Abstract
A high-throughput sequencing approach was utilized to carry out a comparative transcriptome analysis of Trichoderma atroviride IMI206040 during mycoparasitic interactions with the plant-pathogenic fungus Rhizoctonia solani. In this study, transcript fragments of 7,797 Trichoderma genes were sequenced, 175 of which were host responsive. According to the functional annotation of these genes by KOG (eukaryotic orthologous groups), the most abundant group during direct contact was “metabolism.” Quantitative reverse transcription (RT)-PCR confirmed the differential transcription of 13 genes (including swo1, encoding an expansin-like protein; axe1, coding for an acetyl xylan esterase; and homologs of genes encoding the aspartyl protease papA and a trypsin-like protease, pra1) in the presence of R. solani. An additional relative gene expression analysis of these genes, conducted at different stages of mycoparasitism against Botrytis cinerea and Phytophthora capsici, revealed a synergistic transcription of various genes involved in cell wall degradation. The similarities in expression patterns and the occurrence of regulatory binding sites in the corresponding promoter regions suggest a possible analog regulation of these genes during the mycoparasitism of T. atroviride. Furthermore, a chitin- and distance-dependent induction of pra1 was demonstrated.
INTRODUCTION
Due to hazardous chemical fungicides affecting human health (4) and the environment (9), the application of biological control agents like Trichoderma spp. is a promising alternative for plant protection (17). Members of the genus Trichoderma (teleomorph Hypocrea) are potent mycoparasitic fungi that not only compete for nutrients but also secrete cell wall-degrading enzymes (CWDEs) such as chitinases, glucanases, and proteases, among others, and excrete secondary metabolites that are active against a number of plant-pathogenic fungi (15, 28, 29, 41, 58). In addition to an increase in enzyme secretion during interactions with host fungi, it was shown previously that differentiation processes occur, leading to morphological changes and the formation of penetration structures in mycoparasitic Trichoderma spp. (7, 22). Interestingly, mycoparasitism is not a mere result of physical contact but may be proceeded by an early recognition process that leads to the induction of gene expression of hydrolytic enzymes (e.g., prb1 [18] and ech42 [67]).
In recent years, a number of studies have identified enzymes and effectors involved in host recognition and the mycoparasitic responses of Trichoderma (40, 46, 51); however, their modes of action and the mechanisms determining host specificity remain poorly understood. Mainly expressed-sequence-tag (EST) libraries of different Trichoderma strains obtained under various growth conditions have contributed significantly to the large-scale identification of active genes (63, 64). Additionally, diverse DNA array experiments have determined that an expansin-like protein, aspartyl proteases, and hydrophobins, among others, are involved in the biocontrol and mycoparasitism of Trichoderma spp. (8, 51).
Comparative transcriptome analyses have shown the advantages of high-throughput 454 sequencing (6, 11) over standard array technologies (59). The availability of complete genomes of various Trichoderma species (including T. atroviride) in combination with high-throughput methods has enabled the investigation of global cellular mechanisms under different conditions.
Based on the hypothesis that T. atroviride specifically recognizes host fungi of different phyla and therefore activates a host-specific response, we intended to identify genes specifically involved in mycoparasitism, both host and/or distance dependent. In the present study, we used 454 pyrosequencing to obtain transcriptional profiles of T. atroviride IMI206040 prior to physical contact as well as during direct interactions with Rhizoctonia solani. These profiles were compared to those at same stages of confrontation of T. atroviride with itself. We produced and analyzed a total of 277,769 reads, representing over 65% of the predicted genes (7,797 out of 11,863) for this organism. One hundred seventy-five genes were found to be differentially expressed during mycoparasitic interactions. Subsequently, relative transcript levels of differentially expressed genes were investigated during mycoparasitism with the plant-pathogenic oomycete Phytophthora capsici and the host fungi Botrytis cinerea and R. solani, respectively. We show that during the physical contact of T. atroviride with R. solani, differentially expressed genes functionally shift toward an involvement in metabolic processes. Furthermore, enzymes involved in cell wall degradation exhibit similar changes in gene expression patterns during mycoparasitic interactions with the distinct plant-pathogenic fungi. Additionally, we identified a novel promoter binding sequence in these genes, which may have a regulatory function.
This study represents the first comprehensive transcriptome-wide analysis of Trichoderma spp. during mycoparasitic interactions with plant-pathogenic host fungi. Furthermore, this study investigates the physiological changes at the transcript level in Trichoderma due to the physical proximity of other Trichoderma colonies.
MATERIALS AND METHODS
Strains and cultivation.
Confrontation assays using T. atroviride strain IMI206040 (teleomorph Hypocrea atroviridis), R. solani AG-4, B. cinerea, and P. capsici Aym2 were performed with petri dishes and minimal medium (12) supplemented with 0.2% (wt/vol) glucose. Agar plugs of T. atroviride and the corresponding confrontation partner were placed onto the agar plates, overlaid with cellophane, and inoculated at a distance of 9 cm in total darkness at either 28°C (for confrontation with R. solani and P. capsici) or room temperature (for confrontation with B. cinerea). When the growing front of the colonies reached a distance of 1 cm before direct contact (bc) or 1 cm of contact (c), a 1-cm piece of T. atroviride mycelium or the interaction zone was collected under red light and immediately frozen in liquid nitrogen.
cDNA library construction and sequencing.
For RNA preparation, mycelia were homogenized in liquid nitrogen, and total RNA was extracted with Trizol (Invitrogen) and cleaned by using the RNeasy kit (Qiagen). cDNA synthesis and GS20-454 sequencing were performed according to a protocol described previously by Vega-Arreguin et al. (60). In order to sequence all cDNA libraries in the same sequencing run, Multiple Identifiers (Roche) were fused to the different libraries according to the manufacturer's instructions.
Sequencing data analysis.
454 sequencing reads were aligned to the Frozen Gene Catalogue of T. atroviride IMI206040 (http://genome.jgi-psf.org/Triat2/Triat2.home.html) using stand-alone BLAST software (1) from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/); the cutoff was set to an E value of 10e−5. Positive hits were subjected to further analysis to make sure that only sequences of T. atroviride and not those of the confrontation partner R. solani were being used. The following sequence selection criteria were used: 1 mismatch or 1 gap was allowed per 100 bp of alignment (point A), the percentage of alignment should be at least 50% for each read when BLAST searched against the filtered gene models of T. atroviride (point B), and for sequences where 1 mismatch or gap was allowed due to the length of the alignment (point B), the percentage of this alignment should be >70% (point C); the same rule was applied for short-read sequences (≤40 bp). Considering the use of the same cutoff criteria for all libraries, artifacts in the appearance of differentially expressed genes due to host contact were avoided. Subsequently, the transcriptome of confrontation during a direct mycoparasitic interaction (TR c) was aligned to the metagenomic draft version of R. solani (http://www.rsolani.org) using BLAST in order to test the chosen cutoff criteria. This analysis resulted in BLAST hits showing greater similarities to the genes of T. atroviride than to those of R. solani. Therefore, the sequencing read counts reported in Table 1 are derived from only those reads aligning to T. atroviride with high confidence.
Table 1.
