Abstract
MgtC is important for the survival of several bacterial pathogens in macrophages and for growth under magnesium limitation. Among eukaryotes, a gene homologous to mgtC was found only in the pathogenic fungus Aspergillus fumigatus. Our data show that the A. fumigatus MgtC (AfuMgtC) protein does not have the same function as the bacterial MgtC proteins.
TEXT
Although Aspergillus fumigatus has become the most common and dangerous airborne fungal pathogen of humans, the molecular determinants responsible for its virulence remain largely unknown (19). A current hypothesis is that in A. fumigatus, specific genes would code for essential virulence factors for this fungal species. One of these genes, AFUA_7G05060, was of major interest since it was homologous to an essential bacterial virulence factor, MgtC (for magnesium transporter C).
MgtCp has been shown to be a virulence factor important for the proliferation of several intracellular bacterial pathogens in macrophages (1, 4, 27). The function of MgtCp is unknown, but it seems to facilitate or regulate Mg2+ transport since it is required for growth at low Mg2+ concentrations (11). MgtC-like proteins are found in a limited number of bacterial genomes, and phylogenetic analysis suggests that MgtC has been acquired by horizontal gene transfer (HGT) repeatedly throughout bacterial evolution.
Like bacterial pathogens, conidia of A. fumigatus are also phagocytosed by macrophages and they have to germinate intracellularly to establish disease. In addition, divalent cations such as Zn2+ and Fe3+ are required for A. fumigatus growth (30, 31). Magnesium is the most abundant divalent cation in cells and is involved in many cellular functions as a cofactor in numerous enzymatic reactions, as well as being necessary for the stability of plasma membranes. Until now, the growth of A. fumigatus under magnesium limitation has not been investigated and magnesium transporters have not been identified.
Genomic analysis of the mgtC homologs in A. fumigatus.
BLAST analysis (http://blast.ncbi.nlm.nih.gov/) identified a unique MgtC-like protein in A. fumigatus. A. fumigatus MGTC (AfuMGTC) is 840 bp long and encodes a 280-amino-acid protein with a theoretical molecular mass of 31 kDa. MGTCp has four predicted transmembrane helices and one putative N-glycosylation site. The amino acid sequence is 33% identical to the protein of Salmonella enterica serovar Typhimurium and 30% identical to the protein of Mycobacterium tuberculosis. Like its bacterial orthologs (5), the MgtC protein of A. fumigatus contains the conserved hydrophobic “MgtC domain” located in the N-terminal part of the protein (Fig. 1) whereas the C-terminal region of the A. fumigatus protein is not conserved.
Fig. 1.
Alignment of the MgtC domains of MgtC proteins. Proteins were aligned with ClustalX2, and the alignment was refined with the Genedoc program. The MgtC domain is indicated above the alignment.
Interestingly, this homolog of bacterial MgtC is unique in the fungal kingdom since no MGTC homolog was identified by a BLAST search in other eukaryotic species or in Aspergillus species (with the exception of Neosartorya fischeri, which is the taxon closest to A. fumigatus [29]).
AfuMGTC is located on chromosome 7 between a gene coding for a rhamnosidase and a gene coding for a flavin adenine dinucleotide-dependent oxidoreductase (see Fig. S1 in the supplemental material). The gene organization around MGTC in N. fisheri was very similar (Fig. S1). In contrast, in A. clavatus, which does not contain an MGTC homolog but is taxonomically close to A. fumigatus and N. fischeri, the genes that are homologs of the genes around AfuMGTC are located in different chromosomes, indicating an overall rearrangement of the genomes in this area.
Phylogenetic analysis.
