Abstract
Entomopathogenic fungi have been used for biocontrol of insect pests for many decades. However, the efficacy of such fungi in field trials is often inconsistent, mainly due to environmental stresses, such as UV radiation, temperature extremes, and desiccation. To circumvent these hurdles, metabolic engineering of dihydroxynaphthalene (DHN) melanin biosynthetic genes (polyketide synthase, scytalone dehydratase, and 1,3,8-trihydroxynaphthalene reductase genes) cloned from Alternaria alternata were transformed into the amelanotic entomopathogenic fungus Metarhizium anisopliae via Agrobacterium-mediated transformation. Melanin expression in the transformant of M. anisopliae was verified by spectrophotometric methods, liquid chromatography/mass spectrometry (LC/MS), and confocal microscopy. The transformant, especially under stresses, showed notably enhanced antistress capacity and virulence, in terms of germination and survival rate, infectivity, and reduced median time to death (LT50) in killing diamondback moth (Plutella xylostella) larvae compared with the wild type. The possible mechanisms in enhancing the stress tolerance and virulence, and the significance and potential for engineering melanin biosynthesis genes in other biocontrol agents and crops to improve antistress fitness are discussed.
INTRODUCTION
Although chemical pesticides can kill a wide range of insect pests rapidly and cost-effectively, their side effects over time are problematic. The selective pressure from long-term application of chemical pesticides ultimately leads to pesticide resistance, harm to the ecosystem, and residual poisonous effects on humans and animals (9). To lessen such adverse effects, the prospect of using the natural enemies of insect pests, such as predators, parasitoids, and pathogenic microbes, is gaining favor. Among the pathogenic microbes, more than 700 species of entomopathogenic fungi have been described worldwide, and specific strains, such as Beauveria bassiana and Metarhizium anisopliae, have been selected, produced, marketed, and applied in the field to control insect pests. Nevertheless, on a global scale, the use of these biocontrol agents against insect pests is still limited, perhaps due to their inconsistent performance, low infection rates, and limited shelf life. Finding a way to overcome these shortcomings thus remains a great challenge.
In nature, various types of pigments fulfill a diversity of roles. For example, chlorophylls capture sunlight; carotenoids impart beautiful hues, serve as antioxidants, and confer protection against UV radiation; hemes transport essential oxygen molecules to cells; and melanins protect organisms from the harmful effects of solar UV radiation and other environmental stressors, such as heavy metals, oxidants, and microbial lytic enzymes and defensins (26, 38). As in other organisms, melanins in fungi confer survival advantages. Certain plant- and animal-pathogenic fungi, including Magnaporthe grisea, Colletotrichum lagenarium, and Cryptococcus neoformans, that contain melanin are in general more virulent than albino mutants (8, 25, 37, 38).
Melanins are red, brown, or black pigments consisting of phenolic or indolic polymers complexed with carbohydrates, proteins, or lipids and are resistant to strong acids and organic solvents but can be bleached by oxidants and degraded by strong alkali. In fungi, several types of melanins, including DOPA, GDHB (γ-glutaminyl-3,4-dihydroxybenzene), catechol, and dihydroxynaphthalene (DHN) melanins, are synthesized via various biosynthetic pathways (3, 8). The yeast human pathogen C. neoformans synthesizes DOPA melanins by oxidizing the dopamine produced by the host cell by wall-bound laccase. GDHB melanins in the cell walls of basidiospores, in the generative mycelium of the button mushroom Agaricus bisporus, and in many species of basidiomycetes are generated from γ-glutaminyl-4-hydroxybenzene (GHB) through the activity of γ-glutaminyltransferase or phenolic oxidase and finally converted to melanins. On the other hand, the melanins deposited at or enmeshed in the fibrillar wall of the corn smut, Ustilago maydis, are polymers of catechol. The catechol monomers are either reduced or oxidized to semiquinone radicals or o-benzoquinone and adducted to form catechol dimers and finally melanins (40). Both catechol and GDHB have been suggested to originate from the shikimic acid pathway. In the past, DHN melanins have been the melanins overwhelmingly studied in fungi, especially ascomycetes and their anamorphs. The biosynthesis of DHN melanins is initiated by the conjoining and cyclization of five ketide subunits by polyketide synthase (PKS) to form 1,3,6,8-tetrahydroxynaphthalene (1,3,6,8-THN), which undergoes reduction by 1,3,6,8-tetrahydroxynaphthalene reductase (4HNR) or 1,3,8-trihydroxynaphthalene reductase (3HNR) and dehydration by scytalone dehydratase (SCD). Finally, the intermediate 1,8-dihydroxynaphthalene (1,8-DHN) is polymerized into melanin by the catalysis of phenol oxidase, peroxidase, laccase, or catalase (Fig. 1A) (3, 8, 32, 56, 64).
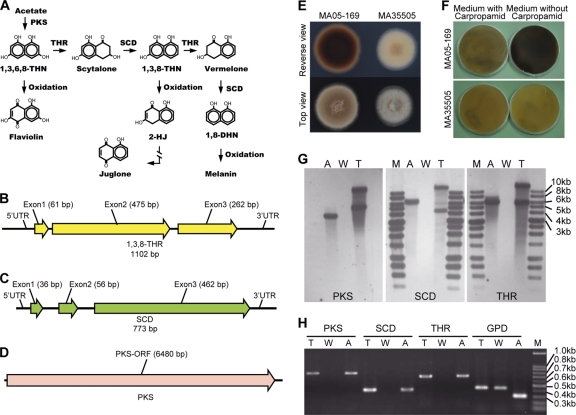
Fig. 1.
The DHN melanin biosynthetic pathway; traits and copy numbers of cloned PKS, SCD, and THR genes in the transformant. (A) DHN melanin biosynthetic pathway. (Adapted from reference 3.) (B to D) Cloned THR, SCD, and PKS genes. (E) Colony phenotype of M. anisopliae transformant MA05-169 (MA05-169) and wild-type BCRC35505 (MA35505). (F) Colony phenotype of M. anisopliae BCRC35505 and MA05-169 affected by carpropamid. (G) PKS, SCD, and THR gene copy numbers for A. alternata (A), M. anisopliae BCRC35505 (W), and MA05-169 (T). (H) PKS, SCD, THR, and GPD gene transcripts revealed by RT-PCR. M, marker; ORF, open reading frame.
Here, experiments were initiated to circumvent the adverse effects of a stressful environment that impairs the efficacy of the well-studied entomopathogen M. anisopliae (19, 50, 51) when applied in the field. Alternaria alternata was chosen as the host for gene cloning, as a previous report indicated that it harbors three DHN melanin synthesis genes in a cluster on a 33-kb chromosomal region (29). Some of these genes have been cloned, and their functions have been characterized in relation to UV resistance or complementation in restoring the virulence of M. grisea or C. lagenarium (27, 28, 54). This information facilitated cloning of the relevant genes. We first cloned the DHN melanin biosynthesis genes (PKS, SCD, and THR) from A. alternata by screening the constructed fosmid library and sequencing the shotgun library of targeted clones. The cloned genes were transformed into M. anisopliae via Agrobacterium tumefaciens-mediated transformation (ATMT). The positive transformants were characterized in terms of melanin gene expression, melanin production, anti-stress capacity (UV, temperature, and desiccation), and virulence toward target insects. The confirmed transformants were promising in enhancing stress tolerance and virulence and merit further field trials to determine their ability to control crucial crop or sanitary insect pests.
MATERIALS AND METHODS
Fungal cultures and shuttle vectors.
A. alternata BCRC30501, which was originally isolated from apple black rot in the United States and has the capacity to synthesize DHN melanin, was obtained from the Bioresource Collection and Research Center (BCRC), Food Industry Research and Development Institute (FIRDI), Hsinchu, Taiwan. M. anisopliae BCRC35505, which we used for transformation, was isolated from the diamondback moth (Plutella xylostella) and was also obtained from the BCRC. C. lagenarium, including the wild-type 104-T strain and the scytalone dehydratase-deficient 8015 mutant strain, were obtained from Y. Kubo, Kyoto Prefecture University, Japan. The binary vector pCAMBIA 1300 (NCBI; GI 7638064) (Cambia Institute, Canberra, Australia), used as a skeleton for constructing the shuttle vector for transformation, was provided by W. H. Hsu, National Chung Hsing University, Taiwan. Details of the cloning of the DHN melanin biosynthesis genes and shuttle vector construction for transformation are included in the supplemental material.