Summary of GS20-454 sequencing and BLASTN resultsa
| Comparison | Total no. of reads | No. of positive BLASTN hitsb | No. of different gene modelsc |
|---|---|---|---|
| TT bc | 53,202 | 43,268 | 4,999 |
| TT c | 104,678 | 85,030 | 6,260 |
| TR bc | 46,009 | 36,899 | 4,554 |
| TR c | 73,880 | 51,462 | 5,359 |
Number of sequences obtained after pyrosequencing and BLASTN analysis of the transcriptomes 1 cm before direct contact of T. atroviride IMI206040 with R. solani (TR bc), during 1 cm of direct contact (TR c), and under the corresponding control conditions of T. atroviride against itself 1 cm before contact (TT bc) and during direct contact (TT c).
Number of reads with hits (E value of <10e−5) against T. atroviride gene models.
Number of distinct T. atroviride gene models represented by transcriptomes during mycoparasitic interactions.
For simplification, identical transcript and protein identifications (IDs) of T. atroviride gene and protein models are displayed as “ID plus number.”
RNA manipulation.
The isolation of total RNA was performed by using the guanidinium thiocyanate method according to a method described previously by Chomczynski and Sacchi (16) and cleaned by using the RNeasy kit (Qiagen).
Quantitative RT-PCR and data analysis.
cDNA synthesis was performed by using the RevertAidTm H Minus first-strand cDNA synthesis kit (Fermentas, Burlington, Canada) according to the manual. An amount of 0.45 μg of each RNA sample was used for reverse transcription (RT), and cDNA samples were diluted 1:100 before quantitative PCR (qPCR). Primer pairs for RT-qPCR were designed by using Primer3 software (version 0.4.0; http://frodo.wi.mit.edu/primer3/) (45) and BLAST searched against fungal genome databases of the confrontation partners R. solani (http://www.rsolani.org/), P. capsici (http://genome.jgi-psf.org/PhycaF7/PhycaF7.home.html), and B. cinerea (http://www.broadinstitute.org/annotation/genome/botrytis_cinerea/Home.html) and additionally against Trichoderma reesei (http://genome.jgi-psf.org/Trire2/Trire2.home.html), Trichoderma virens (http://genome.jgi-psf.org/Trive1/Trive1.home.html), and Neurospora crassa (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html) (cutoff E value of 10e−1). Only primer pairs without predicted amplification in the confrontation partners, and therefore specific for T. atroviride, were chosen for RT-qPCR. Sequences and annealing temperatures of all primers are given in File S3 in the supplemental material. Fungal cultivation conditions were identical to those described above for transcriptome sequencing; however, plant-pathogenic fungi were also grown alone, and mycelia were processed from these pathogenic fungi before and during contact with themselves. Control RT-qPCR of pathogenic fungi grown alone showed no amplification of the genes selected (data not shown).
All RT-qPCRs were performed in triplicates. All reactions were conducted with an Eppendorf Mastercycler Ep Realplex 2.2 system (Eppendorf, Germany) at a final reaction mixture volume of 15 μl containing 7.5 μl of 2× iQ SYBR green mix (Bio-Rad Laboratories), 100 nM forward and reverse primers, and 2 μl cDNA. Amplification was accomplished by using a 3-min initial denaturation step, followed by 50 cycles at 95°C for 15 s, 60°C or 63°C (see File S3 in the supplemental material) for 20 s, and 68°C for 20 s. RT-qPCR products were verified by subsequent melting-curve analyses. PCR efficiencies were calculated by analyzing the linear regression of efficiency (LRE) (47, 48) and were in the range of 100% ± 8% for each reaction. Relative transcript levels were obtained according to methods described previously by Steiger et al. (55), using the relative expression software tool REST2008 (43) with act1 and sar1 as reference genes. These two housekeeping genes were chosen due to unchanged transcription during plate confrontation assays, as described previously by Brunner et al. (10).
Promoter analysis.
A total of 1 kb of the nontranscribed upstream regions of genes of interest was downloaded from the T. atroviride genome project homepage and analyzed with the motif search tool MEME (5; http://meme.nbcr.net). Additionally, promoter sequences were searched for binding sites of regulatory proteins involved in mycoparasitism (MYC1 to MYC4 [18]) and nitrogen and carbon catabolite regulation.
Nucleotide sequence accession number.
GS20-454 sequences were deposited in GenBank under accession number SRA029157.1.
RESULTS
Comparison of GS20-454 sequences to fungal databases.
A total of 277,769 high-quality reads with an average length of 84.44 bp were generated by the de novo sequencing of T. atroviride cultured with itself or R. solani. The reads were aligned to transcripts from the T. atroviride Frozen Gene Catalogue using BLAST, and over 80% of the sequences matched to the predicted transcripts using the criteria outlined in Materials and Methods. For the zone of interaction of T. atroviride with R. solani, only 70% of the sequences obtained (51,462 out of 73,880) could be assigned to Trichoderma (Table 1).
A comparison of the number of gene models expressed during this experiment to the total number of genes postulated for T. atroviride (11,863 genes) revealed that approximately 45% of these genes were transcribed during contact with R. solani (5,359 gene models, which were represented by 51,462 reads), whereas 38.4% (corresponding to 36,899 sequences) of these genes were transcribed prior to contact (Table 1). A diagrammatic representation of the bioinformatics analysis and the corresponding comparisons made is provided in the form of a workflow in File S1 in the supplemental material.
Statistical analysis of the transcriptome during mycoparasitism.
To avoid false positives (differentially expressed genes resulting from different read numbers under each condition), Fisher's exact test was applied to technical repeats of the same RNA samples. Differences between any two conditions were considered significant if a fold change of at least 3 was found for the number of reads for any given gene under one of the conditions compared to the other condition and passed the Fisher's exact test (P value of <10e−5). With respect to the number of differentially expressed genes, T. atroviride showed a weaker response prior to contact with R. solani than during physical interactions compared to the confrontation with itself (comparisons 1, TR/TT bc, and 2, TR/TT c) (Fig. 1 A). When comparisons 4 (TT c/bc) and 3 (TR c/bc) are compared (Fig. 1A), a higher occurrence of altered genes was observed when T. atroviride was challenged with itself (TT) than with R. solani (TR). Considering the use of the same cutoff criteria in all libraries, artifacts in the appearance of differentially expressed genes were avoided.
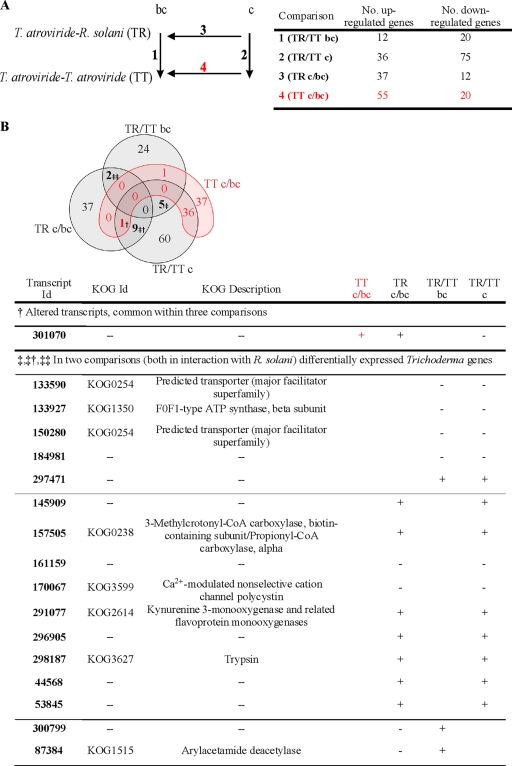
Fig. 1.