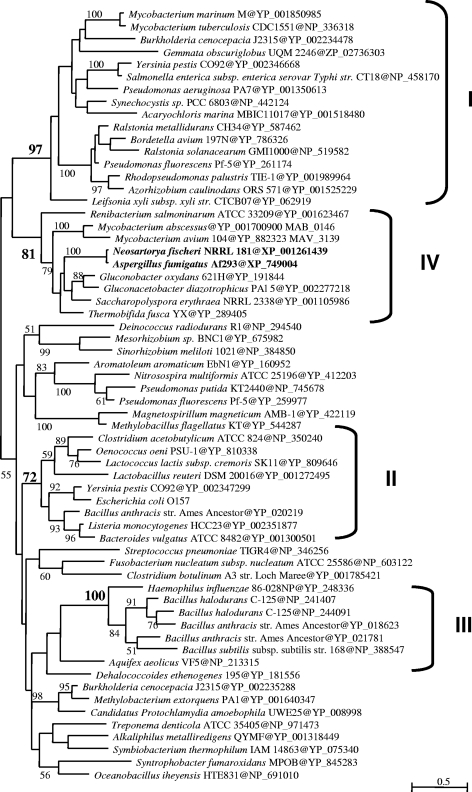
MgtCp homologues were searched for among 62 complete genomes (http://www.ncbi.nlm.nih.gov/). A final data set of 117 amino acid positions was used for maximum-likelihood analysis (Fig. 2). Four clusters were identified. Cluster I contained bacterial species where mgtC has been shown to have a role in pathogenicity. Bacterial mgtC genes have been proposed to be acquired by HGT (5), especially in the cases of S. enterica serovar Typhimurium and S. enterica serovar Typhi, where mgtC was inserted into pathogenicity islands acquired by HGT.
Fig. 2.
Maximum-likelihood tree of MgtC orthologs. Prokaryotic and eukaryotic MgtC sequences were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/). Values at nodes indicate statistical support calculated by nonparametric bootstrapping (only those greater than 50% are shown). The scale bar represents the average number of substitutions per site. The four clusters are indicated. Sequences were aligned by using MUSCLE 3.6 (7, 8). Sixty-two representative MGTC homologs were selected for phylogenetic analysis. Regions where homology was doubtful were manually removed from the alignments before phylogenetic analysis using the NET program of the MUST package (24), providing a final data set of 117 amino acid positions. Phylogenetic analysis was performed with PHYML (13), including a JTT model, a gamma correction to take into account the heterogeneity of evolutionary rates across sites (4 discrete classes of sites, an estimated alpha parameter, and an estimated proportion of invariable sites). The robustness of each branch was estimated by a nonparametric bootstrap procedure implemented in PHYML (100 replicates of the original data set and the same parameters).
In the phylogenetic tree, AfuMGTC belongs to a cluster that was not previously described (group IV), which comprises taxonomically diverse bacteria such as Mycobacterium abscessus and Thermobifida fusca. The mgtC genes of T. fusca and M. abscessus exhibit, respectively, 41% and 38% identity with that of A. fumigatus. T. fusca is a thermophilic bacterium that degrades plant cell walls in compost heaps. Interestingly, A. fumigatus is one of the thermophilic fungal species that are also present in compost due to its capacity to degrade decaying organic material and its thermotolerance (3). Phylogenetic group IV also contains M. abscessus which is a common water contaminant that is an opportunistic pathogen of cystic fibrosis patients (28) and Saccharopolyspora erythraea (which is close to Saccharopolyspora rectivirgula, one of the most important agents involved in farmer's lung disease [26]). Most of the species in clade IV have the same ecology (compost and hay).
Ripoll et al. (28) suggested that mgtC in M. abscessus could have been acquired by horizontal transfer. The specific clustering of AfuMGTC in a subgroup with several bacteria is consistent with HGT from a bacterium in cluster IV (2, 16). HGT has been poorly studied in fungi, and only a few studies have identified prokaryotic genes in fungi (10, 12, 15). Also, two studies of the entire genomes of Saccharomyces cerevisiae and Candida parapsilosis have been undertaken (9, 14). During the course of this study, a global analysis of HGT between prokaryotes and 60 fungal genomes was published and it reveals that the A. fumigatus genome contains 20 putative transferred genes, including AfuMGTC (22). Mallet et al. (21) have also suggested that 3.1% of the genome of A. fumigatus originated from bacteria, other fungi, and viruses.
Phenotypic analysis of an mgtc mutant.
