Abstract
Pseudomonas fluorescens are rhizobacteria known for their biocontrol properties. Several antimicrobial functions are crucial for this process, and the experiments described here investigate the modulation of their expression during the plant-bacterium interaction. The role of a LuxR family regulator in interkingdom signaling has been investigated using genome-scale transcriptome analysis, gene promoter studies in vivo and in vitro, biocontrol assays, and response to plant compounds. PsoR, a LuxR solo or orphan regulator of P. fluorescens, was identified. PsoR is solubilized and activates a lux-box-containing promoter only in the presence of macerated plants, suggesting the presence of a plant molecule(s) that most likely binds to PsoR. Gene expression profiles revealed that genes involved in the inhibition of plant pathogens were affected by PsoR, including a chitinase gene, iron metabolism genes, and biosynthetic genes of antifungal compounds. 2,4-Diacetylphloroglucinol production is PsoR dependent both in vitro and in vivo. psoR mutants were significantly reduced for their ability to protect wheat plants from root rot, and damping-off caused by Pythium ultimum infection. PsoR most likely senses a molecule(s) in the plant and modulates expression of genes that have a role in biocontrol. PsoR and related proteins form a subfamily of LuxR family regulators in plant-associated bacteria.
INTRODUCTION
A large subfamily of LuxR proteins bind the N-acyl homoserine lactone (AHL) quorum-sensing (QS) signaling molecules produced by proteobacteria (38). AHLs interact at quorum concentrations with quorum-sensing LuxR-family proteins, inducing a conformational change that allows LuxR to bind to a specific DNA lux box and activate the transcription of target genes. The QS LuxR family proteins are approximately 250 amino acids long and are modular, composed of two functional domains: an amino-terminal AHL-binding domain and a carboxy-terminal domain containing a helix-turn-helix DNA-binding motif (29). Recent studies have shown that many bacteria possess uncharacterized LuxR proteins with the typical modular structure of QS members but not associated with a cognate LuxI AHL synthase; these proteins have been called LuxR orphans or LuxR solos (17, 32, 45). The sequencing of many proteobacterial genomes has highlighted that many putative LuxR solos are present in bacterial species that contain typical luxI-luxR family pairs, as well as ones that do not.
In most of the few cases studied thus far, LuxR solos have been shown to interact with endogenous and/or exogenously produced AHLs, allowing bacteria to either expand the existing QS network or to respond to AHLs produced by neighboring bacteria (32, 45). For example, QscR of P. aeruginosa responds to endogenous C12-3-oxo-HSL produced by the LasI AHL synthase and controls a regulon distinct from those controlled by the LasI/R or RhlI/R systems (25). The SdiA solos of Salmonella enterica and Escherichia coli, on the other hand, regulate gene expression in response to exogenous AHLs, since neither species produces AHL signal molecules (2, 26).
Two LuxR solos have also been recently reported to bind unknown plant compounds. OryR of Xanthomonas oryzae pv. oryzae and XccR of Xanthomonas campestris pv. campestris are two LuxR solos of plant-pathogenic species that are homologous to each other and are involved in interkingdom signaling as they respond to a plant compound(s) (15, 16, 50). Orthologues of OryR and XccR are also present in the genomes of several other plant-associated species of rhizobia and pseudomonads (16, 50), indicating that they could represent a new subfamily of LuxR solos. However, there is a lack of information regarding their role in plant-associated biocontrol bacteria, as well as the range/nature of their target genes. In an effort to better understand this novel subfamily of proteins, we identified a LuxR solo of Pseudomonas fluorescens Pf-5 and P. fluorescens CHA0 designated here as PsoR that is in the OryR and XccR subfamily. We designated the LuxR solo as PsoR, and we describe here experiments designed to determine its role in P. fluorescens. The closely related strains rhizosphere colonizing P. fluorescens Pf-5 and CHA0 are regarded as models of biological control organisms since they anatgonize deleterious microorganisms through the synthesis of several antimicrobial secondary metabolites, including 2,4-diacetylphloroglucinol (DAPG) and pyoluteorin (PLT) (18–20). We show that PsoR is involved in some of these processes and in the regulation of a number of genes in P. fluorescens CHA0.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All strains, plasmids, and primers used in the present study are listed in Table S1 in the supplemental material. P. fluorescens and E. coli strains were grown in LB (27) medium at 30 and 37°C, respectively. P. fluorescens strains were also grown in M9 minimal medium (39) supplemented with 0.3% Casamino Acids (M9-CAS) at 30°C. Media containing macerated plant material (rice, wheat, or cucumber leaves) were prepared as follows: 20 to 25 g of leaves were macerated in the presence of liquid nitrogen, added to 400 ml of LB or M9-CAS minimal medium, and autoclaved. This medium was filtered and used for experiments. Antibiotics, when required, were at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml (P. fluorescens) or 50 μg/ml (E. coli); tetracycline, 10 μg/ml (E. coli) or 125 μg/ml (P. fluorescens); and gentamicin, 10 μg/ml (E. coli) or 40 μg/ml (P. fluorescens).
A transcriptional fusion construct for the psoR promoter in pMP220 was made by amplifying a 598-bp fragment containing the psoR promoter region from P. fluorescens Pf-5 genomic DNA using the primers 5298promF and 5298promR and cloned into pBluescript (Stratagene), yielding pBS1, and subsequently cloned into pMP220 (HindIII/XbaI), yielding pPsoRprom.
To clone the X. oryzae pv. oryzae pip promoter in pMP220, a 438-bp fragment containing the X. oryzae pv. oryzae proline imino peptidase promoter region was amplified by using the primers PIPPRS and PIPPRR and cloned into pMosblue, yielding pMOSxoopip, and subsequently cloned into pMP220, yielding pXoopipprom.
The psoR gene was cloned into the expression vector pQE30 as follows. A 768-bp fragment the psoR gene of P. fluorescens Pf-5 was amplified using primers PFL5298F1 and PFL5298R1 and cloned into pGEM-T Easy, yielding pGEM2, and subsequently cloned into pQE30(BamHI/HindII), yielding pQEPsoR. The pBBRMCS-5 (24) clone of psoR was constructed as follows. A 1,162-bp fragment containing the psoR gene and promoter region was amplified by using the primers 5298compF and 5298compR and cloned into pMosblue, yielding pMOS4, and subsequently cloned into pBBRMCS-5 (EcoRI, HindIII), yielding pBBRPsoR. The psoR sequence of strains CHA0 and Pf-5 are identical.
Recombinant DNA techniques.
All DNA manipulations and the transformation of E. coli were performed as described previously (39). Southern hybridizations were performed by using N+Hybond membranes (Amersham Biosciences). Plasmids were purified by using Jet Star columns (Genomed GmbH, Löhne, Germany), and total DNA from Pseudomonas was isolated by Sarkosyl-pronase lysis as described previously (5).
Screening P. fluorescens Pf-5 and CHA0 for the production of AHLs.
The two P. fluorescens strains were tested for the production of AHLs in a T-streak analysis, as described by Piper et al. (36) using the AHL biosensors Chromobacterium violaceum CVO26, E. coli MT102(pJBA132), E. coli (pSB1075), and P. putida F117(pKRC12) (43) in LB and with Agrobacterium tumefaciens (NTL4) as described in Farrand et al. (14). AHL production was analyzed by thin-layer chromatography (TLC) after extraction from cell-free spent supernatants of 100 ml of M9-CAS medium cultures as previously described (43). The TLC plates were overlaid with a thin layer of LB or AB top agar (14) seeded with either C. violaceum CVO26, E. coli JM109(pSB401), E. coli(pSB1075), or A. tumefaciens NTL4(pZLR4). AHLs were either commercially acquired from Fluka-Sigma-Aldrich or obtained from P. Williams (University of Nottingham, Nottingham, United Kingdom).