(A) Comparison of genome-wide transcription during confrontations against the host fungus R. solani with the control (T. atroviride against itself). 1 represents the number of differential genes (up- and downregulated) of 1 cm before direct contact (bc) with R. solani compared to the control (TR/TT bc), 2 gives the number of genes regulated during direct contact (c) with R. solani in relation with the corresponding control (TR/TT c), 3 compares gene expression levels of c and bc of T. atroviride with R. solani (TR c/bc), and 4 shows the relationship between genes expressed during c and 1 cm bc in the control (TT c/bc). (B) Four-set Venn diagram with the distribution of differentially expressed genes over the distinct comparisons. Whereas genes in common within TR/TT bc, TR/TT c, or TR c/bc are considered mycoparasitism relevant, genes common for TT c/bc are regarded as being less important during antagonism. The table displays the genes differentially regulated that are shared in three comparisons (†), prior to contact and during contact with R. solani (‡), during direct interactions with R. solani (‡†), and before visible contact with R. solani (‡‡). All these genes are given with their annotations according to the information available on the homepage of the T. atroviride genome project and their transcriptional regulation during different stages of interaction, with + being upregulated and − being downregulated. CoA, coenzyme A.
Interestingly, only a single gene with unknown function (protein/transcript ID 301070) was differentially regulated in three comparisons (Fig. 1B). The lack of common genes in more than two relationships indicates that the response of Trichoderma during mycoparasitism depends primarily on the confrontation partner and might therefore be a result of recognition.
In order to determine similarities in gene expression during different stages of mycoparasitism versus control conditions, differentially expressed genes were annotated, and the results of mutual genes are displayed in Fig. 1B. Interestingly, two of the five genes common to TR/TT bc and TR/TT c were identified as predicted transporters (KOG0254) and were both downregulated in the presence of the host fungus, independent of distance. Currently, only the general function prediction for these two genes is available. We compared gene expression during direct interactions with R. solani (TR c) to gene expression prior to contact (TR bc) or during interactions with the control (TT c) and identified 9 differentially expressed genes. Six of the genes could not be assigned to any eukaryotic orthologous group (KOG), whereas three of the genes were annotated according to gene ontology (GO). IDs 145909 and 296905, showing elevated transcript levels during physical contact, were found to be involved in proteolysis (GO:0006508); ID 145909 is thought to have tripeptidyl-peptidase I activity (GO:0019131), while ID 296905 has proposed aspartic-type endopeptidase activity (GO:0004190). An additional protein (ID 161159), identified as β-xylosidase I (BXL1), is involved in the degradation of xylan to xylose, and in the present study, it was found to be downregulated at the transcriptional level during direct host contact (Fig. 1B).
Functional annotation.
Significant differentially expressed genes were classified according to KOG by using the annotation available for T. atroviride genes from the genome sequencing effort (Table 2 and see File S2 in the supplemental material). It is noteworthy that out of 212 distinct differentially expressed genes, 122 could be assigned to a KOG, which is in accordance with the approximately 50% KOG annotation rate of the Frozen Gene Catalogue.
Table 2.
Numbers of KOG-classified differentially expressed genesa
| KOG category | No. of genes |
|||
|---|---|---|---|---|
| TR/TT bcb | TR/TT cc | TR c/bcd | TT c/bcd | |
| Cellular processes and signaling up | 3 | 2 | 9 | 8 |
| Cellular processes and signaling down | 5 | 11 | 2 | 5 |
| Information storage and processing up | 2 | 1 | 1 | 1 |
| Information storage and processing down | 2 | 2 | 2 | 4 |
| Metabolism up | 1 | 10 | 12 | 20 |
| Metabolism down | 7 | 36 | 4 | 3 |
| Poorly characterized up | 2 | 4 | 3 | 9 |
| Poorly characterized down | 3 | 10 | 1 | 3 |
Functional classification of up- and downregulated T. atroviride genes and assignment to four different KOGs.
T. atroviride before contact with R. solani compared to the control (T. atroviride against itself).
One centimeter of contact of T. atroviride with R. solani in relation to the control.
Confrontation of Trichoderma with R. solani (TR) or with itself (TT) and comparison of the contact stage to the stage before contact.
Interestingly, the distribution of annotated gene products indicated a functionality shift in differential transcripts during mycoparasitism. We found that genes involved in “metabolism” were the most abundant genes during direct contact with R. solani (TR/TT c; 60.5% of the annotated genes). Concurrently, genes involved in metabolism were less represented before contact (TR/TT bc; only 32%) (Table 2).
Genes differentially expressed during the direct interaction of Trichoderma with the host compared to the corresponding control condition (TR/TT c) fell mainly into the following KOG classes: “posttranslational modification”; “signal transduction mechanisms”; “energy production and conversion”; “transport and metabolism of amino acids, carbohydrates, and inorganic ion”; and “secondary metabolite biosynthesis, transport, and catabolism” (see File S2 in the supplemental material). Interestingly, all differentially regulated genes identified in the control (TT c/bc) and assigned to the category “secondary metabolite biosynthesis, transport, and catabolism” were found to be upregulated, whereas all of these genes were downregulated in TR/TT c (see File S2).
Response of T. atroviride to host fungi of different phyla monitored by quantitative RT-PCR.
According to the results obtained by statistical analysis of the sequencing data, the response of T. atroviride to the presence of host fungi is completely different from the response of T. atroviride contacting itself. As Trichoderma has a very broad spectrum of host fungi, we searched for both differences and similarities in the expressions of various genes found to be regulated during confrontation with R. solani compared to confrontation with B. cinerea or P. capsici. We took into account the fact that the plant-pathogenic fungi chosen for this experiment belonged to different phyla. Out of 175 differentially expressed genes from confrontations with R. solani, 13 genes and 2 housekeeping genes (act1 and sar1) were selected for further analysis during mycoparasitic interactions with various host fungi. This selection took into account the following: (i) genes coding for proteins associated with mycoparasitism (e.g., transcript/protein ID 302419, corresponding to PRB1) and (ii) different gene functions and KOG affiliations (i.e., signal transduction, production, and transport of secondary metabolites, amino acids, and inorganic ions and those genes that are either poorly characterized or uncharacterized) (Table 3). The relative transcript level of prb1 (encoding an alkaline protease; ID 302419) was 9.9 times higher in T. atroviride prior to contact with R. solani than after physical contact (TR bc/c). This result confirms the induction of gene expression before the visible contact of the colonies, as previously described (23). Differential expression during mycoparasitic interactions of the remaining 12 genes identified by pyrosequencing was also verified by RT-qPCR (Table 4).
Table 3.
