cDNA was first digested with TaqI and, as a consequence, the EcoRI site was lost. Hence, in this particular case the predicted TDF was not amplified.
ESTs GE1349 and GE1483 were predicted to produce four TDFs. All four TDFs of GE1349 were located on gels at the predicted size. For GE1483 one band was found, the other three being smaller than the cut-off size of the gel.
In summary, from a total of 52 TDFs predicted to be produced by E+AN just one, from EST GE1133, could not be located at the predicted size and primer extensions. This minor discrepancy between the GenEST prediction and the bands displayed on the gel may be caused by a PCR or sequencing error. It is concluded that predicted TDFs from ESTs can always be traced back on cDNA-AFLP gels, when taking PCR and sequencing errors into account.
Validation of virtual TDFs by sequencing the matching bands
As has been described above, sequencing of >100 bands excised from cDNA-AFLP gels showed that the marker-based size estimation was highly accurate. This accuracy was further confirmed by sequencing 24 bands that matched virtual TDFs. Sequencing of these matching bands revealed no inconsistencies with the computed TDFs. It is therefore concluded that three identifiers, the restriction enzyme recognition sites, the primer extensions and the size of the band, are sufficient to find the corresponding real TDFs on cDNA-AFLP gels.
Expression patterns of virtual TDFs derived from ESTs with putative housekeeping functions
We chose several ESTs with putative housekeeping functions and investigated whether we could find TDFs from these genes on cDNA-AFLP gels at the appropriate positions and with the expected constitutive expression pattern.
EST sequences GE1373, GE1659 and GE1699 share high homology (BLASTX expect value < e–30) with elongation factor 1-β from various organisms, GE99 shares high homology (E value = e–35) with 40S ribosomal protein S20 and GE1409 is likely to be a ribosomal protein L20 homolog (E value = e–41). These proteins are essential components in protein synthesis and are constitutively expressed in most eukaryotic organisms. For all individual ESTs, GenEST predicted the generation of at least one TDF. Examination of cDNA-AFLP gels showed discrete bands at the right positions and virtually equal band intensities were observed in the five developmental stages. From one of the developmental stages the amplification products were cloned and sequenced. The resulting sequences were found to perfectly match the corresponding EST sequences.
These results show that the expression profiles were in accord with the predicted functions of the ESTs and that it is feasible to discard or select ESTs by analyzing the expression patterns of the predicted TDFs (see below).
EST to cDNA-AFLP: discarding ESTs on the basis of expression profiles
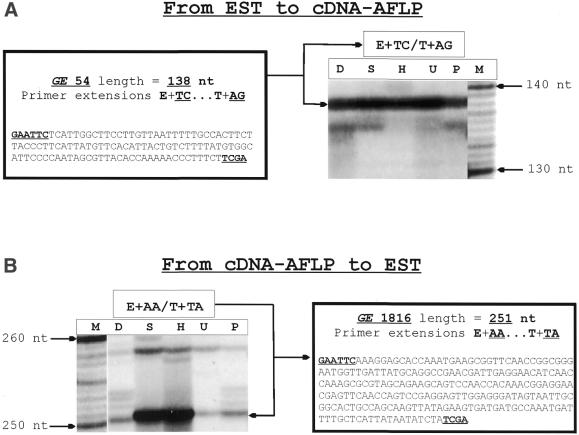
For many ESTs no function could be inferred from homology searches. About 40% of the ESTs obtained from G.rostochiensis were categorized as unknown and many of these genes seemed to be nematode-specific. Proteins encoded by these nematode-specific genes are presumably important in nematode physiology and a few among them may be related to parasitism of host plants (9). Examination of the expression profiles of the virtual TDFs provides valuable information on whether such ESTs deserve further investigation or not. This is exemplified by ESTs GE54 and GE1084. GE54 was predicted to produce a TDF with extensions E+TC/T+AG and a size of 138 nt and GE1084 with extensions E+AC/T+AC and a size of 85 nt. Their corresponding TDFs were readily identified on cDNA-AFLP gels (Fig. 1). Both bands displayed a constitutive expression pattern throughout the five developmental stages. This argues against a direct function of the proteins encoded by these two genes in plant parasitism.
Figure 1.
Overview of the bidirectional link between ESTs and cDNA-AFLP expression profiles established by GenEST. (A) From EST to cDNA-AFLP expression profiles. The predicted TDF of EST GE54 with primer extensions E+TC/T+AG and a size of 138 nt was readily identified on a cDNA-AFLP gel. (B) From cDNA-AFLP to EST. A band on the gel was amplified with primer extensions E+AA/T+TA with a size of 251 nt. These identifiers were used to search the virtual TDF list generated by GenEST. The corresponding EST was identified. M, molecular ladder. D, S, H, U and P represent the five different developmental stages of the potato cyst nematode: D, unhatched J2 in diapause; S, unhatched J2 after diapause, rehydrated for 2 days in water; H, freshly hatched J2 in PRD; U, developing nematodes (J1) in gravid females 2 months post-inoculation; P, developing nematodes (J2) in gravid females 3 months post-inoculation.