Agrobacterium-mediated transformation and characterization of transformants.
The cotransformation protocol was performed according to the method of de Groot et al. (14) with minor modifications. The binary vectors pCAMBIA THR, pCAMBIA SCD, and pCAMBIA PKS-ORF (see Fig. S1A to C in the supplemental material) were transformed into A. tumefaciens EHA105 by electroporation (1,550 V, 150 Ω, 50 μF) and grown in 10 ml of LB broth containing 50 μl ml−1 of kanamycin with shaking at 220 rpm for 18 h. After centrifugation at 5,900 × g for 5 min, the pellet was rinsed three times with autoclaved induction medium (IM) [10 mM K2HPO4, 10 mM KH2PO4, 2.5 mM NaCl, 2 mM MgSO4, 0.7 mM CaCl2, trace FeSO4, 4 mM (NH4)2SO4, and 40 mM morpholineethanesulfonic acid (MES) buffer; the pH was adjusted to 5.3, and then 0.5% glycerol, 5 mM glucose and 200 mM acetosyringone were added]. The acetosyringone was first dissolved in dimethyl sulfoxide (DMSO) or 100% ethanol to prepare the 200 mM stock solution and then filtered through a 0.4-μm Millipore filter for use. The A. tumefaciens competent cells were washed with IM three times and then resuspended in IM and diluted to an optical density at 600 nm (OD600) of 0.30. The diluted IM, which contained 50 μg ml−1 of kanamycin and acetosyringone, was incubated at 28°C with shaking at 220 rpm to an OD600 of 0.6 to 0.8. The entomopathogenic fungus M. anisopliae BCRC35505 was grown in potato dextrose broth (PDB) (Difco) at 25°C in the dark for 5 days. The mycelium was removed by filtration through Miracloth, the spores were precipitated by centrifugation at 2,300 × g for 5 min, and the filtrate was discarded. The spore suspension was adjusted to a concentration of 106 spores ml−1. The M. anisopliae spore suspension was combined with the A. tumefaciens (PKS), A. tumefaciens (SCD), and A. tumefaciens (THR) suspensions at a 1:1:1:1 ratio. One hundred microliters of the mixture was smeared evenly onto cellophane membranes and overlaid with cocultivation medium, which consisted of IM supplemented with 5 mM glucose and 2% agar. After culture at 28°C for 2 days, the membrane was transferred, applied upside down onto Czapek (CPZ) medium (Difco) containing 250 μg ml−1 of cefotaxime and hygromycin (100 μg ml−1), and incubated for another 7 days. The putative transformed clones with higher growth rates and large colony sizes were selected and processed for molecular verification and characterization.
The DNA of the selected transformants growing in CPZ medium was extracted using Maxwell 16 Genomic DNA purification kits (Promega). PCR was conducted with the following specific primers: PKS-TE-sen, PKS-TE-ant, Scy-N-2-1, Scy-N-2-2, 1,3,8-tri(A), 1,3,8-tri(B), HygR-ide-S1, and HygR-ide-A1 (see Table S1 in the supplemental material). The insertion of the melanin biosynthesis genes and the HygR gene in the transformants was verified by electrophoresis. The positive transformants were cultivated on potato dextrose agar (PDA) (Difco) containing hygromycin (100 μg ml−1) for five successive generations and verified by PCR once to prove the inherent genetic stability of the transformant. One of the stable transformants was designated M. anisopliae MA05-169 and chosen for use in experiments.
For Southern blotting, digoxigenin (DIG)-labeled probes were amplified by PCR using the specific primers PKS-TE-sen, PKS-TE-ant, Scy-N-2-1, Scy-N-2-2, 1,3,8-tri(A), and 1,3,8-tri(B) (see Table S1 in the supplemental material), with the plasmids containing SCD, THR, and PKS as templates. For verification of the presence of melanin biosynthesis genes in the wild-type (BCRC35505) and transformant (MA05-169) M. anisopliae and A. alternata (BCRC30501), the genomic DNA (1) was digested with HindIII for the SCD and THR genes and PstI for the PKS gene. The digested genomic DNAs (gDNAs) were subjected to electrophoresis on 0.8% agarose gels at 50 V for 6 h; each well was loaded with 10 μg of DNA. After electrophoresis, the gel was treated with 0.2 N HCl for 20 min, washed with double-distilled water (ddH2O) 2 or 3 times, and incubated in 0.5 N NaOH and 1.5 M NaCl for 45 min with gentle shaking to denature the DNA into single strands. The gel was washed with ddH2O for 5 min and neutralized with 1.5 M Tris buffer, pH 7.4, containing 1.5 M NaCl for 45 min. The gel was again washed with ddH2O for 5 min and vacuum blotted onto Hybond nylon membranes for Southern blotting according to the protocol of Sambrook and Russell (45) with minor modifications. Briefly, the hybridization was performed in reaction mixtures at 50°C for 15 h, followed by rinsing with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature two times for 5 min each and then washed with 0.2× SSC-0.1% SDS at 60°C two times for 15 min each time.
Expression of melanin biosynthesis genes.
RNA from A. alternata BCRC30501 and the M. anisopliae wild-type strain BCRC35505 and mutant strain MA05-169 was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. The extracted RNAs were incubated with Turbo DNase (Ambion) at 37°C for 30 min. Then, 0.1-fold Turbo inactivation agents were added, and the samples were incubated for 2 min, followed by centrifugation at 9,300 × g for 1.5 min. The supernatants were transferred to fresh Eppendorf tubes, and the RNA quality and concentration were determined using a NanoPhotometer (Implen, Germany) and checked by denatured gel electrophoresis at 60 V for 200 min (47). The purified RNA was converted to cDNA by two-step reverse transcription (RT)-PCR using a cDNA Synthesis kit (Clontech) under the following conditions: 95°C for 1 min and 30 cycles of 95°C for 5s, an annealing step of 5s at 65°C, and 68°C for 3 min for extension. The RT-PCR products were purified with Wizard Gel and a PCR Clean-Up system (Promega) and analyzed by PCR using the specific primers PKS-TE-sen, PKS-TE-anti, Scy_N_2-1, Scy_N_2-2, 1,3,8-tri(A), and 1,3,8-tri(B) (see Table S1 in the supplemental material) to detect PKS, SCD, and THR. The glyceraldehyde-3-phosphate dehydrogenase (GPD) gene was generated using degenerate primers from the M. anisopliae GPD mRNA sequence (NCBI; GI 115607610), pAN-7-1 (NCBI; GI 475166), and the A. alternata GPD mRNA sequence (NCBI; GI 31747098) and used as a positive control (see Table S1 in the supplemental material).
Q-PCR quantification of melanin biosynthesis gene transcripts in transformants.
To absolutely quantify the copy number of the expressed DHN melanin synthesis genes, PKS, SCD, and THR, the three genes were amplified by PCR using PKS-, SCD-, and THR-specific primers with pCAMBIA PKS-ORF, pCAMBIA SCD, and pCAMBIA THR, respectively, as templates (see Table S1 in the supplemental material). Each of the specific amplicons was cloned into the pGEM-T Easy vector. The mass of the constructed plasmids was calculated based on the following formula: m = (n) (1.096 × 10−21 g bp−1), where m represents mass and n represents the plasmid size. For example, at 30,000 copies as a standard copy number, the mass of PKS will be 12.55 pg. By using a Bio-Rad iQ5 Real Time PCR and TaqMan probe (Bio-Rad), a standard quantitative-PCR (Q-PCR) curve could be plotted by successive dilution from 3 × 106 copies μl−1 to 3 × 105, 3 × 104, 3 × 103, and 3 × 102 copies μl−1 (see Table S1 in the supplemental material). Reactions were run using 12.5 μl of 2× Q-PCR buffer, 50 ng of A. alternata BCRC30501, M. anisopliae BCRC35505, and M. anisopliae MA05-169 cDNA templates; 2.5 μl of 5 μM primers; and 2 μl of 2.5 μM TaqMan probe. PCR was conducted under the following conditions: 95°C for 3 min and 40 cycles of 95°C for 10 s and 55°C for 30 s. The exact copy number of the genes transcripts was calculated by plotting against the standard curves (data not shown).