Gene selection for RT-qPCR
| Transcript ID/ protein ID | KOG ID | KOG description | KOG function(s) | KOG category |
|---|---|---|---|---|
| 302419 | KOG1153 | Subtilisin-related protease/vacuolar protease B | Posttranslational modification, protein turnover, chaperones | Cellular processes and signaling |
| 297887 | KOG1339 | Aspartyl protease | Posttranslational modification, protein turnover, chaperones | Cellular processes and signaling |
| 80187 | KOG4157 | Beta-1,6-N-acetylglucosaminyltransferase; contains the WSC domain | Posttranslational modification, protein turnover, chaperones | Cellular processes and signaling |
| 286914 | KOG0251 | Clathrin assembly protein AP180 and related proteins; contain the ENTH domain | Signal transduction mechanisms | Cellular processes and signaling |
| 149807 | KOG0519 | Sensory transduction histidine kinase | Signal transduction mechanisms | Cellular processes and signaling |
| 223683 | KOG1517 | Guanine nucleotide binding protein MIP1 | Cell cycle control, cell division, chromosome partitioning | Metabolism |
| 211243 | KOG0054 | Multidrug resistance-associated protein/mitoxantrone resistance protein, ABC superfamily | Secondary metabolite biosynthesis, transport, and catabolism | Metabolism |
| 298187 | KOG3627 | Trypsin | Amino acid transport and metabolism | Metabolism |
| 318101 | KOG0039 | Ferric reductase, NADH/NADPH oxidase, and related proteins | Inorganic ion transport and metabolism | Metabolism |
| 132506 | KOG1227 | Putative methyltransferase | General function; prediction only | Poorly characterized |
| 302587 | ||||
| 84753 | ||||
| 22871 |
Table 4.
Relative transcription ratios under mycoparasitic conditions
| ID | Ratio (95% confidence interval)d |
Ratio (95% confidence interval)d |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Before contact (bc) |
Contact (c) |
Contact (c) |
Contact/contact before (c/bc) |
|||||||
| TPa/TT | TBb/TT | TRc/TT | TP/TT | TB/TT | TR/TT | TP | TB | TR | TT | |
| 297887 | 0.09 (0.04, 0.18) | 0.14 (0.02, 1.27) | 0.11 (0.04, 0.23) | 0.03 (0.01, 0.06) | 0.06 (0.03, 0.18) | 0.05 (0.03, 0.11) | 1.63 (0.88, 2.75) | 2.67 (0.33, 24.05) | 3.10 (2.08, 4.18) | 5.67 (1.84, 15.46) |
| 80187 | 107.72 (90.0, 128.8) | 84.18 (72.8, 98.0) | 98.16 (81.0, 120.9) | 2.19 (1.79, 2.67) | 0.78 (0.60, 1.04) | 1.36 (0.97, 1.78) | 1.22 (1.08, 1.38) | 0.46 (0.39, 0.56) | 0.68 (0.51, 0.84) | 46.55 (36.3, 59.9) |
| 286914 | 1.04 (0.86, 1.28) | 2.13 (1.56, 3.01) | 2.18 (1.77, 2.80) | 2.19 (1.17, 4.09) | 9.53 (5.21, 16.55) | 5.85 (3.23, 10.06) | 0.87 (0.76, 1.06) | 1.54 (1.19, 1.93) | 1.09 (0.93, 1.21) | 0.38 (0.20, 0.74) |
| 149807 | 0.23 (0.13, 0.39) | 0.43 (0.33, 0.58) | 0.26 (0.07, 0.63) | 0.82 (0.55, 1.23) | 0.63 (0.36, 1.13) | 0.94 (0.62, 1.37) | 1.97 (1.20, 4,07) | 0.75 (0.41, 1.46) | 1.98 (0.70, 8.25) | 0.54 (0.45, 0.71) |
| 223683 | 1.28 (0.77, 2.01) | 0.54 (0.28, 0.93) | 0.46 (0.26, 0.89) | 0.58 (0.25, 1.34) | 0.62 (0.23, 1.53) | 0.31 (0.08, 0.99) | 0.16 (0.12, 0.24) | 0.39 (0.19, 0.75) | 0.23 (0.08, 0.48) | 0.34 (0.14, 0.91) |
| 211243 | 0.36 (0.21, 0.71) | 0.44 (0.15, 1.27) | 0.52 (0.32, 0.94) | 0.89 (0.40, 2.04) | 1.96 (0.98, 3.82) | 0.88 (0.33, 2.49) | 0.94 (0.61, 1.65) | 1.64 (0.74, 3.86) | 0.63 (0.34, 1.26) | 0.37 (0.2, 1.0) |
| 298187 | 16.21 (10.96, 22.4) | 18.82 (12.0, 28.57) | 8.92 (5.74, 13.60) | 0.77 (0.45, 1.29) | 215.19 (132, 344.7) | 14.44 (6.16, 31.80) | 0.09 (0.07, 0.12) | 22.78 (17.1, 29.67) | 3.17 (1.58, 5.77) | 1.98 (1.07, 3.65) |
| 318101 | 0.02 (0.01, 0.05) | 0.74 (0.43, 1.37) | 2.96 (1.67, 5.54) | 0.03 (0.02, 0.05) | 0.23 (0.16, 0.34) | 0.92 (0.63, 1.39) | 10.20 (3.48, 29.93) | 1.95 (1.40, 2.74) | 1.77 (1.23, 2.61) | 5.81 (3.23, 11.03) |
| 132506 | 0.16 (0.05, 0.37) | 0.33 (0.15, 0.56) | 0.92 (0.37, 2.21) | 0.32 (0.21, 0.55) | 0.50 (0.34, 0.79) | 2.12 (1.49, 3.30) | 0.98 (0.48, 2.44) | 0.71 (0.50, 0.99) | 1.07 (0.57, 1.62) | 0.48 (0.21, 0.83) |
| 302587 | 13.95 (12.3, 15.82) | 3.86 (2.22, 5.14) | 2.39 (1.95, 2.99) | 2.98 (0.36, 3.84) | 0.24 (0.18, 0.31) | 0.42 (0.28, 0.61) | 0.17 (0.15, 0.19) | 0.05 (0.04, 0.09) | 0.15 (0.10, 0.21) | 0.78 (0.60, 0.96) |
| 84753 | 137.08 (98.9, 172.2) | 96.32 (66.0, 126.3) | 59.04 (43.2, 73.8) | 8.06 (6.06, 9.93) | 1.33 (1.04, 1.69) | 1.56 (0.96, 2.59) | 0.60 (0.49, 0.71) | 0.13 (0.11, 0.17) | 0.27 (0.18, 0.42) | 10.32 (7.15, 14.39) |
| 22871 | 1.14 (0.90, 1.33) | 1.93 (1.46, 2.40) | 0.84 (0.50, 1.17) | 1.36 (1.02, 1.98) | 0.95 (0.68, 1.50) | 2.00 (1.35, 3.17) | 1.17 (1.04, 1.28) | 0.48 (0.38, 0.61) | 2.21 (1.49, 3.66) | 0.91 (0.57, 1.28) |
Plate confrontation assays of T. atroviride with P. capsici.
Plate confrontation assays of T. atroviride with B. cinerea.