Generation of psoR mutants in Pf-5 and CHA0.
In order to generate a psoR mutant in strains Pf-5 and CHA0, we constructed pKNOCKpsoR as follows. A 369-bp internal fragment of the P. fluorescens Pf-5 psoR gene was amplified by PCR using the primers pKNOCK5298F and pKNOCK5298R and cloned into pMOSblue, yielding pMOS5 and subsequently cloned into pKNOCK-Km(XbaI/KpnI) to yield pKNOCKpsoR. pKNOCKpsoR was then used to create knockout mutants of psoR by homologous recombination in P. fluorescens strains Pf-5 and CHA0. The mutants were designated P. fluorescens Pf-5PSOR and P. fluorescens CHA0PSOR, respectively. The fidelity of the marker exchange events was confirmed by Southern analysis of the mutants.
β-Galactosidase-based reporter gene fusion assay.
β-Galactosidase activities were determined during growth in M9-CAS as described by Miller (27), with the modifications of Stachel et al. (41). All experiments were performed in triplicate, and the mean values are given. The β-galactosidase activities were determined at various times after a 20-ml M9-CAS culture was started with an initial inoculum of 5 × 106 CFU.
Total RNA isolation.
RNA isolations were carried out from cultures of P. fluorescens strains carrying pBBRMCS-5 or pBBRPsoR grown in M9-CAS. The cultures were incubated (30°C, 180 rpm) until they reached an optical density at 600 nm (OD600) of approximately 1.2 to 1.3. RNA isolation was carried out from 2 × 109 cells using the RiboPure-Bacteria RNA isolation kit (Ambion, Inc., Austin, TX) according to manufacturer's instructions. Isolated RNA was treated twice with DNase at 37°C for 1 h and purified following directions of the manufacturer (Ambion). The purity of the RNA was assessed by PCR on total RNA (250 ng) with GoTaq polymerase (Promega) using primers specific for P. fluorescens Pf-5 psoR. The RNA quality was assessed by spectrophotometric measurement at 260 and 280 nm, and its intact nature was verified on an agarose gel.
Semiquantitative RT-PCR and analysis.
Reverse transcription (RT) was performed in a 20-μl reaction mixture containing 2.5 μg of total RNA, 200 ng of random primers/μg of RNA (Promega, Madison, WI), and 30 U of AMV reverse transcriptase according to the manufacturer's instructions. The conditions used for RT were 65°C for 3 min, 25°C for 10 min, 42°C for 90 min, and 70°C for 10 min. The Pf516For and Pf516Rev primers were used to measure the transcription of 16S rRNA. Second-strand synthesis was performed using GoTaq Flexi polymerase (Promega) with 1 μl of undiluted (any test gene) or a 1:100-diluted (16S rRNA) cDNA reaction as the template. The number of PCR cycles for each gene was standardized so that the product amplification is in the linear range. A portion (10 μl) of the PCR was analyzed by agarose gel electrophoresis. The intensity of the bands was measured and normalized to that of 16S rRNA by using ImageJ software (1) to obtain the fold difference. Each gene was validated twice by RT-PCR analysis of RNA samples from two independent isolations.
Determination of psoR expression levels was carried out using equal amounts of total RNA isolated from late logarithmic-phase cultures of P. fluorescens wild-type CHA0 carrying the empty pBBRMCS-5 vector and CHA0 carrying pBBRPsoR for overexpression of psoR.
Affinity column chromatography and Western blotting.
E. coli M15(pRep4) containing pQEPsoR was inoculated into 100 ml of either LB alone or LB containing 10 g of macerated rice wheat or cucumber leaves at an initial OD600 of 0.1. When the culture reached an OD600 of 0.6, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside)was added, and the culture was shifted to 30°C (45 min, 180 rpm). The culture was then chilled on ice, and the cell pellet obtained by centrifugation was stored at −80°C. The cell pellets containing overexpressed His6-PsoR were then lysed under native conditions according to the supplier's instructions (Qiagen). The lysate was subjected to Hi-Trap affinity column chromatography as described by Ferluga et al. (15). The flowthrough sample, as well as eluates at 10, 20, and 50% concentrations of buffer B, were collected and analyzed by Western blotting as described previously (15).
E. coli M15(pRep4) containing pQEPsoR was grown at 37°C and also inoculated into 10 ml of either LB alone or LB containing a 2 mM concentration of one of the following AHLs: C4-, C6-, C8-, C10-, and C12-HSL; C4-3-oxo-, C6-3-oxo-, C8-3-oxo-, C10-3-oxo-, and C12-3-oxo-HSL; or C4-3OH-, C6-3OH-, C8-3OH-, C10-3OH-, and C12-3OH-HSL. The cell pellets were processed and analyzed by Western blotting with anti-His6 monoclonal antibody as described above.
Biocontrol, root colonization, and gene promoter assays of antifungal loci.
The biocontrol assays were performed as previously described (4). Seeds of cucumber (Cucumis sativus cv. Chinesische Schlange) and winter wheat (Triticum aestivum cv. Arina) were surface sterilized (18 min in 5% NaClO [vol/vol]), rinsed with sterile water, and germinated on soft agar (Oxoid technical agar at 8.5 g/liter for 90 h at 20°C in the dark). For the biocontrol assays, 200-ml Erlenmeyer flasks with wide openings were partially filled with 60 g of natural sandy loam soil from Eschikon, Switzerland (23). When appropriate, the soil was artificially infested with 2.5 g (cucumber) or 18.75 g (wheat) of a 5-day-old millet-seed inoculum of Pythium ultimum isolate 67-1/kg (23). Three sterile-grown seedlings were then placed in each flask. P. fluorescens strains were added as a suspension (5 ml per flask) of washed cells to give 107 CFU/g of soil. Seedlings were covered with nontreated soil, and the flasks were sealed and incubated as specified by Baehler et al. (4).
The expression of a gfp-based reporter fusion to the DAPG biosynthetic gene phlA carried by pME9417 (see Table S1 in the supplemental material) was monitored on roots of cucumber and winter wheat using a flow cytometry approach detailed previously (10). For the construction of pME9417, the phlA-gfp fusion carried by reporter plasmid pME7100 (3) was transferred into the pVS1 derivative pPROBE-KT (28) to allow for compatibility with the pBBR1-based plasmid used for psoR overexpression. Strains CHA0(pBBRMCS-5), CHA0PSOR(pBBRMCS-5), and CHA0(pBBRPsoR) carrying pME9417 were inoculated onto 3-day-old seedlings (108 bacterial cells per seedling) in sterile plant growth pouches (Mega International, West St. Paul, MN) containing a moist filter paper. The growth pouches, each holding three inoculated plants, were incubated for 5 days in a growth chamber as described previously. Roots from each growth pouch were then pooled in 10 ml of sterile water in 50-ml Falcon tubes and agitated vigorously, and the resulting suspension was analyzed by fluorescence-activated cell sorting (FACS) using a FACSCalibur flow cytometer equipped with a 15-mW, air-cooled argon ion laser excitation light source (488 nm; Becton Dickinson, San Jose, CA). Green fluorescent protein (GFP) fluorescence emitted by the phlA-gfp reporter was detected in the range of 515 to 545 nm with an FL1-H detector set at a photomultiplier tube voltage of 505 V with logarithmic gain. The data were collected using Becton Dickinson CellQuest software version 3.5 and analyzed with the WinMDI software version 2.8 (http://facs.scripps.edu/software.html). Gating was done by defining regions on the density plot and by setting a marker above the autofluorescence background exhibited by strain CHA0(pBBRMCS-5) without GFP reporter as described in detail by de Werra et al. (10).