EST to cDNA-AFLP: selection of ESTs on the basis of expression profiles
GE1156 was predicted to produce three TDFs (E+CA/T+TT/65 nt, E+CT/T+GG/73 nt and E+GC/T+AT/82 nt). A band could be found at each of the predicted positions. The bands in the hatched J2 stage showed the highest intensity. Sequence alignment revealed that GE1156 was similar to the dorsal gland-specific gene GR-dgl-2 from the potato cyst nematode (10). GR-dgl-2 was previously shown to be specifically expressed in PRD-hatched J2. In situ hybridization revealed specific expression of GR-dgl-2 in the dorsal gland of the nematode. The proteins produced by this gland may be involved in induction of a feeding site, a so-called syncytium, in the host plant (13). The protein conceptually translated from the cDNA was predicted to be preceded by a signal peptide for secretion, indicating that this protein might be secreted by the nematode during the infection process.
GE1867 appeared to be identical to GR-eng-2, one of the β-1,4-endoglucanases that is secreted by cyst nematodes. In situ hybridization showed that GR-eng-2 was specifically expressed in the subventral gland (12). Unlike GE1156, the GE1867-derived TDF (E+TT/T+TG/399 nt) showed high expression not only in the H but also in the (earlier) S stage. This points to an earlier transcription activation of subventral gland-specific genes. The proteins encoded by these genes may be important in the early infection process, namely penetration of and migration in the plant root.
cDNA-AFLP to EST: finding (near) full-length cDNAs corresponding to TDFs with relevant expression patterns
The extensions of the EcoRI primer, the extensions of the TaqI primer and the size of a band on a cDNA-AFLP gel constitute a unique identifier for a TDF. These parameters can be used to search in the EST database to find an EST that can produce such a TDF. In this way TDFs with S/H or H stage-specific expression (i.e. gene expression just prior to invasion of the plant) were used to search the list of virtual TDFs generated from the EST database.
One TDF specifically expressed at the H stage, with extensions E+CC/T+CT/ and 137 nt in length, perfectly matched the parameters of the predicted TDF from EST GE2075. This band was subsequently cloned and sequenced. The sequence showed 99% match to EST GE2075. With the help of GenEST the gene sequence representing this H stage-specific band was extended from 137 to 477 bp (Table 2). By sequencing the original plasmid of EST GE2075 from the 3′-end, the cDNA sequence was extended to 685 bp.
Table 2. Genes selected on the basis of a combinatorial use of EST sequences and cDNA-AFLP data from the potato cyst nematode G.rostochiensis.
| Starting point |
Corresponds to |
Expression pattern on gel |
Homology |
| E+AA/T+TA/251 nt | GE1816 | ↑ in S and H | Unknown, predicted to have a signal peptide for secretion |
| E+CC/T+CT /137 nt | GE2075 | ↑ in H | Nematode dorsal gland-specific gene GR-dgl-2 |
| GE1156 | E+CA/T+TT/65 nt; E+CT/T+GG/73 nt; E+GC/T+AT/82 nt | ↑ H (three TDFs, same pattern observed) | Nematode dorsal gland-specific gene GR-dgl-2 |
| GE1867 | E+TT/T+TG/399 nt | ↑ in S and H stages | GR-eng-2 from potato cyst nematode |
In each direction the bidirectional program GenEST allowed for selection of putative pathogenicity-related genes out of hundreds of EST sequences and expression profiles of thousands of genes.
Another band displaying high expression in the S and H stages, with extensions E+AA and T+TA and a size of 251 nt, perfectly matched the predicted TDF from EST GE1816 (Fig. 1). Analysis of this EST sequence revealed that the longest open reading frame (ORF) contained 107 amino acids. This gene had no significant homology with existing genes in public databases. Use of the SignalP program (14) predicted that the protein had a cleavable signal sequence at its N-terminus that presumably targets the mature peptide for secretion. Hence, the combination of cDNA-AFLP and EST analysis has allowed us to identify this gene as worthy of further study for its potential role in nematode parasitism of plants.
These two examples illustrate another benefit of combining cDNA-AFLP and EST, which is to facilitate cloning of full-length cDNA sequences from which interesting TDFs are derived. The corresponding gene can be readily identified from the EST database even without cloning and sequencing of the TDF displayed on a gel. Once the corresponding EST is identified, obtaining a (nearly) full-length sequence is relatively simple by sequencing the entire cDNA insert from which the EST was originally derived.