Plate confrontation assays of T. atroviride with R. solani.
Numbers in parentheses indicate 95% confidence intervals for the relative expression levels calculated by REST2008. PCR amplification efficiencies of all reactions were in the range of 100% ± 8%.
The gene corresponding to transcript/protein ID (further simply cited as “ID”) 297887, showing 89% amino acid (aa) identity to Trichoderma asperellum PAPA (62), was strongly repressed during mycoparasitism (0.025- to 0.139-fold) compared to the Trichoderma control (TP/TT, TB/TT, and TR/TT contact and precontact). Furthermore, the papA homolog was more highly expressed during physical contact with fungi containing chitin in their cell walls (Table 4).
Two of the genes tested were highly induced before the contact of T. atroviride with all host fungi (Table 4). swo1 (ID 80187) was induced 107.7 (TP/TT bc), 84.2 (TB/TT bc), 98.1 (TR/TT bc), and 46.5 (TT c/bc) times. The second gene (ID 84753) with high expression levels before contact, especially before interactions with P. capsici (TP/TT bc; 137-fold), is functionally similar to axe1 from T. reesei.
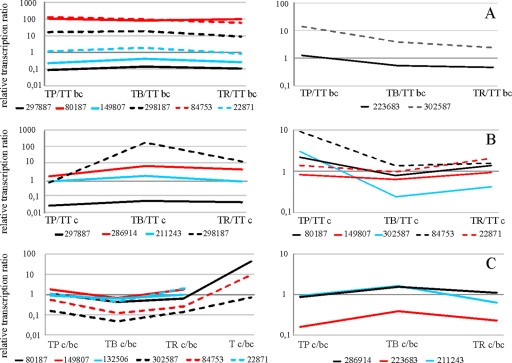
In general, only minor differences in gene inducibility were observed during the precontact stage regardless of the host fungus tested and independent of its phylogenetic affiliation (e.g., oomycete, basidiomycete, or ascomycete) (Fig. 2 A). Interestingly, all enzymes with a potential supportive function in fungal and plant cell wall degradation (SWO1, AXE1, ID 297887, and ID 298187) showed similar gene expression patterns with all host fungi tested. During direct interactions, the gene encoding a trypsin (ID 298187) showed stronger induction when in contact with B. cinerea (Table 4 and Fig. 2B). Particularly similar expression patterns were observed with axe1 and the gene coding for an unknown protein (ID 302587); both genes were more highly expressed during contact with P. capsici (Fig. 2B), indicating host-specific regulation. Additionally, a coherency was found between proteins with predicted functions in signal transduction and multidrug resistance when transcript ratios of precontact to direct contact with different host fungi were analyzed (Fig. 2C). Plotting of the relative transcript ratios further revealed correlations in host-dependent gene expression patterns of swo1, axe1, and a histidine kinase (ID 149807) related to the two-component response regulator SSK1 of Saccharomyces cerevisiae. Only one gene tested by RT-qPCR showed a pattern not comparable to that of any other gene during mycoparasitism with the plant-pathogenic fungi tested: ID 318101 (Table 4). This gene, encoding a ferric reductase, has 23% and 22% amino acid identities to the ferric/cupric reductase transmembrane components FRE7 and FRE1, respectively, of S. cerevisiae. Therefore, this protein is probably involved in the movement of electrons across the plasma membrane.
Fig. 2.
Similarities in gene expression patterns during mycoparasitic interactions presented in a qualitative form of “cluster analysis.” (A) Relative expression ratios of different genes (indicated as ID) before contact (bc) of T. atroviride with P. capsici (TP), B. cinerea (TB), or R. solani (TR) relative to the control (T. atroviride with itself [TT]) before contact. (B) Confrontations of T. atroviride in contact (c) with the host fungi. (C) Expression levels of genes during direct interactions with the host (TP c, TB c, TR c, and TT c) are shown relative to gene expression levels before contact (TP bc, TB bc, TR bc, and TT bc). Results were obtained by RT-qPCR.
Locating a novel putative regulatory motif by comparing nontranscribed 5′-end regions.
A 1-kb upstream region of each gene analyzed by RT-qPCR was extracted from the T. atroviride genome project homepage and searched for binding sites of known regulatory elements. These included recognition sites for the carbon catabolite repressor CreA (5′-SYGGRG-3′) (34, 54); nitrogen derepression and repression (5′-GATA-3′) (24, 30, 39); putative binding sites for proteins with regulatory functions during mycoparasitism (18), such as MYC1 (5′-GCTTCA-3′), MYC2 (5′-TTGGCAA-3′), MYC3 (5′-GGGCAC-3′), and MYC4 (5′-GGCAWTCGGCAT-3′); and the major regulators of cellulase and xylanase expression: Xyr1 (5′-GGCTAA-3′) (37), the AceI core sequence (5′-AGGCA-3′) (49), and AceII (5′-GGCTAATAA-3′) (2) (see File S4 in the supplemental material). No similarities between the promoter sequences of known, early, mycoparasite-induced genes (prb1 and ech42) and the promoter sequences of differentially expressed genes identified in this study were found for the binding of MYC1 to MYC4. Only three of these mycoparasitism-related elements were found in the upstream region of a gene coding for a multidrug resistance-associated protein belonging to the ABC superfamily (ID 211243). However, the expression pattern of the corresponding transcript during host interactions was not comparable to that of prb1. Furthermore, this gene was slightly downregulated in Trichoderma before physical contact with the host compared to the control. In total, surprisingly few MYC binding sites were found in the nontranscribed 5′ regions. Therefore, additional sequence similarities were investigated by using MEME, version 4.4.0 (5). A novel motif (5′-GARAAAAAARG-3′) was identified in the promoter regions of the papA (ID 297887), swo1 (ID 80187), and axe1 (ID 84753) homologs; a protein of the clathrin assembly or a related protein involved in signal transduction (ID 286914); a putative methyltransferase (ID 132506); a putative fungal transcription factor (ID 22871) (see File S4 in the supplemental material); and the 5′-untranslated region (UTR) of prb1. The sequence was searched against the DNA motif database TRANSFAC using the motif comparison tool TOMTOM, version 4.4.0 (26). Only a marginal correlation with already known binding motifs could be found: M01136 Dof (Pearson correlation coefficient, 0.00034), M00163 HSF (Pearson correlation coefficient, 0.00037), and M00022 Hb (Pearson correlation coefficient, 0.00057). Dof proteins represent a family of transcription factors with important roles in specific phenomena, i.e., defense responses in plants (65), but do not appear to have homologs in fungi (57). However, no evidence exists that this motif actually has a regulatory function during mycoparasitism. Therefore, further studies are required to elucidate the significance of this putative binding site.
DISCUSSION
In this study, we performed high-throughput transcriptome-wide sequencing to obtain a comprehensive overview of the mycoparasitic response processes in T. atroviride IMI206040. Genes found to be differentially expressed during interactions with R. solani were annotated, and a subset of genes differentially regulated during mycoparasitism was verified by RT-qPCR. Additionally, relative expression levels of these genes were examined during confrontation assays with host fungi from different phyla.