For the expression of the phlA-gfp reporter on pME9417, cultures were grown in 20 ml of M9-CAS medium as described for RNA isolation. Bacterial growth and expression of the reporters were monitored with a FLUOstar fluorescence microplate reader (BMG Labtech, Offenburg, Germany) throughout the exponential- and stationary-growth phases, as detailed by Baehler et al. (4).
Microarray experiment and analysis.
Because strains CHA0 and Pf-5 are very similar to one another and genes sequenced in both strains are also very similar, the effect of psoR overexpression on the transcriptome of CHA0 was assessed by using an extant genomic microarray for Pf-5 (22). The microarray is composed of 70mer DNA oligonucleotides designed to represent each of the protein coding genes annotated from the genomic sequence of Pf-5, which were spotted onto Aminosilane-coated UltraGAPS glass slides (Corning, Action, MA). Three replicate cultures of P. fluorescens CHA0 and strain CHA0(pBBRPsoR) were grown in 20 ml in M9-CAS medium until the end of the logarithmic phase (OD600 of 1.2). RNA extractions, the removal of DNA contamination, and RNA quality and quantity determination were carried out as described above. cDNA labeling was conducted using an indirect labeling method as previously described (34). Briefly, cDNA was synthesized in the presence of either cyanine 3 (Cy3)- or cyanine 5 (Cy5)-coupled nucleoside triphosphate analogues. Portions (6 to 12 μg) of total RNA were used for each indirect labeling reaction to produce ∼20 μg of cDNA containing ∼200 pmol of Cy3 or Cy5 dye per μg. Spectrophotometric absorbance readings of the labeled cDNA were conducted prior to hybridization to ensure optimal dye incorporation for adequate sample intensity. Each microarray experiment included two technical repeats comprising a dye-flip experiment for each of the three replicate RNA samples.
Hybridizations were conducted as previously described (34). Slides were scanned at 10-μm resolution using an Axon 4000B scanner with GenePix 4.0 software. Tiff images of hybridized arrays were processed by using TIGR-SpotFinder, and the datasets were normalized by applying the Lowess algorithm, using block mode and a smooth parameter of 0.33 in the TIGR-MIDAS package. Statistical analysis was performed on log2-transformed signal ratios of the replicate spots using statistical analysis of microarrays (SAM) algorithms (47). Genes whose transcript levels were influenced by psoR overexpression exhibited fold differences of at least 1.4 that were found to be significant using a false discovery rate of <5%, unless otherwise indicated.
All microarray data presented are in accordance with the Microarray Gene Expression Data Society's minimum information about microarray experiment (MIAME) recommendations (7). Descriptions of the microarray experiment, quantification data, and array design have been deposited into GEO (www.ncbi.nlm.nih.gov/geo/).
RESULTS
PsoR is a LuxR solo belonging to a novel subgroup of plant-responding regulators.
The genome of P. fluorescens Pf-5 does not include genes with annotated functions as LuxI or LuxM type of AHL synthases. We have tested P. fluorescens Pf-5 and CHA0 for AHL production using four different biosensors that are able to detect structurally different AHLs. No AHL production was detected in our experiments; P. fluorescens CHA0 was already reported previously not to produce AHLs (12). However, in silico analysis of P. fluorescens Pf-5 revealed the presence of two open reading frames (ORFs; PFL_3627 and PFL_5298) that code for QS LuxR type proteins. One of these, PFL_5298 was termed psoR and characterized in the present study. The psoR gene is flanked by pepQ (PFL_5299; coding for creatinase) upstream and by pip (PFL_5297; coding for proline imino peptidase) downstream (see Fig. S1 in the supplemental material). Both psoR and the pip gene are transcribed from the complementary strand of genomic DNA. The psoR–pip gene organization is mostly conserved in all bacterial genomes where orthologues of both genes were identified by BLASTP analysis. The predicted protein encoded by psoR is 252 amino acids long, and domain analysis revealed the presence of an N-terminal autoinducer-binding domain (PF03472) and a C-terminal DNA-binding domain typical of QS LuxR family proteins (PF00196). PsoR shows a high degree of similarity (>55% at amino acid level) to QS LuxR regulators in several plant-associated bacteria belonging to the genera Pseudomonas, Agrobacterium, Rhizobium, Sinorhizobium, and Xanthomonas. BLASTP reveals considerable similarity to functionally characterized LuxR solo regulators, including OryR of X. oryzae pv. oryzae (XOO1268; 45% identity and 59% similarity, YP_199907), XccR of X. campestris pv. campestris (XCC2818; 45% identity and 57% similarity, NP_638166), and NesR of S. meliloti Rm1021(SMc04032; 41% identity and 61% similarity, NP_386921). Multiple sequence alignment revealed that seven of nine amino acids that are strictly conserved in typical QS LuxR type regulators are present in PsoR; the exceptions are the positions corresponding to Tyr53 and Tyr61 of TraR which are substituted by Thr and Trp, respectively (Thr58 and Trp66 in PsoR; see Fig. S1 in the supplemental material). Due to their high sequence similarity and the substitution of conserved amino acids at the positions corresponding to Tyr53, Trp57, or Tyr61 (of TraR), these proteins are thought to belong to a separate class of LuxR regulators that recognize plant-derived molecules but not AHLs (50). PsoR (as well as PsoR-like proteins in other P. fluorescens and P. syringae strains) seems to be unlike other members of this group of proteins since all the other orthologs of PsoR have a Met substitution at the position corresponding to Trp57 (of TraR), whereas PsoR retains Trp (Trp62 in PsoR), indicating that the ligand may be different.
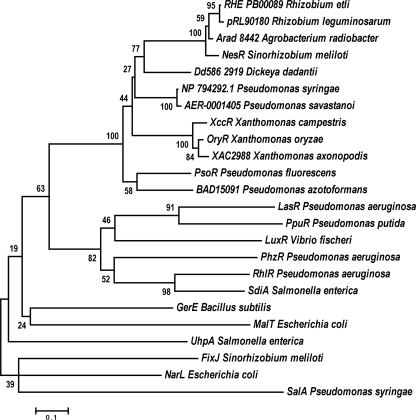
Together, both sequence features of PsoR and the presence of similar proteins in various plant-associated bacteria led us to speculate that PsoR possibly exerts its regulatory effects by responding to plant compounds similar to OryR of X. oryzae and XccR of X. campestris (see below and Fig. 6).
Fig. 6.
Dendrogram showing the members of the PsoR subfamily of LuxR family regulators. The clustering of all of the members of the PsoR subfamily is depicted and, in addition, other LuxRs are included to further highlight the subfamily. The ORFs in the tree are found in the following sequenced strains: Pseudomonas fluorescens Pf-5, Pseudomonas syringae pv. tomato T1, Pseudomonas azotoformans IAM 1603, Agrobacterium radiobacter K84, Rhizobium etli CFN 42, Dickeya dadantii Ech586, Rhizobium leguminosarum bv. viciae 3841, Sinorhizobium meliloti 1021, Xanthomonas campestris pv. campestris 8004, Xanthomonas oryzae pv. oryzae MAFF 311018, Xanthomonas axonopodis pv. citri 306, Pseudomonas aeruginosa PAO1, Pseudomonas putida PCL1445, Vibrio fischeri ES114, Salmonella enterica subsp. enterica serovar Typhimurium, Bacillus subtilis subsp. subtilis strain 168, and Escherichia coli K-12. The dendrogram was assembled with the neighbor-joining method with bootstrap as a test of inferred phylogeny using the MEGA 4 program (46).