Verification of melanin and its precursors synthesized by the transformant.
For extraction and purification of melanin, the method of Selvakumar et al. (47) was used. Briefly, A. alternata BCRC30501, M. anisopliae BCRC35505, and M. anisopliae MA05-169 were grown in PDB in a 5-liter fermentor. After incubation for 4 days, the mycelium, harvested by filtration through Miracloth, was homogenized and treated with 2 M NaOH, pH 10.5, for 36 to 48 h. The suspension was centrifuged at 4,000 × g for 15 min to precipitate the coarse debris. The supernatant was acidified with 2 M HCl to pH 2.5, after which the melanin became insoluble and was suspended in situ for 2 h. The suspension was centrifuged at 4,000 × g for 15 min to remove the supernatant, and the precipitate was suspended in 6 M HCl and boiled at 100°C for 2 h to hydrolyze and remove carbohydrate and protein. The precipitate was successively rinsed with chloroform, ethyl acetate, and ethanol to remove lipids upon centrifugation at 10,000 rpm for 15 min. After air drying, the precipitate was redissolved in 2 M NaOH and centrifuged at 4,000 × g for 15 min. The supernatant was treated with 1 M HCl, and the precipitated melanin was rinsed with ddH2O and freeze-dried by lyophilization. The chemical properties of the purified melanin were verified by UV-visible light (UV-Vis) and Fourier transfer infrared (FTIR) spectrometry as well as by electron paramagnetic resonance (EPR), using A. alternata DHN melanin and DOPA melanin (Sigma) as standards.
For UV-Vis spectrometry, the melanin was dissolved in 0.1 M boracic acid buffer, pH 8.0, with an adjusted concentration of 0.002% (wt/vol) and scanned using a NanoPhotometer (Implen, Germany) at a wavelength of 200 to 500 nm (47). The log-converted absorption values of the melanin were linearized by regression to calculate the slope.
The EPR of the melanin (ca. 10 μg) was performed with an X-band Spectrometer EMX-10/12 (Bruker, Germany) at 77 K, 9.48 GHz, and 100 KHz modulation. The spectrum, including G values (gyromagnetic ratio) for the DHN melanin and DOPA melanin, was plotted using WinEPR version 2.11 software (Bruker, Germany).
For FTIR, the melanin and potassium bromide (KBr; Sigma) were dried in an oven at 60°C for 1 h. The melanin and KBr were combined at a ratio of 1:19 (vol/vol), homogenized, and scanned with an FTIR 4100 (Jasco, Japan) at a wavelength of 4,000 to 400 nm. The spectrum of the melanin after transformation of the unit of wavelength to μm was plotted using OriginPro 7.5 SRI software and subjected to further analysis for functional groups.
For melanin precursor determination, the method of Wheeler and Klich (61) was followed. Briefly, the C. lagenarium wild-type strain 104-T and M. anisopliae BCRC35505 and MA05-169 were grown on PDA for 7 days. The mycelium scraped from plates was segmented by homogenization, smeared evenly onto 15-cm PDA plates, and incubated at 25°C for 10 days. After the surface mycelium was scraped, the agar plates underneath were cut into 0.5-cm3 pieces. Approximately 80 g agar blocks was added to 50 ml of ethyl acetate, acidified with glacial acetic acid, and agitated at 50 rpm for 2 h. The agar blocks were discarded, and the supernatant was vacuum vaporized with an Eyela Aspirator A-3S (Tokyo, Japan). The dried residue was dissolved in 1 ml of methanol and stored at −20°C until it was used. The same procedure was also applied to M. anisopliae strains BCRC35505 and MA05-169, A. alternata BCRC30501, and C. lagenarium strains 104-T and 8015, which were previously grown on PDA plates amended or not with 200 ppm carpropamid, an inhibitor of scytalone dehydratase. The extracts were subjected to thin-layer chromatography (TLC) and liquid chromatography/mass spectrometry (LC/MS) analyses.
For TLC, the TLC silica gel phase and a mixture of chloroform-methanol (9:1) were used as the mobile phases. The extracts mentioned above were spotted on the baseline 0.5 cm above the bottom, with each spot approximately 3 mm in diameter. One hour prior to chromatography, a 20- by 2-cm Whatman No. 2 filter paper was dipped in the mobile liquid and rested along the glass chamber wall to equilibrate the chamber humidity. The chromatographed TLC plate was examined for scytalone or melanin precursors after being sprayed with FeCl3, which enables visualization of the compounds as light-brown spots under visible light or as bright-yellow fluorescent spots under 365-nm UV and which become black after being quenched under 254-nm UV (49).
For LC/MS analysis, the intermediate metabolites or shunt products described above were scraped from the TLC plate and analyzed at room temperature with Waters CapLC Micromass Q-TOF using C18 columns with the following mobile phases at a speed of 9 μl min−1: (i) 95% H2O, 5% acetonitrile, 1% formic acid and (ii) 95% acetonitrile, 5% H2O, 1% formic acid.
Bioassay of transformant and wild-type sporulation capacity and virulence.
To determine the sporulation capacity of the M. anisopliae wild-type strain BCRC35505 and the transformant strain MA05-169, 2-mm2 agar discs were excised from the margin of a fresh colony, inoculated onto 9-cm PDA plates, and incubated at 28°C for 21 days. The spores harvested by vortexing the agar blocks in sterile 0.1% Tween 80 for 2 min were filtered through Miracloth and counted under a light microscope with a hemocytometer. Each experiment was conducted using three replicates, and each replicate consisted of three agar plates. To assess the pathogenicity of M. anisopliae BCRC35505 and M. anisopliae MA05-169 for the diamondback moth, the synchronized 3rd-instar larvae were dipped in the spore suspension (2 × 107 conidia ml−1) for 30 s, and the excess moisture was absorbed with filter paper. The inoculated larvae were reared on fresh canola leaflets and incubated at 27°C for 3 days, and the mortality rate was determined. Experiments were repeated seven times. Each test was conducted using three replicates, with each replicate consisting of 50 larvae. Noninoculated larvae were used as negative controls. To measure the median time to death (LT50) of Plutella larvae over 24 to 72 h caused by wild-type or transformant M. anisopliae, experiments were performed with three replicates, with each replicate containing 50 larvae. Time-mortality data were subjected to Probit regression analyses with a 95% significance limit.
To determine the effect of UV radiation on the virulence of M. anisopliae MA05-169 and M. anisopliae BCRC35505, 5 ml of the spore suspension (2 × 107 conidia ml−1 in 5.5-cm plastic petri dishes) was exposed to UV radiation at 23.4 mJ cm−2 and 46.8 mJ cm−2, respectively. The spore suspensions with or without UV radiation were used as inocula to test the pathogenicity for the diamondback moth larvae according to the protocol described above. The experiment was repeated three times with three replicates, and each replicate contained 50 larvae.
For confocal laser scanning microscopy, the moth larvae that were killed by the fungi were surface disinfected with 1% sodium hypochlorite for 1 min, excessive moisture was absorbed, and the larvae were incubated in a moistened petri dish at 100% relative humidity (RH) for 7 days. The cadavers were embedded in Tissue-Tek OCT, frozen to −40°C, and sectioned at −20°C with a Cryotome FE (Thermo) at 20-μm thickness. The sections were stained with 0.1% Calcofluor (Sigma) for 10 min, destained with ddH2O, and examined with a confocal scanning light microscope (CTR 6500; Leica, Germany) for blue light (420- to 460-nm wavelength) emission excited by UV at a 250-nm wavelength and green light (510- to 550-nm wavelength) emission from green fluorescent protein (GFP) excited by blue light at a 488-nm wavelength.
Stress tolerance assay.