Recently, an increasing number of reports have been published regarding genome-wide transcriptome analyses. A few studies have also investigated the transcriptional responses of various Trichoderma strains under different biocontrol, mycoparasitism, as well as secretion stress conditions (3, 53, 64). However, all of these approaches utilized cDNA cloning strategies or array technologies. Here, we used de novo sequencing to avoid cloning artifacts and overcome the limitations of microarrays, which are susceptible to the detection of false-positives at low expression levels derived from cross-hybridization. In this context, van Bakel et al. previously found a high correlation between sequencing data and array intensities for the most highly transcribed regions; this was only marginal for low-level-expressed transcripts in human total brain tissue (59).
In comparison to the data available for the T. atroviride genome, the number of genes found in the present study covered between 38.4% (before contact with R. solani) and 52.8% (Trichoderma in confrontation with itself) of the total number of transcribed genes predicted for this organism. In this study, a total of 92 genes were found to be upregulated, 81 were downregulated, and 39 were either up- or downregulated depending on the conditions. Therefore, 175 differentially expressed transcripts (74 upregulated, 98 downregulated, and 3 up- or downregulated) can be related to precontact and mycoparasitic interactions with R. solani.
Of note, genes assigned to the functionality group “metabolism” represented the majority of genes that were upregulated during mycoparasitic interactions with R. solani. Such a finding provokes the assumption that the lysis of the host fungus improves the availability of nutrients, consequently triggering the growth and fitness of T. atroviride. In contrast, all genes involved in the biosynthesis, transport, and catabolism of secondary metabolites found to be differentially regulated were downregulated under the same conditions. This result is somewhat astounding, since antagonistic Trichoderma strains produce metabolites with antibiotic functions against bacteria and fungi. Additionally, it was shown previously that their action is synergistic with cell wall-lysing enzymes (CWDEs) (33, 35, 52). However, so far, no information is available on the regulation of nonribosomal peptide synthetases, which are responsible for the synthesis of peptaibols, the most prominent group of secondary metabolites in Trichoderma, at the transcript level during mycoparasitism.
In agreement with the current model for mycoparasitism (for a review, see reference 61), enzymes involved in the degradation of plant and fungal cell walls were identified in this study. In addition to prb1 (ID 302419), nag1 (ID 136120, encoding an N-acetyl-beta-d-glucosaminidase) was represented by different numbers of reads under distinct conditions. However, like other genes which did not pass Fisher's exact test, they were not included in the list of transcripts that were differentially expressed. In this context, the genes presented in this study with a P value of <10e−5 were regarded as being more relevant.
A gene that passed the selection criteria was swo1, encoding a protein with sequence similarities to plant expansins. The transcription of this gene was remarkably upregulated before visible contact with the respective host fungi. Similarly to swo1, a gene coding for the T. reesei AXE1 homolog (38) was strongly induced (up to 137-fold in experiments with P. capsici) before the direct contact of the colonies. Acetyl xylan esterases are hydrolases that deacetylate xylan and xylooligosaccharides and play an important role in the degradation of plant material. Interestingly, both SWO1 and AXE1 were previously shown to have elevated transcript levels in fungi grown on cellulose (31, 50). Both proteins consist of a cellulose binding domain (CBD) as well as an expansin domain for SWO1 or a separate catalytic domain for AXE1. Generally, cellulases and xylanases are composed of a catalytic domain connected to a CBD, which can also reversibly bind with low affinity to chitin (32). This, however, does not explain the role of both gene products during the early phase of mycoparasitism by Trichoderma. With the exception of the Oomycetes, fungal cell walls are composed predominantly of α(1,3)- and β(1,3)-glucans, chitin, galactomannan, and β(1,3),(1,4)-glucan but do not contain cellulose. A comparison of the 5′ upstream regulatory regions of both genes revealed the presence of binding sites for the global nitrogen regulator AreA, Xyr1, and the putative novel motif described here. A possible role for Xyr1 leading to the early induction of CWDEs still remains unclear in mycoparasitic interactions. In a previous proteome study that supports our findings, xylanase activity was detected in the culture supernatant of Trichoderma harzianum when grown in medium containing 1% deactivated B. cinerea mycelia (66). Unfortunately, no explanation for the presence of xylanases in the fraction of secreted proteins was found.
When relative expression ratios in the presence of distinct host fungi were compared, a gene encoding a sensory transduction histidine kinase (ID 149807) showed an expression pattern similar to those of swo1 and axe1. This type of kinase is part of a two-component phosphorylation system, which responds to environmental signals by autophosphorylation. Phosphorylation activates a response regulator protein, which further activates a mitogen-activated protein (MAP) kinase cascade (13, 14). For this protein, only a C-terminal response regulator receiver domain (IPR001789) has been identified. Not all functions for the nine different groups of histidine kinases found in fungi are known. Interestingly, no regulatory element was found in the upstream region of ID 149807, and thus, no information is known about the inducibility of this gene. Until now, no indications for the transcriptional regulation of a gene involved in signal transduction during mycoparasitic interactions have been provided; previous investigations revealed constitutive transcript formation for such components (44, 68). Consequently, that study did not detect any of the currently described genes encoding signaling proteins related to mycoparasitism.
Interestingly, as hydrolytic enzymes involved in cell wall degradation were reported to be upregulated during Trichoderma antagonism, transcript levels of papA (ID 297887) during confrontation with plant-pathogenic fungi were reduced compared to those of the control. Secreted proteases of mycoparasitic agents are important factors in the degradation of fungal cell walls and plasma membrane proteins and also facilitate the penetration of the plant root epidermis. papA was found previously to be expressed in planta in experiments with cucumber and even 4-fold induced in plate confrontation assays before contact with R. solani (62). Additionally, when T. asperellum T203 was grown alone, a basal expression level was detectable but not elevated during confrontation with itself. In contrast to previous findings, in this study, papA was induced during the confrontation of T. atroviride with itself, which therefore influenced the relative expression levels of this gene compared to those of the self-confrontation control. However, when transcript levels from the interaction zone with R. solani were compared directly to those of confrontation before direct contact (TR c/bc), papA was induced in T. atroviride. Previous studies of papA gene expression were carried out with T. harzianum, where it was shown that papA is repressed by ammonium, glucose, and glycerol but induced by organic nitrogen sources. Additionally, binding sites for AreA and MYC were found in the promoter region (19). In contrast, our promoter analysis identified only one MYC site (MYC1) and multiple binding sites for Xyr1, Cre1, AceI, and a novel putative motif. In T. harzianum, constitutive expression was lost when the fungus was grown on minimal medium supplemented with 2% glucose. Genes coding for chitinolytic and glucanolytic enzymes were repressed in T. atroviride grown in medium with a high glucose concentration but were highly expressed following glucose depletion and starvation conditions obtained with 0.1% glucose added as a carbon source (20, 36). In order to not influence gene transcription in a glucose-dependent manner, in the present study, a concentration of 0.2% glucose was used, as described previously (27, 62).