Identification of the PsoR regulon by genome-scale transcriptome analysis.
In order to gain insights into the function of PsoR, a whole-genome oligonucleotide microarray of P. fluorescens Pf-5 was used to identify differentially expressed genes in a PsoR-overexpressing derivative of P. fluorescens CHA0 compared to the wild type. Many molecular studies have shown that these two closely related model P. fluorescens biocontrol strains share highly similar genes and phenotypes (18, 20); in addition, PsoR from strain CHA0 is identical to PsoR of strain Pf-5.
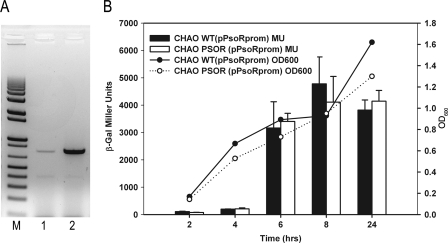
The stable pBBRMCS-5 cloning vector harboring psoR with its own promoter was conjugated into P. fluorescens CHA0 creating the PsoR overexpressing the CHA0PsoR+ strain. It was then determined by RT-PCR that in the late logarithmic phase, psoR mRNA levels were significantly higher in CHA0PsoR+ compared to the wild-type strain harboring the empty pBBRMCS-5 vector (Fig. 1 A). In addition, psoR expression levels were monitored for wild-type CHA0 and CHA0PSOR by using a psoR promoter-lacZ reporter fusion. The expression of psoR in both strains was found to increase with growth reaching maximum levels when the culture density reached an OD of 1.2 (Fig. 1B). The levels of expression of psoR was also determined in CHA0PSOR and CHA0PsoR+. The results clearly showed that psoR expression was growth phase dependent, reaching high levels under all of the conditions tested (data not shown). Comparison of psoR promoter expression in CHA0 and CHA0PSOR in the presence or absence of rice plant extracts indicated that there was no autoregulation or any effect by the presence of the plant extract (data not shown). It was therefore concluded that higher levels of psoR transcripts in the CHA0PsoR+ strain (Fig. 1A) were due to the multicopy effects of psoR harbored in the pBBR plasmid. The CHA0PsoR+ strain had a slightly reduced growth in M9-CAS (Fig. 1B) but displayed growth profiles comparable to that of the wild type when grown in rich media (data not shown). Transcriptomic profiles of the wild-type and CHA0PsoR+ strains were then determined for cultures harvested at OD of 1.2 as described in Materials and Methods.
Fig. 1.
Determination of psoR expression levels. (A) Lanes: M, 1 kb + DNA ladder (Invitrogen); 1, RT-PCR product obtained for CHA0(pBBRMCS-5); 2, RT-PCR product obtained for CHA0(pBBRPsoR). (B) β-Galactosidase assays showing the expression profile of psoR in P. fluorescens CHA0 wild type and its psoR mutant CHA0PSOR, both harboring a psoR–lacZ reporter fusion. The graph was plotted using SigmaPlot version 10.0. β-Gal, β-galactosidase; OD600, optical density at 600 nm; MU, Miller units.
Overexpression of PsoR had a global effect, affecting positively and negatively the transcription of many genes. Transcript levels of 227 ORFs, representing 3.7% of the total ORFs in the genome of P. fluorescens Pf-5 differed significantly by at least 1.4-fold between the CHA0PsoR+ and wild-type strains. Of these, 162 ORFs were negatively regulated, while 65 ORFs were positively regulated by PsoR (see Tables S2 and S3 in the supplemental material, respectively).
Positive regulation by PsoR.
The gene showing the highest induction levels (12-fold) was psoR itself, and this confirmed that in our experimental conditions psoR was overexpressed, as also noted above (Fig. 1A).
The expression of genes coding for chitin-binding protein and chitinase (PFL_2090 and PFL_2091) was significantly increased (2.1- and 2.7-fold, respectively) in the CHA0PsoR+ strain, indicating positive regulation. Chitinase is an enzyme that degrades chitin, an important constituent of fungal cell walls and insects; this points to a role for PsoR for the biocontrol of P. fluorescens CHA0. PFL_2090 and PFL_2091 have been predicted to constitute an operon using computational approaches (http://www.microbesonline.org/operons/ [37]). The transcript levels of the chitinase gene (PFL_2091) were >5-fold higher in the CHA0PsoR+ strain, as confirmed by an RT-PCR analysis of independent RNA samples from wild-type and CHA0PsoR+ strains ( Student t test, P < 0.0001; Fig. 2 and data not shown).
Fig. 2.
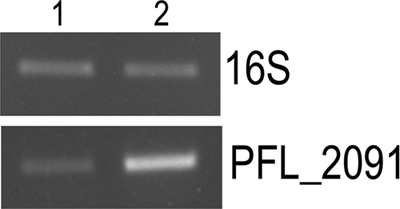
RT-PCR analysis to validate expression of the chitinase gene in P. fluorescens CHA0. RT-PCR analysis was performed with RNA obtained from two independent isolations, and the figure shows the results of one such experiment. The agarose gel shows the RT-PCR product for PFL_2091. As a control, RT_PCR for 16S rRNA was carried out for the same RNA samples to ensure that equal amounts of RNA were taken for the reaction. Lanes: 1, RT-PCR on RNA sample from CHA0 harboring the empty vector pBBRMCS-5; 2, RT-PCR on RNA sample from CHA0 harboring the plasmid overexpressing psoR, pBBRPsoR.
P. fluorescens CHA0 is known to produce multiple antifungal compounds, in particular DAPG, HCN, pyrrolnitrin, and PLT, that are important for its biocontrol properties. PLT and HCN biosynthetic clusters were not affected by PsoR. The transcript levels of three genes in the pyrrolnitrin biosynthetic cluster (PFL_3604, PFL_3606, and PFL_3607) were 1.5-fold higher in the PsoR+ strain (see Table S2 in the supplemental material); these three genes (prnA, prnC, and prnD) are most likely part of an operon that also includes PFL_3605 (prnB). PFL_3607, another gene in this cluster, showed only a 1.3-fold increase in the CHA0PsoR+ strain. Seven genes in the DAPG biosynthetic cluster were upregulated by an average of 2.5- to 8.2-fold in the CHA0PsoR+ strain, but differences in gene expression were observed in only two of three replicate cultures (four of these genes [phlA to phlD] are most likely part of an operon). To follow up on the effects of PsoR on the expression of DAPG biosynthesis genes observed in two replicates, we assessed a phlA-gfp transcriptional fusion in CHA0PsoR+, CHA0PSOR, and CHA0 (see below).
Negative regulation by PsoR.
Among the negatively regulated loci, a gene coding for a putative proline imino peptidase (pip) located adjacent to psoR exhibits >2.2-fold downregulation in the CHA0PsoR+ strain. Interestingly, pip has been reported to be transcriptionally regulated by homologues of PsoR, namely, XccR and OryR in the plant pathogens X. oryzae pv. oryzae and X. campestris pv. campestris (15, 16, 50). In contrast to P. fluorescens CHA0, pip in both of these pathogens is regulated positively by the PsoR homologues. It must be noted that the pip gene of P. fluorescens, unlike the other pip gene, does not contain a lux box.