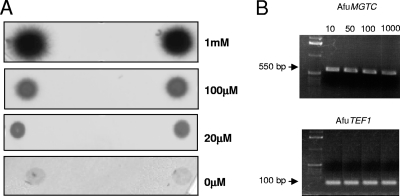
The strategy described in Fig. S2 in the supplemental material was used to produce an A. fumigatus strain with a nonfunctional copy of AfuMGTC. Psp1-MgtC was used to transform protoplasts of A. fumigatus CEA17Δku80 (6). To complement the mgtc mutant, the AfuMGTC gene was cloned into SK+ and used with plasmid pAN8.1 (23), containing a phleomycin resistance marker, to cotransform the mgtc mutant. The ectopic integration of an intact copy of the AfuMGTC gene was verified by PCR (Fig. S2C). No growth difference between the parental, the mgtC mutant and the complemented mgtc mutant strain was observed on usual media like minimal medium, RPMI 1640 (Gibco), and Sabouraud liquid or solid medium with 3% glucose and 1% yeast extract at 37°C or 50°C. The conidial and hyphal morphology of the mutant was identical to that of the parental strain, whatever the temperature. No growth or very limited growth of the parental and mutant strains was observed in the absence of magnesium or at a low concentration of magnesium (0 or 20 μM), showing that magnesium is essential for A. fumigatus growth (Fig. 3 A). In addition, the expression of AfuMGTC was not dependent on the magnesium concentration of the medium since similar reverse transcription-PCR profiles were obtained from cultures grown for 16 h at 37°C in medium supplemented with 10 μM, 50 μM, 100 μM, or 1 mM magnesium (Fig. 3B). AfuMGTC did not contribute to adaptation to a low-magnesium environment.
Fig. 3.
(A) Growth of the parental strain and the mgtc mutant for 24 h at 37°C on Czapek agar medium without magnesium or supplemented with 20 μM, 100 μM, or 1,000 μM magnesium. (B) Expression levels of the AfuMGTC gene and the AfuTEF1 control gene after 32 h of growth at 37°C in Czapek liquid medium supplemented with 10 μM, 50 μM, 100 μM, or 1 mM magnesium.
The role of AfuMGTC in A. fumigatus pathogenicity was investigated as described previously by Lambou et al. (18). No significant difference was seen in the survival rates of cohorts of 10 mice infected intranasally at 105 conidia/mouse with the parental strain, the mgtc mutant, and the complemented mgtc mutant strain. Survival was analyzed by the Kaplan-Meier test (chi square, 0.022) (see Fig. S3A in the supplemental material). Moreover, the conidial survival of the parental strain, the mgtc mutant, and the complemented mgtc mutant strain in the lungs of immunocompetent mice was similar, as estimated by a Student test (P < 0.0001) (Fig. S3B). These results indicated that AfuMGTC is not associated with fungal virulence.
Rang et al. (25) identified several amino acid residues involved in the loss-of-function phenotype in macrophages or in growth by complementation of the Salmonella mgtC mutant. E27, E84, N92, E193, and W226 were shown to be important for growth, whereas N92 and C99 were important for survival in macrophages. In A. fumigatus, M. abscessus, and M. avium, only E27 is conserved. These results suggested that point mutations in the sequences of members of the MgtC protein family belonging to subgroup IV are responsible for the loss of function of MgtC. Accordingly, it was not possible to complement the Salmonella mgtC gene with the AfuMGTC gene for growth in a magnesium-depleted medium, whereas other bacterial mgtC genes are able to complement Salmonella gene mutants (A. B. Blanc-Potard, data not shown; 25).
Because A. fumigatus growth is dependent on the presence of magnesium and AfuMGTC does not have any role in magnesium metabolism, other transporters that remain to be identified must play a role in magnesium metabolism in A. fumigatus. In eukaryotes, a family of proteins (2-TM-GxN proteins) carrying a GMN tripeptide motif between two transmembrane domains has been shown to be involved in magnesium transport (17). A member of this family is the Alr1p protein of S. cerevisiae (20). Several putative magnesium transporters are present in A. fumigatus, since we identified three genes homologous to the S. cerevisiae ALR1 gene in the A. fumigatus (strain AF293) genome, AFUA_5G05830 (identity, 53%), AFUA_2G08070 (identity, 46%), and AFUA_4G00930 (identity, 38%). Their functional analysis is currently under way.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 20 May 2011.