To determine the effect of UV on the viability and spore germination of M. anisopliae MA05-169 and M. anisopliae BCRC35505, a UV radiation source (Philips UV-B TL 20W/12RS; Holland) without a filter barrier was used; it consisted primarily of UV-B and a trace of UV-C and UV-A. The spectrum and energy of UV radiation were measured using a USB2000 Miniature Fiber Optic Spectrophotometer (Ocean Optics). Approximately 200 μl of each of the M. anisopliae MA05-169 and M. anisopliae BCRC35505 spore suspensions (2 × 107 conidia ml−1) was evenly spread onto PDA plates without hygromycin B (HygB) and received UV radiation dosages of 0 mJ cm−2, 23.4 mJ cm−2, and 46.8 mJ cm−2, with each treatment performed with three replicates. After exposure to UV, the plates were incubated for 6, 10, 23, 28, 48, and 72 h at 25°C in the dark. The agar plate was covered with glass and examined under light microscopy at ×400 magnification. A conidium with a germ tube that exceeded its length was counted as germinated. In each of the three replicates tested, at least 100 spores were counted. The germination rate was calculated based on the summation of germinated and nongerminated spores.
To determine the effect of temperature on the growth of M. anisopliae MA05-169 or M. anisopliae BCRC35505, approximately 1.5-mm2 pieces of the agar blocks cut from the margin of a colony grown on PDA at 25°C for 7 days were inoculated onto PDA plates without HygB. After incubation at 10, 15, 20, 25, 30, and 35°C, the colony was measured and compared to the size of the original inoculation agar block. Each test was performed three times with three replicates, and each replicate contained five plates. Alternatively, 150 μl of a spore suspension (106 conidia ml−1) grown for 21 days was evenly smeared onto the PDA plates and incubated at 10, 15, 20, 25, 30, and 35°C. The spore germination rate was estimated as described above after incubation for 15, 24, 39, 48, 63, and 72 h.
To determine the effect of water activity (aw) (the water pressure of a liquid divided by that of pure water at the same temperature; pure distilled water has a water activity of exactly 1) on the conidial germination of M. anisopliae MA05-169 and M. anisopliae BCRC35505, the water activity of PDA was adjusted with 0, 4, 6, 10, 12, 20, 30, and 40 g of glycerol. The exact water activity after adjustment was measured using an AquaLab 3TE Water Activity Meter (Washington) and showed values of 1, 0.997, 0.988, 0.980, 0.971, 0.967, 0.938, and 0.895, respectively. The inoculated plates were incubated at 25°C, and the spore germination rate after each treatment was determined after incubation for 6, 13, 17, 24, 48, and 72 h.
Statistics.
Statistics Package for Social Science (SPSS) was used for analysis of the significant differences in the experiments described above.
RESULTS
Cloning of the melanin biosynthesis genes of A. alternata.
The genes encoding PKS, SCD, and 3HNR of A. alternata BCRC30501 were amplified by PCR using the following specific degenerate primers: KS1 and KS2, scyA and scyB, and 1,3,8-tri(A) and 1,3,8-tri(B) (see Table S1 in the supplemental material). The amplified gene products (700 bp, 250 bp, and 750 bp, respectively) were cloned and used for DIG probe synthesis. To isolate the genomic region of A. alternata that contained the gene cluster for DHN melanin biosynthesis, a fosmid library was constructed and screened using a THR and a PKS gene probe. After verification by PCR, one of the recovered clones, aaf01018E, was chosen for shotgun library construction and sequencing. The 41-kb fosmid clone harbored six genes, including the full-length open reading frames of PKS (6,480 bp; GenBank accession number HM486910) and THR (1,102 bp; GenBank accession number HM 486909) and four hypothetical protein-encoding genes. The anticipated SCD gene, previously reported to be located between the PKS and THR genes (29), was absent from aaf01018E. However, five independent fosmid clones that showed positive signals for the SCD gene probe by Southern blotting were obtained. One of the clones, aaf01005I10, was selected for further sequence analysis. The size of the full-length SCD gene was determined to be 773 bp (GenBank accession number HM 486908) (Fig. 1B to D).
Characteristics of M. anisopliae transformants.
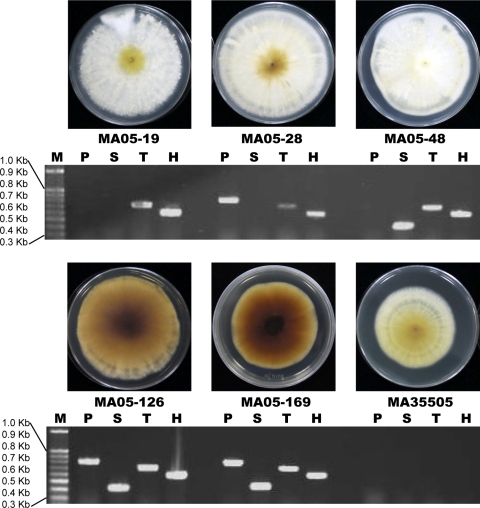
The shuttle vectors used to transform A. tumefaciens EHA105 competent cells each contained only one target gene (PKS, SCD, or THR) and one selection marker (HygR) or a reporter (GFP) gene. The vectors, consisting of PKS plus GFP, SCD plus GFP, or THR plus HygR, were used to transform M. anisopliae. The putative transformants, which usually grew faster on selective medium than wild-type controls, were subcultured for further molecular verification. Of the 153 total putative transformants that were selected, 79 were positive. Twenty-six transformants harbored a complete set of PKS, SCD, and THR genes, in contrast to 22 transformants that contained two genes (20 with PKS plus THR and 2 with SCD plus THR) and 33 transformants that contained one gene (27 with THR and 3 each with SCD or PKS). Phenotypically, there was a sharp distinction among the transformants; however, a deep-brown to black colony phenotype was observed only in the transformants containing the three crucial DHN melanin synthesis genes (Fig. 2). One of the transformants, M. anisopliae MA05-169, possessed a higher growth rate but a distinctly lower sporulation capacity than the wild-type strain, M. anisopliae BCRC35505 (1.53 × 107 ± 0.24 × 107 versus 19.33 × 107 ± 4.06 × 107/plate with significance at 95% confidence) (Fig. 1E) and was therefore chosen for subsequent study. The M. anisopliae MA05-169 colonies were characterized by a velvety brown color changing to deep brown and then to black, with olive-brown to deep-brown droplets appearing on aerial mycelium with age. In contrast, M. anisopliae BCRC35505 colonies were white and floccose, changing to yellowish to pale orange, without the deep olive-brown exudation droplets developing with age (Fig. 1E). The conidia and mycelia of M. anisopliae MA05-169 were pale brown, and the culture filtrate was deep brown to black, whereas the M. anisopliae BCRC35505 conidia were hyaline or pale greenish, and the filtrate was pale yellow in PDB.
Fig. 2.
Phenotypes of M. anisopliae transformants. M, 100-bp ladder; P, PKS; S, SCD; T, THR; H, HygR; M. anisopliae BCRC05-19, -28, -48, -126, and -169, transformants; M. anisopliae BCRC35505, wild type.
PCR or Southern blot analysis revealed the presence of the integrated HygR gene (Fig. 1G and H and 2), as well as two copies of each of the PKS, SCD, and THR genes in M. anisopliae MA05-169 (Fig. 1G and Table 1). The gene transcript number and gene copy number amplified by RT-PCR (Fig. 1H) or absolute Q-PCR can be calculated by plotting against the standard curves (data not shown). The data indicated that all of the incorporated genes were expressed in the transformant, M. anisopliae MA05-169 (Table 1).
Table 1.
Transcript copy numbers and gene copy numbers of melanin biosynthesis genes, PKS, SCD, and THR, in A. alternata BCRC30501, M. anisopliae BCRC35505 (wild type) and M. anisopliae MA05-169 (transformant) estimated by absolute Q-PCR and Southern blotting, respectively
| Strain | Transcript copy no.a |
Gene copy no. |
||||
|---|---|---|---|---|---|---|
| PKS | SCD | THR | PKSb | SCDc | THRc | |
| A. alternata BCRC30501 | 22.2 × 104 | 1.9 × 104 | 4.5 × 104 | 1 | 1 | 1 |
| M. anisopliae BCRC35505 | NA | NA | NA | 0 | 0 | 0 |
| M. anisopliae MA05-169 | 4.0 × 104 | 7.9 × 104 | 1.2 × 106 | 2 | 2 | 2 |
NA, not applicable.
gDNAs digested with PstI.
gDNAs digested with HindIII.
Verification of melanin in transformants.