Genes involved in taurine metabolism and sulfur utilization were also negatively regulated by PsoR (see Table S3 in the supplemental material). Several genes involved in iron uptake and homeostasis were differentially regulated by PsoR. Several other genes show reduced the levels of expression, as reported in Table S3 in the supplemental material. We analyzed the promoters of genes identified in the microarray experiment to identify well-conserved possible PsoR lux-box-binding sites; this resulted in the identification of three potentially very well conserved lux boxes (highly identical to the pip boxes). These putative lux boxes were in the promoters of (i) PFL_2090, a putative chitin-binding protein (ACCTGTCTGTAACGCAGAGG); (ii) PFL_3064, the pyrollnitirn prnA gene (ACCTACGCAATATTTTCCTT); and (iii) PFL_5953, the DAPG phlF gene (ACCTGCAGGCCAGAGCGGTT). It remains to be determined whether any of these elements are functional in the PsoR-mediated regulation of these genes.
PsoR responds to plant molecules.
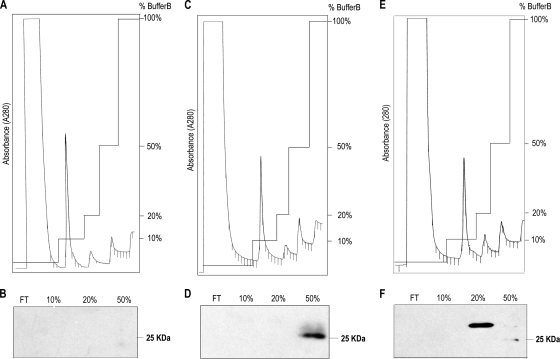
The primary structure of PsoR indicated that it most probably belongs to a subfamily of QS-related LuxR family proteins that interact with low-molecular-weight plant-derived molecules (see above). QS LuxR family proteins are known to be mostly insoluble when highly overexpressed; however, when bound to their cognate AHL, they become soluble (40, 48, 49). In addition, QS LuxR family proteins unbound to AHLs are more unstable and prone to proteolytic attack (35). The PsoR protein was abundantly overexpressed in E. coli as a His6-PsoR using the pQEPsoR plasmid, and it was established that PsoR was highly insoluble (data not shown). The His6-PsoR protein was then overexpressed in E. coli in the presence of many structurally different AHLs (C4-, C6-, C8-, C10-, and C12-HSL; C4-3-oxo-, C6-3-oxo-, C8-3-oxo-, C10-3-oxo-, and C12-3-oxo-HSL; C4-3OH-, C6-3OH-, C8-3OH-, C10-3OH-, and C12-3OH-HSL), and in all cases no PsoR solubilization was detected (data not shown). As a positive control, solubilization of the QS LuxR family protein BraR (Burkholderia kururiensis) as His6-BraR (pQEBRAR) (44) was carried out as described above. BraR was solubilized upon exogenous addition of cognate C12-3-oxo-HSL (data not shown). It was concluded that PsoR under the conditions we tested was not binding AHL molecules. PsoR protein solubility was then studied in the presence of macerated plant material (cucumber, wheat, or rice leaves) in the media. After affinity chromatography, pure His6-PsoR in native soluble form was detected by Western blot analysis when rice and wheat plant material was supplied to the media (Fig. 3 D and F). When LB medium was used alone or in the presence of macerated cucumber extract, no PsoR solubilization was observed (Fig. 3B). We also tested spent supernatants of other bacteria in a similar solubilization assay of PsoR since it would suggest response to signaling molecules produced by other plant-associated bacterial species. Spent supernatants were extracted with the same procedure as extracting AHLs from P. carotovorum, P. syringae pv. syringae, A. tumefaciens, B. subtilis, and B. cepacia cultures, and none of these extracts resulted in PsoR solubilization (data not shown). These results therefore indicated that most likely there is a plant produced molecule(s) in rice and wheat and not in cucumber that most likely bound to and solubilized PsoR. Alternatively, it cannot be excluded that the plant molecules inhibit proteases, which consequentially result in higher levels of PsoR soluble protein.
Fig. 3.
PsoR binds plant molecules. Soluble His6-PsoR can be detected only in the presence of macerated plant material (see Materials and Methods for details). Affinity chromatography (A, C, and E) and Western blot analysis of each elution peak (B, D and F) using samples prepared under native conditions for E. coli containing pQEPsoR and pREP4 grown in LB supplemented with macerated cucumber leaves (A and B), macerated rice leaves (C and D), and macerated wheat leaves (E and F) were performed. Western blot analysis was carried out with anti-His6 monoclonal antibody. FT, flowthrough. The 10, 20, and 50% refer to the percentages of buffer B used for elution, 50% buffer B corresponds to 135 mM imidazole.
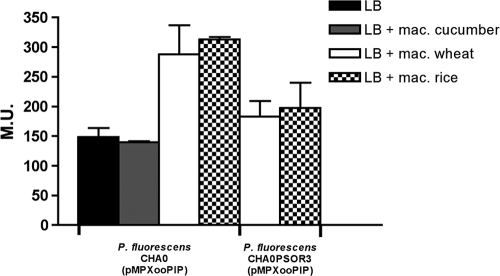
We previously established that the related protein OryR of X. oryzae pv. oryzae was solubilized by rice extract and that activated transcription of the neighboring proline iminopeptidase target gene pip, which contains a well-conserved lux box in its promoter region, in a rice-dependent manner (15, 16). Similarly, in P. fluorescens CHA0 and Pf-5, pip is located adjacent to the psoR gene; unlike the pip of X. oryzae pv. oryzae, however, it does not possess a lux box. The activity of the pip promoter was analyzed in strain CHA0, in the psoR mutant in the presence or absence of macerated plant material. This promoter was active; however, no significant difference in expression was observed under all of the conditions tested (see Fig. S2 in the supplemental material). In addition, we also verified pip gene promoter activity in a background of psoR overexpression. Again, no significant differences were observed (see Fig. S2 in the supplemental material). We also tested the activity of the X. oryzae pv. oryzae pip promoter, harbored as a lacZ transcriptional fusion in pMPXooPIP, in P. fluorescens CHA0 in LB medium and in LB medium supplemented with macerated leaves of cucumber, rice, or wheat. The pip promoter of X. oryzae pv. oryzae in P. fluorescens CHA0 was found to be activated in the presence of macerated rice and wheat but not upon the addition of macerated cucumber leaves (Fig. 4). We then performed the same experiments, but this time the pMPXooPIP was harbored in the psoR mutant P. fluorescens CHA0PSOR. In this case, the activation of the promoter by the macerated rice and wheat leaves was significantly attenuated. These findings are in accordance with the PsoR solubility data presented above, underscoring the existence of a molecule(s) in rice and wheat, and not in cucumber, which affects PsoR activity. In conclusion, PsoR regulates the X. oryzae pv. oryzae pip promoter which contains a lux box.
Fig. 4.
Gene promoter activity in P. fluorescens CHA0 and its psoR mutant CHA0PSOR harboring plasmid pMPXooPIP, which contains the X. oryzae pv. oryzae pip promoter fused to a promoterless lacZ gene. In the presence of macerated wheat and rice leaves, there was a significant increase in promoter activity which was less so when the gene promoter fusion was harbored in CHA0PSOR. Macerated cucumber leaves, on the other hand, did not alter gene promoter activity. All measures were performed in biological triplicates, and the mean value is given.