REFERENCES
- 1. Alix E., Blanc-Potard A. B. 2007. MgtC: a key player in intramacrophage survival. Trends Microbiol. 15:252–256 [DOI] [PubMed] [Google Scholar]
- 2. Anaissie E. J., et al. 2003. Pathogenic molds (including Aspergillus species) in hospital water distribution systems: a 3-year prospective study and clinical implications for patients with hematologic malignancies. Blood 101:2542–2546 [DOI] [PubMed] [Google Scholar]
- 3. Beffa T., et al. 1998. Mycological control and surveillance of biological waste and compost. Med. Mycol. 36:137–145 [PubMed] [Google Scholar]
- 4. Blanc-Potard A. B., Groisman E. A. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO. J. 16:5376–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanc-Potard A. B., Lafay B. 2003. MgtC as a horizontally-acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J. Mol. Evol. 57:479–486 [DOI] [PubMed] [Google Scholar]
- 6. da Silva Ferreira M. E., et al. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fitzpatrick D. A., Logue M. E., Butler G. 2008. Evidence of recent interkingdom horizontal gene transfer between bacteria and Candida parapsilosis. BMC Evol. Biol. 8:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Vallvé S., Romeu A., Palau J. 2000. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol. Biol. Evol. 17:352–361 [DOI] [PubMed] [Google Scholar]
- 11. Garcia Véscovi E., Soncini F. C., Groisman E. A. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174 [DOI] [PubMed] [Google Scholar]
- 12. Gojković Z., et al. 2004. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Genet. Genomics 271:387–393 [DOI] [PubMed] [Google Scholar]
- 13. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 14. Hall C., Brachat S., Dietrich F. S. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryotic. Cell 4:1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall C., Dietrich F. S. 2007. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177:2293–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayette M. P., et al. 2010. Filamentous fungi recovered from the water distribution system of a Belgian university hospital. Med. Mycol. 48:969–974 [DOI] [PubMed] [Google Scholar]
- 17. Knoop V., Groth-Malonek M., Gebert M., Eifler K., Weyand K. 2005. Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol. Genet. Genomics 274:205–216 [DOI] [PubMed] [Google Scholar]
- 18. Lambou K., Lamarre C., Beau R., Dufour N., Latgé J. P. 2010. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol. Microbiol. 75:910–923 [DOI] [PubMed] [Google Scholar]
- 19. Latgé J. P., Steinbach W. (ed.) 2009. Aspergillus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 20. MacDiarmid C. W., Gardner R. C. 1998. Overexpression of the Saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J. Biol. Chem. 273:1727–1732 [DOI] [PubMed] [Google Scholar]
- 21. Mallet L. V., Becq J., Deschavanne P. 2010. Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus. BMC Genomics 11:171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcet-Houben M., Gabaldón T. 2010. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26:5–8 [DOI] [PubMed] [Google Scholar]
- 23. Mattern I. E., Punt P. J., Van den Hondel C. A. M. J. J. 1988. A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newsl. 35:25 [Google Scholar]
- 24. Philippe H. 1993. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 21:5264–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rang C., et al. 2007. Dual role of the MgtC virulence factor in host and non-host environments. Mol. Microbiol. 63:605–622 [DOI] [PubMed] [Google Scholar]
- 26. Reboux G., et al. 2007. Assessment of four serological techniques in the immunological diagnosis of farmers' lung disease. J. Med. Microbiol. 56:1317–1321 [DOI] [PubMed] [Google Scholar]
- 27. Retamal P., Castillo-Ruiz M., Mora G. C. 2009. Characterization of MgtC, a virulence factor of Salmonella enterica serovar Typhi. PLoS One 4:e5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ripoll F., et al. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS. One 4:e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samson R. A., Varga J., Dyer P. S. 2009. Morphology and reproductive mode of Aspergillus fumigatus, p. 7–13 In Latgé J.-P., Steinbach W. J. (ed.), Aspergillus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 30. Schrettl M., et al. 2004. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200:1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vicentefranqueira R., Moreno M. A., Leal F., Calera J. A. 2005. The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot. Cell 4:837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.