Another component of the hydrolytic enzyme system is the acidic protease PRA1, which was previously characterized in T. harzianum CECT 2413 (56) and has 74% amino acid identity to ID 298187. The transcription of pra1 was previously described to be induced by chitin and cell walls of the Ascomycetes. This was also found for the corresponding gene in T. atroviride, since induction occurred during physical contact with host fungi, which contain chitin in their cell walls. On the other hand, the pra1 homolog was repressed in fungi encountering P. capsici, indicating a cell wall composition-dependent regulation of gene transcription. Proteomics analysis supportively identified a trypsin-like protease in T. atroviride cultures after growth on R. solani cell walls (25). Unfortunately, this proteome analysis of Trichoderma was carried out before the release of the respective genomes, and consequently, most differentially expressed proteins remained unidentified. In general, peptidases of the chymotrypsin (S1) family can be linked to fungal pathogenicity (21). A homolog in T. atroviride (ID 298187) has the potential to act as a marker for pathogenicity and/or host specificity.
Trichoderma mycoparasitism is complex, and it was proposed previously that the penetration of the host hyphae is accomplished by appressorium-like structures (7). The formation of these structures is also essential for plant-pathogenic fungi to infect plant cells. Oh et al. (42) recently reported a transcriptome analysis of Magnaporthe oryzae during appressorium formation. In that study, the authors demonstrated that genes encoding proteins with CFEM (common in fungal extracellular membrane proteins) domains are significantly upregulated during such morphological changes. CFEM (IPR008427) is described as an eight-cysteine-containing domain found in fungal extracellular membrane proteins. Such a domain is also present in the transcript encoding protein ID 302587. The respective gene is significantly upregulated before contact with the host compared to levels under control conditions (confrontation of T. atroviride with itself). Since no functional annotation of this protein in T. atroviride is available, the putative interaction partner(s) of this protein is still unknown. Due to this possible biological function, ID 302587 certainly warrants further investigation.
Genome-wide expression studies are useful tools to identify genes associated with particular biological processes like mycoparasitism. The information obtained makes it possible to choose interesting genes for functional characterization in order to obtain further insight into the physiological processes taking place in the presence of host fungi. In addition, we showed that the mycoparasitic fungus T. atroviride not only reacts to the presence of plant-pathogenic fungi by changes in gene expression but also reacts to itself. This remarkable result will be subjected to further investigation and might help us to understand processes involved in the recognition and host specificity of this fungus.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Austrian Science Fund with an Erwin Schrödinger fellowship (FWF J2674) and a stand-alone project (FWF P21584).
We thank Araceli Fernández-Cortés and Rodrigo Echegoyén-Nava for their assistance with the bioinformatics analysis.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 April 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Aro N., Saloheimo A., Ilmén M., Penttilä M. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309–24314 [DOI] [PubMed] [Google Scholar]
- 3. Arvas M., et al. 2006. Common features and interesting differences in transcriptional responses to secretion stress in the fungi Trichoderma reesei and Saccharomyces cerevisiae. BMC Genomics 7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atreya K. 2008. Health costs from short-term exposure to pesticides in Nepal. Soc. Sci. Med. 67:511–519 [DOI] [PubMed] [Google Scholar]
- 5. Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28–36 In Altman R., Brutlag D., Karp P., Lathrop R., Searls D. (ed.), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Stanford, CA: [PubMed] [Google Scholar]
- 6. Barakat A., et al. 2009. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol. 9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benhamou N., Chet I. 1997. Cellular and molecular mechanisms involved in the interaction between Trichoderma harzianum and Pythium ultimum. Appl. Environ. Microbiol. 63:2095–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brotman Y., Briff E., Viterbo A., Chet I. 2008. Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol. 147:779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brun G. L., MacDonald R. M., Verge J., Aubé J. 2008. Long-term atmospheric deposition of current-use and banned pesticides in Atlantic Canada; 1980-2000. Chemosphere 71:314–327 [DOI] [PubMed] [Google Scholar]
- 10. Brunner K., et al. 2008. Trichoderma G protein-coupled receptors: functional characterisation of a cAMP receptor-like protein from Trichoderma atroviride. Curr. Genet. 54:283–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantacessi C., et al. 2010. Differences in transcription between free-living and CO2-activated third-stage larvae of Haemonchus contortus. BMC Genomics 11:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carsolio C., Gutierrez A., Jimenez B., van Montagu M., Herrera-Estrella A. 1994. Characterization of ech-42, a Trichoderma harzianum endochitinase gene expressed during mycoparasitism. Proc. Natl. Acad. Sci. U. S. A. 91:10903–10907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Catlett N. L., Yoder O. C., Turgeon B. G. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang C., Stewart R. C. 1998. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117:723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chet I. 1987. Trichoderma—application, mode of action, and potential as a biocontrol agent of soilborne pathogenic fungi, p. 137–160 In Chet I. (ed.), Innovative approaches to plant disease control. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 16. Chomczynski P., Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 17. Copping L. G., Menn J. J. 2000. Biopesticides: a review of their action, applications and efficacy. Pest Manag. Sci. 56:651–676 [Google Scholar]
- 18. Cortés C., et al. 1998. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol. Gen. Genet. 260:218–225 [DOI] [PubMed] [Google Scholar]
- 19. Delgado-Jarana J., Rincon A. M., Benitez T. 2002. Aspartyl protease from Trichoderma harzianum CECT 2413: cloning and characterization. Microbiology 148:1305–1315 [DOI] [PubMed] [Google Scholar]
- 20. Donzelli B. G. G., Harman G. E. 2001. Interaction of ammonium, glucose, and chitin regulates the expression of cell wall-degrading enzymes in Trichoderma atroviride strain P1. Appl. Environ. Microbiol. 67:5643–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dubovenko A. G., et al. 2010. Trypsin-like proteins of the fungi as possible markers of pathogenicity. Fungal Biol. 114:151–159 [DOI] [PubMed] [Google Scholar]
- 22. Elad Y., Chet I., Boyle P., Henis Y. 1983. Parasitism of Trichoderma spp. on Rhizoctonia solani and Sclerotium rolfsii—SEM studies and fluorescence microscopy. Phytopathology 73:85–88 [Google Scholar]
- 23. Flores A., Chet I., Herrera-Estrella A. 1997. Improved biocontrol activity of Trichoderma harzianum by over-expression of the proteinase-encoding gene prb1. Curr. Genet. 31:30–37 [DOI] [PubMed] [Google Scholar]
- 24. Fu Y. H., Marzluf G. A. 1987. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol. Cell. Biol. 7:1691–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grinyer J., Hunt S., McKay M., Herbert B. R., Nevalainen H. 2005. Proteomic response of the biological control fungus Trichoderma atroviride to growth on the cell walls of Rhizoctonia solani. Curr. Genet. 47:381–388 [DOI] [PubMed] [Google Scholar]