In Table 2 four examples of putatively interesting ESTs and their corresponding TDFs are given. In Figure 1 an overview of the bidirectional link between ESTs and cDNA-AFLP expression profiles established by GenEST is given.
DISCUSSION
In this paper we present an efficient and bidirectional link between (partial) cDNA sequences and gene expression profiles as generated by cDNA-AFLP. A program called GenEST establishes this link. The added value of combining cDNA sequence information and cDNA-AFLP profiles is illustrated for one particular case, namely the search for putative pathogenicity factors from a plant parasitic nematode, G.rostochiensis. On the one hand, GenEST enabled us to find the expression profile of a given EST among the profiles of thousands of genes. The other way round, it allowed quick extension of TDFs by searching for the corresponding EST(s). As we have shown, the restriction enzyme recognition sites, the primer extensions and the size of the band displayed on a cDNA-AFLP gel constitute a unique set of identifiers for a TDF, the corresponding (nearly) full-length cDNA can be identified even without cloning and sequencing of the TDF of interest. In this way, the bottleneck of identifying the (near) full-length cDNAs in high throughput functional genomics studies using gel-based gene expression monitoring systems can be overcome. Since database similarity searches are more robust when using longer sequence fragments, the possibility of moving directly from a short TDF to a much longer EST may be very useful in further characterizing the putative function of a gene.
Use of GenEST for the selection of putative pathogenicity factors
Selection on the basis of expression profiles of the 228 virtual TDFs that were produced by in silico restriction of 985 ESTs with EcoRI and TaqI revealed four putative pathogenicity-related genes. One was a known gene encoding a cellulase (12). GE2075 and GE1156 displayed strong homology with a nematode secretory gland-specific gene GR-dgl-2, indicating a possible role in the parasitism of host plants. GE1816 is a novel gene. Its function will be studied further to reveal its role in the nematode infection process. It should be noted that this is the result of a small-scale pilot experiment only. Even on this scale, the value of GenEST, which combines two high throughput technologies, is evident: four putative pathogenicity-related genes were selected out of hundreds of EST sequences and expression profiles. The applicability of this freely available tool is broad as long as the expression of genes of interest is strictly limited, either spatially or temporarily.
Further applications of GenEST
Contrary to EST approaches, the cDNA-AFLP technique is not biased towards abundant transcripts and does not involve selection on insert size. Moreover, there is no unwanted selection due to intolerance of Escherichia coli to a subset of the inserts. To estimate the fraction of genes not tagged by ESTs, Penn et al. (15) have spotted 10 000 predicted ORFs from the human genome on a cDNA array and monitored expression of these ORFs under various conditions. They concluded that potentially up to 30% of the genes in the human genome will not be discovered by an EST approach. A similar experiment could be performed by linking cDNA-AFLP and EST data with GenEST. Failure to find a good match for a TDF shown on a cDNA-AFLP gel in a large-scale EST database is informative. The corresponding gene is presumably a novel gene expressed at a low level, a small gene or a gene refractory to cloning in E.coli. An advantage of our approach is that ESTs and cDNA-AFLP are not linked physically, as is the case for cDNA arrays. This avoids the amplification and spotting of thousands of EST clones, saving huge logistical efforts.
Besides generating restriction patterns of sequences, GenEST can also be used to find other sequence motifs in a large data set, a process which is often too time consuming to be done manually. To illustrate this application, GenEST was used to predict the occurrence of trans-spliced leader sequences from a database composed of ∼1000 ESTs of the root knot nematode Meloidogyne incognita. In many nematode species up to 70% of the mature mRNAs are trans-spliced with a 22 nt leader sequence on the 5′-end of the mRNAs (16). When the EcoRI recognition sequence in the command file is replaced by the trans-spliced leader sequence all the ESTs containing this sequence can be quickly identified using GenEST. This information can be used to estimate the fraction of full-length cDNAs present in a library and to check whether the encoded ORFs start with a peptide signal for secretion. This latter process could be further streamlined by establishing a link between GenEST and search algorithms such as SignalP.
AFLP techniques have been used extensively in genetic mapping in various organisms and a large number of AFLP markers associated with genes of interest have been identified (17,18). Such markers combined with a fully sequenced genome (e.g. Arabidopsis thaliana; 19) could facilitate efficient cloning of target genes. To this end, GenEST can be adapted to assist in the identification of the physical locus of an interesting gene by using the identifiers of appropriate AFLP markers.