PsoR is required for biocontrol capacity of P. fluorescens.
In order to determine the role of PsoR in the biocontrol capacity of P. fluorescens CHA0, plant assays were set up in natural soil microcosms involving either cucumber or wheat as the host plant and P. ultimum as the pathogen causing root rot and damping-off disease. Using wheat as the host plant, as expected, addition of strain CHA0 to pathogen-infested soil, significantly enhanced plant health (Table 1). The psoR mutant CHA0PSOR, however, was significantly impaired in its capacity to protect wheat from the root disease, as was evident from the root fresh weights that were >30% lower than those protected by the wild-type strain CHA0. Overexpression of psoR in CHA0(pBBRPsoR) resulted in biocontrol activities similar to the level provided by wild-type CHA0. Root colonization levels were similar for all bacterial strains used in this experiment (Table 1). A similar experiment was set up using cucumber as the plant host and, as expected, the P. ultimum infection levels were higher, and the addition of strain CHA0 also significantly enhanced plant health (Table 2). In this case, however, the psoR mutant behaved just like the wild-type strain by providing the same levels of biocontrol activity. It was therefore concluded that PsoR played an important role in the biocontrol of P. ultimum in wheat but not in cucumber. This observation was consistent with solubilization of PsoR in wheat but not in cucumber extract containing medium.
Table 1.
Contribution of PsoR to the suppression of Pythium damping-off disease of wheat by Pseudomonas fluorescens CHA0 in natural soila
| Bacterial strain added | Pythium added | Fresh wt per flask (g) |
Colonization (mean log CFU of inoculants/g of roots ± SD) | |
|---|---|---|---|---|
| Shoot | Root | |||
| None | − | 0.60A | 0.33A | ND |
| CHA0(pBBRMCS-5) | − | 0.55A | 0.32AB | 6.91 ± 0.24 |
| CHA0PSOR(pBBRMCS-5) | − | 0.58A | 0.33AB | 7.42 ± 0.30 |
| CHA0(pBBRPsoR) | − | 0.60A | 0.37A | 6.94 ± 0.14 |
| None | + | 0.53A | 0.13D | ND |
| CHA0(pBBRMCS-5) | + | 0.58A | 0.28B | 7.42 ± 0.14 |
| CHA0PSOR(pBBRMCS-5) | + | 0.56A | 0.19C | 7.49 ± 0.36 |
| CHA0(pBBRPsoR) | + | 0.52A | 0.27B | 7.42 ± 0.17 |
The data represent means from 10 replicate flasks (each containing three wheat plants) per treatment. Means within the same column followed by different superscript capital letters are significantly different (P ≤ 0.05) according to Fisher's protected LSD test. The experiment was repeated once with similar results. ND, not detected. P. fluorescens strains were added at 107 CFU per g of natural soil contained within 200-ml flasks (60 g of soil per flask), after planting three 90-h-old, sterile-grown wheat seedlings per flask. P. ultimum was added (+) or not added (-) as a millet-seed inoculum to soil before planting. Plants were harvested after 14 days.
Table 2.
Contribution of PsoR to the suppression of Pythium damping-off disease of cucumber by Pseudomonas fluorescens CHA0 in natural soila
| Bacterial strain added | Pythium added | Surviving plants per flask (%) | Fresh wt per flask (g) |
|
|---|---|---|---|---|
| Shoot | Root | |||
| None | − | 100A | 1.64B | 0.41B |
| CHA0 | − | 100A | 2.06A | 0.47A |
| CHA0PSOR | − | 100A | 1.94A | 0.44AB |
| None | + | 38B | 0.70C | 0.17C |
| CHA0 | + | 92A | 1.80AB | 0.40B |
| CHA0PSOR | + | 100A | 1.90A | 0.42B |
The data represent the means from eight replicate flasks (each containing three cucumber plants) per treatment. Means within the same column followed by different superscript capital letters are significantly different (P ≤ 0.05) according to Fisher's protected LSD test. The experiment was repeated once with similar results. P. fluorescens strains were added at 107 CFU per g of natural soil contained within 200-ml flasks (60 g of soil per flask), after planting three 90-h-old, sterile-grown cucumber seedlings per flask. P. ultimum was added (+) or not added (-) as a millet-seed inoculum to soil before planting. Plants were harvested after 7 days.
PsoR regulates DAPG production in vitro and in vivo.
A primary mechanism for the biological control of fungal pathogens by P. fluorescens CHA0 is by direct antagonism via the production of diffusible antimicrobial compounds, in particular DAPG (20). In order to determine whether the production of DAPG was regulated by PsoR, the expression levels of a GFP-based transcriptional reporter fusion to biosynthetic genes for DAPG carried by pME9417 were monitored in M9-CAS in vitro and on cucumber and wheat roots in vivo. After 40 h of incubation, the expression of the phlA-gfp reporter in CHA0PsoR+ was almost five times higher than in the wild type, whereas the expression of the reporter was three times lower in CHA0PSOR (Fig. 5). The growth of the bacterial strains did not significantly differ. These findings confirm microarray data on positive regulation of DAPG biosynthetic gene expression by PsoR. The analysis of expression of the phlA-gfp reporter on roots by FACS-based flow cytometry revealed a drastically higher percentage of cells of CHA0PsoR+ expressing the antifungal reporter on roots of wheat, whereas the percentage of cells expressing phlA on roots of cucumber remained at wild-type levels (see Fig. S3 in the supplemental material). Virtually no cells of CHA0PSOR strain expressed the phlA-gfp reporter on roots of wheat. Together, these data further support the importance of PsoR in DAPG gene regulation and underscore the plant specificity of the effect.
Fig. 5.
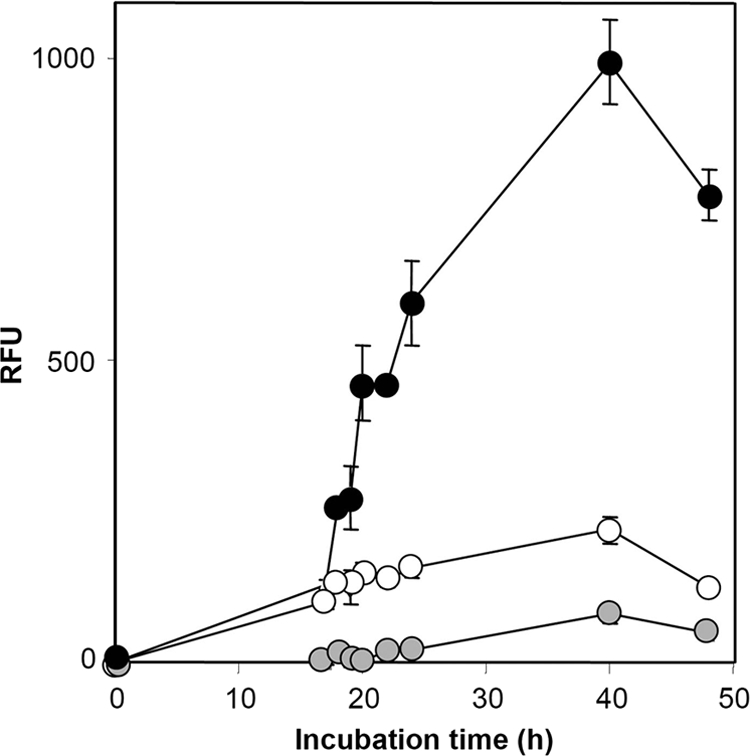
PsoR-mediated control of the expression of DAPG biosynthetic genes in P. fluorescens CHA0. Expression of a phlA-gfp reporter fusion (pME9417) in growing cultures of strain CHA0(pBBRMCS-5) (open circles), the psoR mutant CHA0PSOR(pBBRMCS-5) (gray-shaded circles), and the psoR-overexpressing strain CHA0(pBBRPsoR) (solid black circles). Strains were grown in M9-CAS minimal medium at 30°C. Means ± the standard deviations from three replicate cultures are shown. The experiment was repeated twice with highly similar results. RFU, relative fluorescence units.