The colony colors of wild-type M. anisopliae BCRC35505 grown on PDA with or without addition of carpropamid, a competitive inhibitor of SCD (59), were very similar. In contrast, the transformant, M. anisopliae MA05-169, exhibited a drastic color change when grown on PDA without carpropamid (deep brown to black) compared to growth on PDA with carpropamid (pale light brown) (Fig. 1F). The change in colony phenotype indicated that the induced melanin biosynthesis pathway in M. anisopliae BCRC05-169 was partly blocked by carpropamid.
In TLC chromatography, extracts of C. lagenarium 104-T (wild type), C. lagenarium 8015 (SCD−), A. alternata BCRC30501, and M. anisopliae MA05-169, but not BCRC35505, grown on PDA treated with carpropamid displayed scytalone, unidentified melanin intermediate precursors, or shunt metabolites. The three strains exhibited brown spots under visible light after being stained with FeCl3 and quenched spots under 254-nm UV irradiation (see Fig. S2A and B in the supplemental material). The brown spots indicated the presence of scytalone, as reported previously (59). The additional nine spots (compounds) revealed by FeCl3 staining or under 254-nm UV irradiation were of unknown nature (see Fig. S2A and B in the supplemental material). LC/MS analysis of the extracts from agar blocks excised from M. anisopliae MA05-169 and C. lagenarium 104-T culture plates without carpropamid indicated the presence of four melanin precursors, scytalone, vermelone, 1,3,6,8-THN, and 1,3,8-trihydroxynaphthalene (1,3,8-THN), and two shunt metabolites, 2-hydroxyjugulone (2-HJ) and flaviolin, whereas neither 1,8-DHN nor juglone was observed. Additionally, neither melanin precursors nor shunt products were found in M. anisopliae BCRC35505 (Table 2; see Fig. S3 in the supplemental material).
Table 2.
LC/MS analysis of DHN melanin or its precursors in M. anisopliae BCRC35505, M. anisopliae MA05-169, and C. lagenarium 104-T
| Compound | Samplea |
||||
|---|---|---|---|---|---|
| MF | MW | CL | W | T | |
| Scytalone | C10H10O4 | 194.184 | + | − | + |
| Vermelone | C10H10O3 | 178.186 | + | − | + |
| 1,3,6,8-THN | C10H8O4 | 192.168 | + | − | + |
| 1,3,8-THN | C10H8O3 | 176.169 | + | − | + |
| 1,8-DHN | C10H8O2 | 160.172 | − | − | − |
| 2-Hydroxyjuglone | C10H6O4 | 190.15224 | + | − | + |
| Flaviolin | C10H6O5 | 206.15164 | + | − | + |
| Juglone | C10H6O3 | 174.15284 | − | − | − |
MF, molecular formula; MW, molecular weight; CL. C. lagenarium 104-T; W. wild- type (M. anisopliae BCRC35505); T, transformant (M. anisopliae MA05-169); +, present; −, absent.
Spectrophotometric analysis of melanins.
UV-Vis (200 to 500 nm) spectrophotometric analysis revealed that melanin from M. anisopliae MA05-169 and DHN melanin shared a common absorption peak at 210 nm when A. alternata (BCRC30501) DHN melanin or DOPA melanin was used as a standard (see Fig. S4A in the supplemental material). The absorption values of DOPA melanin and melanin from BCRC30501 and M. anisopliae MA05-169 at different UV-Vis wavelengths were converted into log algorithms, revealing that the slopes of the plotted regression lines for the three types of melanins were parallel and had values of −0.00242, −0.00199, and −0.00246, respectively (see Fig. S4B in the supplemental material). The range of values shown here was comparable to values shown by melanins from other organisms (17). The signature G value, 2.0037, derived from the EPR spectra of DOPA melanin, was almost identical to that of DHN melanin from M. anisopliae MA05-169 and BCRC30501, suggesting the existence of an unpaired free electron radical with a G value of 2.0023 (see Fig. S4C in the supplemental material). Furthermore, the FTIR spectra of DOPA melanin and M. anisopliae MA05-169 DHN melanin also exhibited very similar patterns, with an absorption peak at 3.2 μm, which likely represents an NH3 or OH bond, and an absorption peak at 5.8 μm, which might represent a carbonyl group (C O) or C
O) or C C bond (see Fig. S4D in the supplemental material). Additionally, there was a unique absorption peak in M. anisopliae MA05-169 melanin at 3.3 μm, which might represent a CH3 or CH2 functional group (see Fig. S4D in the supplemental material).
C bond (see Fig. S4D in the supplemental material). Additionally, there was a unique absorption peak in M. anisopliae MA05-169 melanin at 3.3 μm, which might represent a CH3 or CH2 functional group (see Fig. S4D in the supplemental material).
Transformant virulence assay.
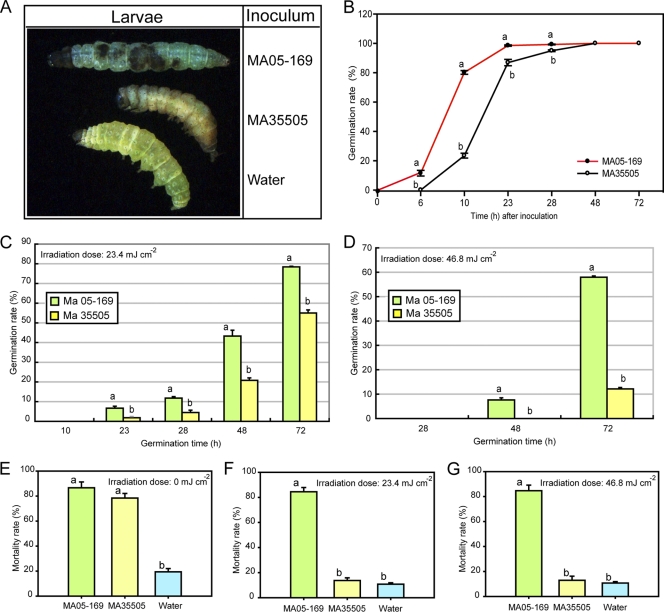
As M. anisopliae BCRC35505 was originally isolated from a diamondback moth (P. xylostella), the insect species was selected as the target host for determining its susceptibility to wild-type M. anisopliae BCRC35505 and transformant M. anisopliae MA05-169. The virulence assay is based on the host mortality rate and was repeated seven times. Both M. anisopliae BCRC35505 and MA05-169 infected the host insect normally. Diamondback moth larvae infected with wild-type M. anisopliae BCRC35505 spores exhibited a change in cuticle color after the second day, turning from green to yellowish and eventually to brown before succumbing to death (Fig. 3A). In contrast, larvae infected with spores from M. anisopliae MA05-169 often displayed distinct black patches on the cuticle after the second day (Fig. 3A). Both M. anisopliae MA05-169 and M. anisopliae BCRC35505 sporulated on the infected and dead larvae when they were incubated in moistened petri dishes, although the former exhibited less sporulation than the latter (data not shown). Additional measurement of the mortality rate over time (24, 40, 48, 65, and 72 h) indicated that the transformant had a reduced LT50 (43.46 h) compared to wild-type M. anisopliae BCRC35505 (56.05 h), with significance at the 95% confidence level of 44.01 to 48.88 and 54.08 to 58.16, respectively. The mortality of diamondback larvae was 19.4% when a water solution was used as the inoculum. When spore suspensions were used to inoculate moth larvae, the mortality caused by the transformant spores was higher than that caused by the wild-type spores (86.7% versus 78.4%), but the difference was not statistically significant (Fig. 3E). However, subjecting the spore suspensions to UV-B radiation at dosages of 23.4 mJ cm−2 or 46.8 mJ cm−2 prior to inoculation greatly enhanced the difference in virulence between M. anisopliae MA05-169 and M. anisopliae BCRC35505 strains. The transformant was more tolerant of UV-B, and hence, it was more virulent than the wild type. The natural mortality of larvae inoculated with water solution as a negative control was 10.7%. After exposure to a dosage of 23.4 mJ cm−2 of UV-B, the transformant caused 84.4% mortality versus 13.7% caused by the wild type; at a dosage of 46.8 mJ cm−2 of UV-B, the level of larval mortality was 84.7% caused by the transformant versus 12.9% caused by the wild type (Fig. 3F and G). Under confocal laser scanning microscopy, calcofluor-stained sections of larvae infected with M. anisopliae BCRC35505 or M. anisopliae MA05-169 were observed as blue fluorescence in the larval hemocoel (data not shown). These results were anticipated because calcofluor incorporates into glucan or chitin of the fungal cell wall no matter what type of fungus infects the larvae. However, green fluorescence was visible only in the hemocoel of insects infected with the transformant, M. anisopliae MA05-169, because its hyphae express a GFP gene (data not shown).