Further improvement of the EST coverage
Only 16% (159/985 × 100%) of the 985 ESTs were digested in silico by EcoRI and TaqI. To increase the percentage of ESTs from which virtual TDFs are obtained a set of alternative rare cutters, including NcoI, KasI and AseI, are currently being used in combination with TaqI. With three additional primer combinations, more than half of the EST sequences [1 – (1 – 0.16)4 = 50.2%] will produce at least one virtual TDF, which could be identified on cDNA-AFLP gels. To further increase the coverage of the EST population, cDNA-AFLP can be performed with two frequent cutters. Alternatively, cDNAs could be digested with a frequent cutter only and ligated to the corresponding adapter. Subsequently, 3′-anchored cDNA-AFLP could be performed using an oligo(dT) primer in combination with the rare cutter adapter primer. This approach may be especially useful with organisms for which the entire genome has been sequenced or for which large-scale 3′-end EST sequencing has been performed. Moreover, by increasing the fraction of full-length cDNA sequences, the chance of finding at least one corresponding TDF on a gel would improve significantly.
As shown in this study, the ability to switch between sequence data and expression profiles revealed by cDNA-AFLP and vice versa is a very powerful approach to select genes for further research. This novel link provided by GenEST will be useful for functional genomics studies and is applicable to any organism where differentially expressed genes are of interest. The source code of the GenEST program is freely downloadable. The user is free to modify the existing program according to his/her own wishes.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by European Community grants BIO4-CT96-0318 (L.Q.), QLK5-1999-01501 (L.Q.) and QLK5-1999-01462 (H.P.) and by Scottish Executive Rural Affairs Department project no. SCR/514/98 (J.T.J.).
References
- 1.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 563–567. [DOI] [PubMed] [Google Scholar]
- 2.The C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C.elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 3.Blaxter M. (1998) Caenorhabditis elegans is a nematode. Science, 282, 2041–2046. [DOI] [PubMed] [Google Scholar]
- 4.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 5.Schena M. (1996) Genome analysis with gene expression microarrays. Bioessays, 18, 427–431. [DOI] [PubMed] [Google Scholar]
- 6.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 7.Liang P. and Pardee,A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. [DOI] [PubMed] [Google Scholar]
- 8.Bachem C.W., van der Hoeven,R.S., de Bruijn,S.M., Vreugdenhil,D., Zabeau,M. and Visser,R.G. (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J., 9, 745–753. [DOI] [PubMed] [Google Scholar]
- 9.Popeijus H., Blok,V.C., Cardle,L., Bakker,E., Phillips,M.S., Helder,J., Smant,G. and Jones,J.T. (2000) Analysis of genes expressed in second stage juveniles of the potato cyst nematodes Globodera rostochiensis and G. pallida using the expressed sequence tag approach. Nematology, 2, 567–574. [Google Scholar]
- 10.Qin L., Overmars,H., Helder,J., Popeijus,H., Rouppe van der Voort,J.N.A.M., Groenink,W., van Koert,P., Schots,A., Bakker,J. and Smant,G. (2000) An efficient cDNA-AFLP-based strategy for the identification of putative pathogenicity factors from the potato cyst nematode Globodera rostochiensis. Mol. Plant Microbe Interact., 13, 830–836. [DOI] [PubMed] [Google Scholar]
- 11.Rouppe van der Voort J.N.A.M., van Eck,H.J., van Zandvoort,P., Overmars,H., Helder,J. and Bakker,J. (1999) Linkage analysis by genotyping of sibling populations: a genetic map for the potato cyst nematode constructed using a “pseudo-F2” mapping strategy. Mol. Gen. Genet., 261, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 12.Smant G., Stokkermans,J.P., Yan,Y., de Boer,J., Baum,T.J., Wang,X., Hussey,R.S., Gommers,F.J., Henrissat,B., Davis,E.L. et al. (1998) Endogenous cellulases in animals: isolation of β-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl Acad. Sci. USA, 95, 4906–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson V.M. and Hussey,R.S. (1996) Nematode pathogenesis and resistance in plants. Plant Cell, 8, 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 15.Penn S.G., Rank,D.R., Hanzel,D.K. and Barker,D.L. (2000) Mining the human genome using microarrays of open reading frames. Nat. Genet., 26, 315–318. [DOI] [PubMed] [Google Scholar]
- 16.Blaxter M. and Liu,L. (1996) Nematode spliced leaders—ubiquity, evolution and utility. Int. J. Parasitol., 26, 1025–1033. [PubMed] [Google Scholar]
- 17.Vos P., Hogers,R., Bleeker,M., Reijans,M., van der Lee,T., Hornes,M., Frijters,A., Pot,J., Peleman,J., Kuiper,M. and Zabeau,M. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res., 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buschges R., Hollricher,K., Panstruga,R., Simons,G., Wolter,M., Frijters,A., van Daelen,R., van der Lee,T., Diergaarde,P., Groenendijk,J. et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell, 88, 695–705. [DOI] [PubMed] [Google Scholar]
- 19.Kaul S., Koo,H.L., Jenkins,J., Rizzo,M., Rooney,T., Tallon,L.J., Feldblyum,T., Nierman,W., Benito,M.I., Lin,X.Y. et al. (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]