DISCUSSION
Pseudomonads often live commensally with plants benefiting from the plant-exuded nutrients. In exchange, bacteria provide protection against deleterious microorganisms, improve the availability of vital nutrients, and produce plant growth regulators (6). Despite this association, limited progress has been made in elucidating the most likely interkingdom communication taking place. It has been reported that a small proportion of the isolated plant-associated pseudomonads possess an AHL QS system (8, 13, 42). In the present study, we have investigated a LuxR-family protein of plant-associated P. fluorescens, designated PsoR, which is closely related to QS LuxR proteins and yet does not bind AHLs but responds to plant compounds. PsoR was found to be present in both closely related model biocontrol strains P. fluorescens CHA0 and Pf-5, and the present study focused on CHA0.
PsoR contains seven of nine amino acids that are very well conserved in the AHL-binding domain of most QS LuxR-family regulators (29) (see Fig. S1 in the supplemental material), but it does not bind AHLs. It is likely that the lack of conservation in this residue and most probably of other ones within the AHL-binding domain allows PsoR to bind a different molecule(s) which could be related to AHLs. Importantly, PsoR is closely related to proteins present in several bacteria, which are all plant associated, including Xanthomonas, P. syringae, Agrobacterium, Rhizobium, and Sinorhizobium (Fig. 6). As evident from Fig. 6, we therefore propose that PsoR is part of a subfamily of LuxR-family regulators closely related to QS and most probably binds to a low-molecular-weight plant compound(s), as demonstrated for the closely related OryR protein and due to the high conservation with the AHL-binding domain, indicating that the molecule might be closely related to AHLs. In Xanthomonas spp. two members of this family, XccR of X. campestris pv. campestris and OryR of X. oryzae pv. oryzae, have been shown to bind plant molecules and regulate virulence of these plant pathogens (15, 16, 50). XccR and OryR have been previously shown to regulate promoters that possess very well conserved lux boxes and XccR promoter binding is dependent upon the presence of a plant low-molecular-weight molecule (16, 50). Here, we demonstrated that the lux-box-containing pip promoter of X. oryzae pv. oryzae is regulated by PsoR. Hence, members of this subfamily of proteins, even though they do not respond to AHLs, bind and transcriptionally regulate promoters that are conserved within AHL QS systems. This family of plant-associated LuxR solos have the pip gene genetically linked; interestingly, the pip promoter of P. fluorescens does not have a putative lux box. Similarly, the pip promoter of S. meliloti next to the psoR homolog nesR also does not have a lux box (31), which might indicate that, unlike plant pathogens, in plant beneficial bacteria pip is not regulated by this family of LuxR solos.
The CHA0PsoR+ strain was used to identify gene targets affected by having PsoR abundantly present in the cell, since the precise induction conditions for PsoR-mediated regulation are not known. The expression profiles of a large number of genes were affected, suggesting that PsoR is a global regulator. More ORFs were negatively regulated rather than positively regulated, and several functional categories were particularly abundant in the set of genes that were PsoR regulated. RNA levels of several transcriptional regulators were affected in the CHA0PsoR+ strain, indicating that many of the differentially expressed ORFs identified in the microarray experiment could be regulated indirectly. Some of the differentially expressed ORFs also contained conserved lux-box-like elements in their promoters, suggesting direct regulation by PsoR.
The ORFs positively regulated by PsoR comprise loci encoding a putative chitin-binding protein (PFL_2090) and a chitinase (PFL_2091), which are most probably organized in an operon. Expression of biosynthetic genes of major antifungal metabolites in P. fluorescens CHA0 was regulated by PsoR. Genes in the DAPG biosynthetic cluster showed substantial increases in transcript levels in CHA0PsoR+ strain both in vitro and in vivo. The regulation of the DAPG gene cluster (phlACBD) involves pathway-specific mechanisms, as well as global regulators that respond to environmental stimuli and cell-density-dependent mechanisms (4, 20, 33). Future studies need to take into consideration how PsoR-mediated regulation of DAPG production integrates with the specific and global regulators already known to affect phl transcription. Interestingly, it has recently been reported that several plant-derived compounds affect the expression of the DAPG phl operon (11); it cannot be excluded that some of these compounds act via PsoR. Many genes involved in iron acquisition are regulated by PsoR. Competition for iron has long been known to be an important trait for beneficial rhizosphere colonization and for antagonism of plant deleterious microorganisms (30). In summary, many phenotypes closely associated with rhizosphere colonization and biocontrol are regulated by PsoR.
A further evidence of the function of PsoR in interkingdom signaling is the fact that PsoR plays an important role in biocontrol of a pathogenic oomycete in a wheat experimental system. In contrast, no role for PsoR in biocontrol of the same pathogen was detected on a cucumber experimental setup. The differential role of PsoR in biological control on the two plant hosts corresponds to the observation that cucumber extracts, unlike wheat extracts, did not solubilize PsoR. This possibly indicates that cucumber plants, unlike wheat plants, do not possess the molecule(s) that activate PsoR and consequentially PsoR cannot regulate transcription of target genes involved in biocontrol. Other mechanisms might function in cucumber-bacterium interaction to modulate the expression of genes important for biocontrol, as previously studied and reported (21). Interestingly, it has been previously observed that P. fluorescens rhizosphere colonization varies according to the host plant, most probably indicating that plant-bacterium signaling is dependent on plant genotype (9). The solubilization of PsoR by plant compounds could be a strategy in the future to determine the nature of this compound(s) via copurification with PsoR. Unfortunately, in our initial experiments, very low amounts of soluble protein bound to plant compounds were recovered. We cannot exclude that the effect of the plant molecule(s) on PsoR is indirect by inhibiting proteases that result in higher soluble protein levels. However, the previous work on proteins of this subfamily, such as XccR and OryR, and the functional characteristics of this larger group of LuxR family proteins indicate that the most likely role of the plant extract is to activate PsoR by direct binding (15, 16, 50).
In summary, we demonstrated here that the closely related beneficial P. fluorescens strains CHA0 and Pf-5 possess PsoR, which affects the expression of a large number of genes and most likely responds to a plant compound. PsoR is important for the biocontrol properties of P. fluorescens CHA0, further indicating a role in interkingdom signaling. Importantly, PsoR belongs to a subfamily of LuxR proteins of plant-associated bacteria (Fig. 6). Future studies need to identify the plant signaling molecule(s) for PsoR and common elements in target gene promoters.