Fig. 3.
Effects of UV-B irradiation on the conidial germination rate and infectivity of the M. anisopliae transformant (MA05-169) and wild type (BCRC35505; MA35505) on 3rd-instar larvae of the diamondback moth. (A) Larvae showing distinctive black patches on the cuticles of a carcass inoculated with M. anisopliae MA05-169 versus yellow or light brown caused by M. anisopliae BCRC35505 and greenish caused by water. (B) Germination rates of M. anisopliae BCRC35505 and M. anisopliae MA05-169 at different time intervals without UV-B irradiation. (C and D) Germination rates of M. anisopliae BCRC35505 and M. anisopliae MA05-169 at different time intervals following exposure to UV-B at dosages of 23.4 mJ cm−2 (C) and 46.8 mJ cm−2 (D), respectively. (E to G) Mortality rates of 3rd-instar larvae infected by M. anisopliae BCRC35505 and M. anisopliae MA05-169 without (E) or with UV-B exposure at a dosage of 23.4 mJ cm−2 (F) or 46.8 mJ cm−2 (G), using water as a negative control. The error bars indicate standard deviations. Different characters above each point or bar represent significant differences at a P value of <0.05.
Transformant antistress assay.
Of the whole solar UV spectrum, UV-B radiation is considered to be the most deleterious to organisms (26). In the absence of UV-B irradiation, the transformant M. anisopliae MA05-169 not only grew faster than the wild-type M. anisopliae BCRC35505 strain but also germinated faster (Fig. 1E and 3B). After incubation for 10 h, the germination rate of M. anisopliae MA05-169 had already reached 80%, whereas only 20% of M. anisopliae BCRC35505 spores had germinated (Fig. 3B). Extending the incubation time to 48 h diminished the difference between the two strains, and the germination rates of both strains approached 100% (Fig. 3B). However, a drastic effect on the germination rate was observed when the spores of both strains were exposed to UV-B radiation at a dosage of 23.4 mJ cm−2 (Fig. 3C). Germination of both M. anisopliae MA05-169 and M. anisopliae BCRC35505 was completely retarded at 10 h, reaching only <7% for the transformant and <2% for the wild type after 23 h of incubation compared to their nonirradiated counterparts (Fig. 3C). However, with extended incubation times, the difference in germination rates between the two strains became more obvious; at 48 h, 43% of M. anisopliae MA05-169 spores had germinated, whereas only 21% of M. anisopliae BCRC35505 spores had done so; at 72 h, 79% of M. anisopliae MA05-169 and 55% of M. anisopliae BCRC35505 spores had germinated (Fig. 3C). Exposure of spores to an even higher dose of UV-B radiation (46.8 mJ cm−2) further underscored the superior germination capability of the transformant, which displayed an 8-fold-higher rate at 48 h and a 4.8-fold-higher rate at 72 h of incubation (58% versus 12%) than the wild type (Fig. 3D).
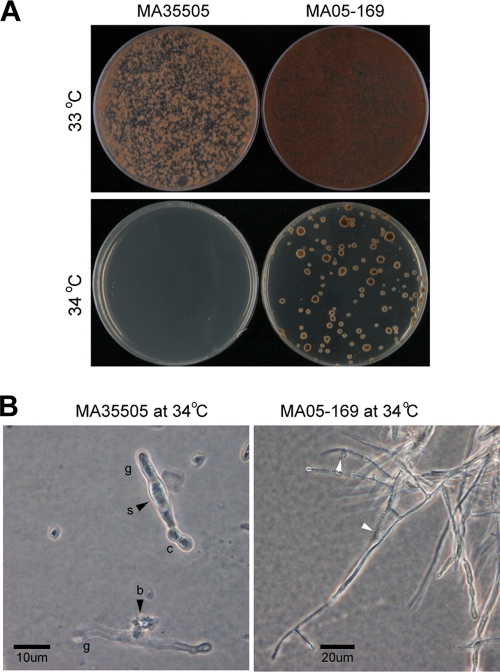
Over a range of temperatures tested, the optimal growth temperature for both M. anisopliae BCRC35505 and M. anisopliae MA05-169 was 25°C, followed by, in descending order, 30°C and 20°C (see Table S2 in the supplemental material). At 10 to 15°C, the growth of both strains was greatly slowed, although M. anisopliae MA05-169 showed a significant growth advantage over M. anisopliae BCRC35505 during the initial 7 to 9 days. However, colony growth of both strains halted at 35°C (see Table S2 in the supplemental material). With regard to the germination rate, except at 10°C, no difference between the strains was observed over the range of 10 to 30°C after incubation for 48 h (see Table S3 in the supplemental material). However, at 35°C, during the first 6 to 15 h, a drastic to moderate difference in the germination rate was observed, but the difference diminished with incubation time to 48 h (see Table S3 in the supplemental material). In addition, at 33°C, the colonies formed by M. anisopliae BCRC35505 were tiny and restricted, even though the germinated conidia continued to grow when incubated for 25 days; in contrast, M. anisopliae MA05-169 showed moderate growth under the same culture conditions (Fig. 4A). At 34°C, the germinated conidia of M. anisopliae BCRC35505 failed to produce visible colonies, whereas the germinated conidia of M. anisopliae MA05-169 formed numerous tiny and restricted colonies (Fig. 4B). Under light microscopy at 34 to 35°C, although the germinated conidia of M. anisopliae MA05-169 developed slowly, they exhibited normal mycelial morphology, in contrast to M. anisopliae BCRC35505, in which the germinated conidia were often crooked, swollen, or even burst (Fig. 4B).
Fig. 4.
Colony and mycelium traits of the M. anisopliae wild type (MA35505) and transformant (MA05-169) on PDA. (A) Incubation on PDA at 33°C and 34°C for 25 days; at 34°C, growth of both strains was restricted, particularly M. anisopliae BCRC35505. (B) Conidia (c) of M. anisopliae BCRC35505 on PDA at 34°C for 7 days germinated, but most germ tubes (g) were abnormal, swollen (s), or burst (b) (arrowheads); in contrast, the conidia of M. anisopliae MA05-169 mostly exhibited normal growth, though occasionally the tip dissolved (arrowheads).
At aw values ranging from 0.996 (equal to 99.6% RH) to 0.971 (equal to 97.1% RH), in the initial 6 h of incubation, M. anisopliae MA05-169 germinated more rapidly than M. anisopliae BCRC35505. A significant difference between the strains still existed at aw values of 0.989 to 0.971 at 17 h but diminished at 24 h. At aw values of 0.996 to 0.971, both strains showed almost equal germination rates at 24 h. However, it is worth noting that at an aw value of 0.967, no germination occurred in either strain prior to 24 h, whereas after 48 to 72 h, M. anisopliae MA05-169 displayed a moderate germination rate of 32.7% to 42.9%, which was significantly higher than that of M. anisopliae BCRC35505, which was 0 to 4.3% (see Table S4 in the supplemental material).
DISCUSSION
The biocontrol of insect pests by entomopathogenic fungi in field tests has often failed, particularly under conditions of environmental stress, such as solar UV radiation, desiccation, or temperature extremes. To overcome these problems, the general strategy has been to obtain entomopathogens of target or nontarget insects from different geographic regions or to induce fungal variants by physical or chemical mutagenesis and then select the mutants with the desired genetic characters by bioassay (5, 7, 36, 42, 43). However, under such circumstances, the underlying mechanisms that contribute to the antistress capacity of the fungi mostly remain undefined. In the present study, we took a more direct approach by cloning the DHN melanin biosynthesis genes PKS, SCD, and THR from A. alternata BCRC30501 (29) and successfully expressing them in M. anisopliae BCRC35505 by ATMT. The incorporation of a complete set of DHN melanin biosynthesis-encoding genes into the genome of wild-type M. anisopliae was a prerequisite for inducing the melanization phenotype (Fig. 2). However, the melanization intensity varied among transformants, such as that observed in M. anisopliae MA05-169 versus M. anisopliae MA05-126 (Fig. 2). Whether the colony phenotype, anti-stress tolerance, and virulence of the transformants were also correlated with the copy number of the integrated DHN melanin genes remains unclear and needs to be clarified.