- 26. Gupta S., Stamatoyannopoulos J., Bailey T., Noble W. 2007. Quantifying similarity between motifs. Genome Biol. 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haran S., Schickler H., Oppenheim A., Chet I. 1996. Differential expression of Trichoderma harzianum chitinases during mycoparasitism. Mol. Plant Pathol. 86:980–985 [Google Scholar]
- 28. Harman G. E., Björkman T. 1998. Potential and existing uses of Trichoderma and Gliocladium for plant disease control and plant growth enhancement, p. 229–266 In Harman G. E., Kubicek C. P. (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom [Google Scholar]
- 29. Hjeljord L., Tronsmo A. 1998. Trichoderma and Gliocladium in biological control: an overview, p. 131–152 In Harman G. E., Kubicek C. P. (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom [Google Scholar]
- 30. Kudla B., et al. 1990. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 9:1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X.-L., Skory C. D., Cotta M. A., Puchart V., Biely P. 2008. Novel family of carbohydrate esterases, based on identification of the Hypocrea jecorina acetyl esterase gene. Appl. Environ. Microbiol. 74:7482–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Linder M., Salovuori I., Ruohonen L., Teeri T. T. 1996. Characterization of a double cellulose-binding domain. J. Biol. Chem. 271:21268–21272 [DOI] [PubMed] [Google Scholar]
- 33. Lorito M., Farkas V., Rebuffat S., Bodo B., Kubicek C. P. 1996. Cell wall synthesis is a major target of mycoparasitic antagonism by Trichoderma harzianum. J. Bacteriol. 178:6382–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lorito M., et al. 1996. Mycoparasitic interaction relieves binding of the Cre1 carbon catabolite repressor protein to promoter sequences of the ech42 (endochitinase-encoding) gene in Trichoderma harzianum. Proc. Natl. Acad. Sci. U. S. A. 93:14868–14872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lorito M., et al. 1996. Synergistic interaction between cell wall degrading enzymes and membrane affecting compounds. Mol. Plant Microbe Interact. 9:206–213 [Google Scholar]
- 36. Mach R. L., et al. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65:1858–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mach R. L., Zeilinger S. 2003. Regulation of gene expression in industrial fungi: Trichoderma. Appl. Microbiol. Biotechnol. 60:515–522 [DOI] [PubMed] [Google Scholar]
- 38. Margolles-Clark E., Ihnen M., Penttilä M. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167–179 [Google Scholar]
- 39. Minehart P. L., Magasanik B. 1991. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol. Cell. Biol. 11:6216–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Monteiro V., et al. 2010. New insights in Trichoderma harzianum antagonism of fungal plant pathogens by secreted protein analysis. Curr. Microbiol. 61:298–305 [DOI] [PubMed] [Google Scholar]
- 41. Nelson E. B. 1991. Current limits to biological control of fungal phytopathogens, p. 327–355 In Arora D. K., Rai B., Mukerji D. G., Knudsen G. R. (ed.), Handbook of applied mycology: soil and plants. Marcel Dekker, New York, NY [Google Scholar]
- 42. Oh Y., et al. 2008. Transcriptome analysis reveals new insight into appressorium formation and function in the rice blast fungus Magnaporthe oryzae. Genome Biol. 9:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pfaffl M. W., Horgan G. W., Dempfle L. 2002. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reithner B., et al. 2005. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 42:749–760 [DOI] [PubMed] [Google Scholar]
- 45. Rozen S., Skaletsky H. J. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 46. Ruocco M., et al. 2009. Identification of a new biocontrol gene in Trichoderma atroviride: the role of an ABC transporter membrane pump in the interaction with different plant-pathogenic fungi. Mol. Plant Microbe Interact. 22:291–301 [DOI] [PubMed] [Google Scholar]
- 47. Rutledge R., Stewart D. 2008. Critical evaluation of methods used to determine amplification efficiency refutes the exponential character of real-time PCR. BMC Mol. Biol. 9:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rutledge R., Stewart D. 2008. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 8:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saloheimo A., Aro N., Ilmén M., Penttilä M. 2000. Isolation of the ace1 gene encoding a Cys2-His2 transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 275:5817–5825 [DOI] [PubMed] [Google Scholar]
- 50. Saloheimo M., et al. 2002. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269:4202–4211 [DOI] [PubMed] [Google Scholar]
- 51. Samolski I., de Luis A., Vizcaino J., Monte E., Suarez M. B. 2009. Gene expression analysis of the biocontrol fungus Trichoderma harzianum in the presence of tomato plants, chitin, or glucose using a high-density oligonucleotide microarray. BMC Microbiol. 9:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schirmböck M., et al. 1994. Parallel formation and synergism of hydrolytic enzymes and peptaibol antibiotics, molecular mechanisms involved in the antagonistic action of Trichoderma harzianum against phytopathogenic fungi. Appl. Environ. Microbiol. 60:4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seidl V., et al. 2009. Transcriptomic response of the mycoparasitic fungus Trichoderma atroviride to the presence of a fungal prey. BMC Genomics 10:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sophianopoulou V., Suárez T., Diallinas G., Scazzocchio C. 1993. Operator derepressed mutations in the proline utilisation gene cluster of Aspergillus nidulans. Mol. Gen. Genet. 236:209–213 [DOI] [PubMed] [Google Scholar]
- 55. Steiger M. G., Mach R. L., Mach-Aigner A. R. 2009. An accurate normalization strategy for RT-qPCR in Hypocrea jecorina (Trichoderma reesei). J. Biotechnol. 145:30–37 [DOI] [PubMed] [Google Scholar]
- 56. Suarez B., Rey M., Castillo P., Monte E., Llobell A. 2004. Isolation and characterization of PRA1, a trypsin-like protease from the biocontrol agent Trichoderma harzianum CECT 2413 displaying nematicidal activity. Appl. Microbiol. Biotechnol. 65:46–55 [DOI] [PubMed] [Google Scholar]
- 57. Takatsuji H. 1998. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54:582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tronsmo A. 1991. Biological and integrated controls of Botrytis cinerea on apple with Trichoderma harzianum. Biol. Control 1:59–62 [Google Scholar]
- 59. van Bakel H., Nislow C., Blencowe B. J., Hughes T. R. 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 8:e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vega-Arreguin J., et al. 2009. Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics 10:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vinale F., et al. 2008. Trichoderma-plant-pathogen interactions. Soil Biol. Biochem. 40:1–10 [Google Scholar]
- 62. Viterbo A., Harel M., Chet I. 2004. Isolation of two aspartyl proteases from Trichoderma asperellum expressed during colonization of cucumber roots. FEMS Microbiol. Lett. 238:151–158 [DOI] [PubMed] [Google Scholar]
- 63. Vizcaino J., et al. 2006. Generation, annotation and analysis of ESTs from Trichoderma harzianum CECT 2413. BMC Genomics 7:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vizcaíno J., et al. 2007. Generation, annotation, and analysis of ESTs from four different Trichoderma strains grown under conditions related to biocontrol. Appl. Microbiol. Biotechnol. 75:853–862 [DOI] [PubMed] [Google Scholar]
- 65. Yanagisawa S. 2002. The Dof family of plant transcription factors. Trends Plant Sci. 7:555–560 [DOI] [PubMed] [Google Scholar]
- 66. Yang H.-H., Yang S. L., Peng K.-C., Lo C.-T., Liu S.-Y. 2009. Induced proteome of Trichoderma harzianum by Botrytis cinerea. Mycol. Res. 113:924–932 [DOI] [PubMed] [Google Scholar]
- 67. Zeilinger S., et al. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26:131–140 [DOI] [PubMed] [Google Scholar]
- 68. Zeilinger S., et al. 2005. Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl. Environ. Microbiol. 71:1591–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.