Supplementary Material
Acknowledgements
We acknowledge ICGEB funding to V.V.'s lab and ICGEB fellowships to S.S. and J.F.G. We acknowledge support from the Swiss National Science Foundation grant 3100A0-105881 to L.R., M.P.-T., and C.K. and from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service Competitive Grant 2006-35319-17427 to I.P. and J.E.L.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 April 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11: 36–42 [Google Scholar]
- 2. Ahmer B. M. 2004. Cell-to-cell signaling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52: 933–945 [DOI] [PubMed] [Google Scholar]
- 3. Baehler E., Bottiglieri M., Pechy-Tarr M., Maurhofer M., Keel C. 2005. Use of green fluorescent protein-based reporters to monitor balanced production of antifungal compounds in the biocontrol agent Pseudomonas fluorescens CHA0. J. Appl. Microbiol. 99: 24–38 [DOI] [PubMed] [Google Scholar]
- 4. Baehler E., et al. 2006. Two novel MvaT-like global regulators control exoproduct formation and biocontrol activity in root-associated Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 19: 313–329 [DOI] [PubMed] [Google Scholar]
- 5. Better M., Lewis B., Corbin D., Ditta G., Helinski D. R. 1983. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell 35: 479–485 [DOI] [PubMed] [Google Scholar]
- 6. Bloemberg G. V., Lugtenberg B. J. 2001. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4: 343–350 [DOI] [PubMed] [Google Scholar]
- 7. Brazma A., et al. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29: 365–371 [DOI] [PubMed] [Google Scholar]
- 8. DeAngelis K. M., Firestone M. K., Lindow S. E. 2007. Sensitive whole-cell biosensor suitable for detecting a variety of N-acyl homoserine lactones in intact rhizosphere microbial communities. Appl. Environ. Microbiol. 73: 3724–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De La Fuente L., Landa B. B., Weller D. M. 2006. Host crop affects rhizosphere colonization and competitiveness of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 96: 751–762 [DOI] [PubMed] [Google Scholar]
- 10. de Werra P., Baehler E., Huser A., Keel C., Maurhofer M. 2008. Detection of plant-modulated alterations in antifungal gene expression in Pseudomonas fluorescens CHA0 on roots by flow cytometry. Appl. Environ. Microbiol. 74: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Werra P., Huser A., Tabacchi R., Keel C., Maurhofer M. 2011. Plant- and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl. Environ. Microbiol. 77: 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubuis C., Keel C., Haas D. 2007. Dialogs of root-colonizing biocontrol pseudomonads. Eur. J. Plant Pathol. 119: 311–328 [Google Scholar]
- 13. Elasri M., et al. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67: 1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farrand S. K., Qin Y., Oger P. 2002. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 358: 452–484 [DOI] [PubMed] [Google Scholar]
- 15. Ferluga S., Bigirimana J., Hofte M., Venturi V. 2007. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 8: 529–538 [DOI] [PubMed] [Google Scholar]
- 16. Ferluga S., Venturi V. 2009. OryR is a LuxR-family protein involved in interkingdom signaling between pathogenic Xanthomonas oryzae pv. oryzae and rice. J. Bacteriol. 191: 890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuqua C. 2006. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188: 3169–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gross H., Loper J. E. 2009. Genomics of secondary metabolite production by Pseudomonas spp. Nat. Prod. Rep. 26: 1408–1446 [DOI] [PubMed] [Google Scholar]
- 19. Haas D., Defago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3: 307–319 [DOI] [PubMed] [Google Scholar]
- 20. Haas D., Keel C. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41: 117–153 [DOI] [PubMed] [Google Scholar]
- 21. Haas D., Keel C., Reimmann C. 2002. Signal transduction in plant-beneficial rhizobacteria with biocontrol properties. Antonie Van Leeuwenhoek 81: 385–395 [DOI] [PubMed] [Google Scholar]
- 22. Hassan K., et al. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12: 899–915 [DOI] [PubMed] [Google Scholar]
- 23. Keel C., Ucurum Z., Michaux P., Adrian M., Haas D. 2002. Deleterious impact of a virulent bacteriophage on survival and biocontrol activity of Pseudomonas fluorescens strain CHAO in natural soil. Mol. Plant-Microbe Interact. 15: 567–576 [DOI] [PubMed] [Google Scholar]
- 24. Kovach M. E., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176 [DOI] [PubMed] [Google Scholar]
- 25. Lequette Y., Lee J. H., Ledgham F., Lazdunski A., Greenberg E. P. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188: 3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Michael B., Smith J. N., Swift S., Heffron F., Ahmer B. M. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183: 5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller J. H. 1972. Experiments in molecular genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [Google Scholar]
- 28. Miller W. G., Leveau J. H., Lindow S. E. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13: 1243–1250 [DOI] [PubMed] [Google Scholar]
- 29. Nasser W., Reverchon S. 2007. New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem. 387: 381–390 [DOI] [PubMed] [Google Scholar]
- 30. O'Sullivan D. J., O'Gara F. 1992. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol. Rev. 56: 662–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patankar A. V., Gonzalez J. E. 2009. An orphan LuxR homolog of Sinorhizobium meliloti affects stress adaptation and competition for nodulation. Appl. Environ. Microbiol. 75: 946–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Patankar A. V., Gonzalez J. E. 2009. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33: 739–756 [DOI] [PubMed] [Google Scholar]
- 33. Pechy-Tarr M., et al. 2005. RpoN (sigma54) controls production of antifungal compounds and biocontrol activity in Pseudomonas fluorescens CHA0. Mol. Plant-Microbe Interact. 18: 260–272 [DOI] [PubMed] [Google Scholar]
- 34. Peterson S. N., et al. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51: 1051–1070 [DOI] [PubMed] [Google Scholar]
- 35. Pinto U. M., Winans S. C. 2009. Dimerization of the quorum-sensing transcription factor TraR enhances resistance to cytoplasmic proteolysis. Mol. Microbiol. 73: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Piper K. R., Beck von Bodman S., Farrand S. K. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362: 448–450 [DOI] [PubMed] [Google Scholar]
- 37. Price M. N., Huang K. H., Alm E. J., Arkin A. P. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33: 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruby E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50: 591–624 [DOI] [PubMed] [Google Scholar]
- 39. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor, NY [Google Scholar]
- 40. Schuster M., Urbanowski M. L., Greenberg E. P. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A. 101: 15833–15839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stachel S. E., An G., Flores C., Nester E. W. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steindler L., Bertani I., De Sordi L., Bigirimana J., Venturi V. 2008. The presence, type and role of N-acyl homoserine lactone quorum sensing in fluorescent Pseudomonas originally isolated from rice rhizospheres are unpredictable. FEMS Microbiol. Lett. 288: 102–111 [DOI] [PubMed] [Google Scholar]
- 43. Steindler L., Venturi V. 2007. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266: 1–9 [DOI] [PubMed] [Google Scholar]
- 44. Suarez-Moreno Z. S., Caballero-Mellado J., Venturi V. 2008. The new group of non-pathogenic plant-associated nitrogen-fixing Burkholderia spp. shares a conserved quorum sensing system, which is stringently regulated by the RsaL repressor. Microbiology 154: 2048–2059 [DOI] [PubMed] [Google Scholar]
- 45. Subramoni S., Venturi V. 2009. LuxR-family ‘solos’: bachelor sensors/regulators of signaling molecules. Microbiology 155: 1377–1385 [DOI] [PubMed] [Google Scholar]
- 46. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 47. Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Urbanowski M. L., Lostroh C. P., Greenberg E. P. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186: 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vannini A., et al. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21: 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L., Jia Y., Wang L., Fang R. 2007. A proline iminopeptidase gene upregulated in planta by a LuxR homologue is essential for pathogenicity of Xanthomonas campestris pv. campestris. Mol. Microbiol. 65: 121–136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.