A. tumefaciens-mediated transformation has opened up the possibility for DNA transfer across various kingdoms of organisms (11, 35). Taking advantage of its simplicity and efficiency, many successful transformations of M. anisopliae mediated by A. tumefaciens have been achieved, mostly with benomyl or ammonium glufosinate as selection markers (4, 16, 18, 20). These transformations have proven that M. anisopliae has innate resistance to HygB, Geneticin, oligomycin, and phleomycin, which can be barriers to successful transformation (4, 20). Nevertheless, depending on the strain of M. anisopliae, the resistance to HygB varies. For example, one M. anisopliae strain isolated from the rhinoceros beetle (Allomyrina dithotomus) showed extremely high resistance to HygB (1,550 μg ml−1), whereas the M. anisopliae BCRC35505 strain used for this study, which was originally isolated from the diamondback moth (P. xylostella), exhibited moderate tolerance for HygB at concentrations ranging from 50 to 300 μg ml−1. Based on preliminary tests, many putative transformants that showed higher growth rates at a concentration of 100 μg ml−1 were selected and subjected to further molecular verification (Fig. 2).
LC/MS analysis of the melanin extracts from M. anisopliae MA05-169 and C. lagenarium 104-T showed that, except for 1,8-DHN, DHN melanin intermediate precursors (1,3,6,8-THN, scytalone, 1,3,8-THN, and vermelone) and two shunt products (flaviolin and 2-HJ) were accessed. Although the dehydration of vermelone leads to the formation of 1,8-DHN, this molecule was absent in the present investigation. The absence of 1,8-DHN may be due to its instability, as it is easily oxidized and degraded when released extracellularly (8). Alternatively, perhaps the released 1,8-DHN was transient and quickly oxidized by phenoloxidase and converted to melanins (3, 8, 60). With regard to the other precursors, even without the blockage of enzymatic activity at specific steps in the biosynthetic pathway by inhibitors, 1,3,6,8-THN and 1,3,8-THN have been found to auto-oxidize into flaviolin and 2-HJ, respectively (3, 8, 31). Therefore, the presence of these shunt products in the present investigation are accounted for. Moreover, our results indicated that only incorporation of PKS, SCD, and THR cloned from A. alternata into the genome of M. anisopliae allowed proper function by promoting the synthesis of the desired DHN melanins. This makes sense based on the recent enzymatic kinetic and protein crystallographic studies on two naphthol reductases (4HNR and 3HNR) and SCD (32, 56, 64). The two enzymes 4HNR and 3HNR have been shown to catalyze the reactions of two physiological substrates, 1,3,6,8-THN and 1,38-THN, but the substrate preferences and Kcat/Km ratios vary (32, 56). Likewise, the capability of SCD to catalyze the reaction of the two physiological substrates, scytalone and vermelone, has been unequivocally proven (64).
Laccase (p-diphenol oxidase) has been suggested to be involved in the polymerization of 1,8-DHN in the last step of DHN melanin synthesis (3, 55). However, this process may not apply to all fungi that produce DHN melanin. For instance, the laccase gene, LAC1, of C. lagenarium has been cloned, characterized, and shown to be the only copy of the laccase gene by Southern blotting. However, mutants in which the laccase gene is disrupted are still able to synthesize DHN melanin, as well as maintain pathogenicity (58). In M. anisopliae, the laccase gene is expressed during isotopic growth and is involved in pigmentation, tolerance for abiotic stress, and virulence. However, pigment production is not blocked by tricyclazole, kojic acid, or glufosinate, suggesting that the pigment in M. anisopliae is not related to DHN melanin or carotenoid pigment (19, 57). This finding is further supported by the findings of Rangel et al. (43); they demonstrated no scytalone dehydratase activity, an essential process for DHN melanin synthesis, in 4-day-old cytoplasmic extracts of M. anisopliae ARSEF 2575. Our TLC experiments also showed the absence of scytalone accumulation in wild-type M. anisopliae after treatment with carpropamid, a fungicide that specifically inhibits the activity of scytalone dehydratase. Taken together, these studies indicated that wild-type M. anisopliae lacked the machinery for DHN melanin synthesis.
DOPA and DHN melanin are polymers of oxidized indolic or phenolic compounds, and the former are occasionally incorporated with thiosulfate residues. Undoubtedly, the chemical and physical properties of DOPA and DHN melanin are very different. Nevertheless, due to enormous molecular compositions, the exact molecular structures of both melanins remain undefined but share some nonspecific traits. (3, 22, 37). For instance, both possess stable free-radical and common functional groups, such as hydroxyl (OH) and carbonyl (C O) groups. These characteristics were observed in the spectra of UV-Vis, EPR, and FTIR and, in a broad sense, could be used as fingerprints to determine whether the compounds investigated contained melanin. To verify this viewpoint, an authentic melanin extracted from A. alternata was simultaneously subjected to the same chemical and physical spectral analysis. The results further demonstrated that M. anisopliae MA05-169 possessed the same characteristic melanins as A. alternata (see Fig. S4 in the supplemental material).
O) groups. These characteristics were observed in the spectra of UV-Vis, EPR, and FTIR and, in a broad sense, could be used as fingerprints to determine whether the compounds investigated contained melanin. To verify this viewpoint, an authentic melanin extracted from A. alternata was simultaneously subjected to the same chemical and physical spectral analysis. The results further demonstrated that M. anisopliae MA05-169 possessed the same characteristic melanins as A. alternata (see Fig. S4 in the supplemental material).
The present data clearly indicated that the increase in brown to deep-brown pigmentation of the transformant, M. anisopliae MA05-169, was due to the de novo synthesis of melanin as a result of the introduction of three key genes (PKS, SCD, and THR) from the A. alternata melanin biosynthesis pathway into M. anisopliae (Fig. 1). Moreover, in addition to an increase in the colony growth rate, the transformant also showed changes in spore germination rates and resistance to heat, desiccation, and UV-B. However, the transformant sporulated significantly less than the wild type. It is reasonable to predict that melanin production could contribute to increased heat, desiccation, and UV-B tolerance, but the possibility that one or more of these characteristics, as well as increased colony growth and spore germination rates, may also be due to the insertion of any one of the three genes, promoters, terminators, selection markers, or reporter genes into the open reading frames of functional genes or to a change in the primary or secondary metabolic or signal transduction pathways cannot be ruled out (39). The additional spots shown on the TLC plate when exposed to 254-nm UV might represent additional melanin intermediate precursors or shunt metabolites, in addition to those detected by LC/MS (Table 2; see Fig. S2 in the supplemental material). However, the nature of these compounds remains unclear, and extraction of an adequate quantity of intermediate metabolites or shunt products from the M. anisopliae MA05-169 or C. lagenarium strains and LC/MS or high-performance liquid chromatography (HPLC) analysis might aid in resolving the identities of these compounds.
Ambient temperature extremes compounded with UV radiation can influence the pathogenesis of entomopathogens, particularly in the tropical and subtropical regions (5, 6). For instance, infection of the desert locust Schistocerca gregaria with M. anisopliae induced a mortality rate of 98 to 100% at 25 to 30°C, 40 to 100% at 35°C, and 0% at 40°C (30). Furthermore, the desert locust and other insects could offset, retard, or eradicate conidial germination and infection of entomopathogens through basking or fever behaviors (9). More recent studies have shown that M. anisopliae conidia exposed to the full spectrum of solar UV radiation for 4 h (weighted dosage, equivalent to ca. 7 to 9 kJ m−2) have reduced relative culturability by approximately 30% for strain ARSEF 324 and 100% for strains ARSEF 23 and 2575. UV-A exposure also exerts negative effects on the relative culturability of conidia and conidial germination, but it is not as pronounced as exposure to the full-spectrum solar radiation (5, 6). In our studies, under UV-B stress, the transformant M. anisopliae MA05-169 germinated much faster and survived much better than the wild-type M. anisopliae BCRC35505 strain, particularly in the early growth stages. The germlings of the transformant developed normally even at 35°C. These traits together might aid the transformant in counteracting the basking or fever behavior of its insect host and also facilitate infection and colonization of the insect host under harsh conditions with intermittent periods conducive to colonization. This hypothesis is supported by the fact that melanins can absorb harmful solar UV radiation and transform energy into harmless heat through a process called ultrafast internal conversion (13).
Melanized C. neoformans at stationary phase is more resistant to heat (42 to 47°C) and cold (−20°C) treatment, but the magnitudes of some of the survival differences are small (44). Currently, the underlying mechanism for the increased resistance of melanized cells to heat or cold is still unknown. A potential explanation is that the accumulated melanin in the cell wall or cytoplasm may help absorb energy and transform and dissipate it slowly as heat. Similarly, melanins may promote heat absorption and dissipate the heat to compensate for the cold stress. Additionally, the melanized cells are generally thicker and more rigid and accumulate larger amounts of osmolites, such as glycerol, than unmelanized cells or albino mutants (63). The osmolites may serve as cold protectants.
Relative humidity is crucial for microbial biocontrol agents to germinate, spread, and infect their insect hosts. For example, lower humidity usually inhibits the ability of M. anisopliae to control the rice green leafhopper, Nephotettix virescens. However, UV-induced mutants able to grow at reduced water activity have shown higher virulence than the parental strains (34). Alternatively, using an emulsifiable adjuvant oil formulation as a carrier to deliver M. anisopliae to control the cattle tick Boophilus microplus resulted in a higher conidial germination rate and a lower average host survival time than a water-based formulation (41). In the present study, the melanized transformant germinated faster and possessed a higher accumulated germination rate, particularly at lower water activity (aw = 0.967), exhibiting a 10- to 30-fold difference compared to the wild type. Although the melanized transformant had a tougher and more rigid cell wall, the cell wall also showed increased turgor pressure. The melanin lining between the inner cell wall and plasma membrane restricts the passage of molecules larger than those of water, such as glycerol. Therefore, in the melanized cells, more and more osmolites accumulate and become hypertonic and will more easily imbibe water from the surrounding environment (15, 63). Additionally, melanins have been shown to be a naturally occurring cation exchange material. The purified melanin can take up a large amount of water, similar to synthetic cationic resin (62). As a consequence, the melanized conidia, unlike unmelanized conidia, may have increased desiccation resistance and absorb more water to facilitate germination.
In plant pathogens, such as C. lagenarium and M. grisea, melanin accumulated between the plasma membrane and the cell wall of an appressorium plays a crucial role in maintaining cellular integrity and osmolarity and in creating turgor pressure. The mechanical force and the secreted lytic enzymes facilitate peg penetration of the host plant epidermal cell for successful infection and colonization (63). In the present study, upon inoculation of the diamondback moth larvae with the transformant M. anisopliae MA05-169, conidia attached to the cuticle, germinated, and produced distinct globose to subglobose rugulose-walled appressoria (data not shown). Although we know that the integrated melanin biosynthesis genes functioned properly, directing the synthesis of melanin and its precursors in the cytosol and perhaps also secreting the melanin into the fibrillar and outermost cell wall, we do not know the exact distribution of the synthesized melanin or the role it plays during the infection process. To clarify these issues, measurement of the appressorium's turgor pressure and immunocytochemical studies using a melanin-specific antibody will be necessary (51, 63).
In the human pathogen C. neoformans, the cell wall composed of melanized conidia is much tougher, and its surface topology is different from that of an albino mutant. Conidial ghosts remain visible even after they are subjected to strong alkali, acid, or organic solvent treatments. These traits may help conceal the fungus from the host immune system, make the fungus more resistant to attack by free radicals generated from oxygen or nitrogen bursts, or prevent the fungus from being engulfed by host macrophages (25, 48). In addition, melanins are radical scavengers, negatively charged, and hydrophobic and act synergistically to protect animal fungal pathogens from attack by the host defense response (3). Whether the transformant M. anisopliae MA05-169 displays similar attributes during infection of its insect hosts is worthy of further study. The cuticles of the diamondback moth larvae that succumbed to death exhibited black patches after infection with the transformant, but not with the wild type. Insects usually activate polyphenol oxidase activity and melanize their cuticles when wounded or infected with microbial pathogens to heal wounds or prevent microbial intrusion (2). Therefore, the black patches on the larval cuticle may be formed by M. anisopliae MA05-169 alone or simultaneously by the oxidization of DOPA or similar compounds by polyphenol oxidases of the insect itself (2).
Interestingly, the genetically transformed M. anisopliae ARSEF1080 has been found to overexpress multiple copies of the homologous toxic protease gene Pr1 under the control of a constitutive promoter in the hemolymph of infected tobacco hornworms (Manduca sexta); Pr1, in addition to exhibiting insecticidal activity, also activated the prophenoloxidase system, which melanized the surrounding tissues and reduced survival by 25% and food consumption by 40% (52). In light of this finding, to increase insect mortality and decrease the food consumption rate, the incorporation of additional protease, chitinase, or ecdysis hormone-regulating genes into the M. anisopliae MA05-169 genome might further enhance virulence and promote biocontrol efficacy.
The release of recombinant M. anisopliae harboring either a green fluorescent protein gene (GFP) alone or additional protease (Pr1) genes into a cabbage field to monitor its survival over time has been conducted (23, 53). The study indicated that the recombinant fungi are genetically stable for at least 1 year, do not interfere with the culturable indigenous fungal microflora, and are not transmitted to nontarget insects. Unexpectedly, it was discovered that the inner rhizosphere can serve as a reservoir for maintaining the recombinants (23). Therefore, rhizosphere competence might be considered a factor for selecting biocontrol agents (53). Likewise, the GTP, PKS, SCD, or THR gene could be used to monitor the performance and fate of the transformed M. anisopliae MA05-169 strain when released into the wild for biocontrol purposes.
Repeat-induced point mutation (RIP) has been reported in several fungi, including Neurospora crassa (10, 46), M. grisea (24), and Podospora anserina (21), during the sexual phase of the life cycle, and RIP has been suggested to maintain the stability of the genome, as well as species diversity for evolution (46). Recently, Fusarium circinatum (teleomorph, Gibberella circinata), an important pathogen of pine, was transformed with HygR via ATMT; the transfer DNA randomly integrated into the genome and remained stable through mitotic and meiotic cell division (12). Given the similar methodology used here with the transformant M. anisopliae MA05-169, the enhanced antistress and virulence capacity due to the introduction of the DHN melanin synthesis gene by ATMT will likely be sustained (33, 63).
In conclusion, the genetically engineered mycoinsecticidal fungus M. anisopliae is able to produce melanin and an array of melanin intermediates. The transformant was more tolerant of UV-B radiation and extreme temperatures and had lower water activity than the wild type. This antistress capacity may well endow the transformant with superior survival ability, resulting in its enhanced control of agricultural insect pests, as well as notorious disease vectors, in harsh environments (9). The cloned melanin biosynthesis genes might also be used to transform other microbial biocontrol agents or crops to improve their ecological fitness. To our knowledge, this is the first study to demonstrate the metabolic engineering of the melanin biosynthetic pathway into a microbial biocontrol agent to enhance its tolerance for environmental stresses and to improve its virulence.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support from the National Science Council (NSC 97-2313-B-002-034-MY3 to S.S.T.) and the Council of Agriculture [98AS-9.3.1-BQ-B2(1) to S.S.T.], Executive Yuan, Taiwan.
We thank Y. Kubo at Kyoto Prefecture University for providing C. lagenarium 8015 and 104-T and W. H. Hseu at National Chung Hsing University, Taichung, Taiwan, for providing the pCAMBIA 1300 plasmid.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 13 May 2011.
REFERENCES
- 1. Al-Samarrai T. H., Schmid J. 2000. A simple method for extraction of fungal genomic DNA. Lett. Appl. Microbiol. 30:53–56 [DOI] [PubMed] [Google Scholar]
- 2. Ashida M., Brey P. T. 1995. Role of the integument in insect defense: pro-phenol oxidase cascade in the cuticular matrix. Proc. Natl. Acad. Sci. U. S. A. 92:10698–10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell A. A., Wheeler M. H. 1986. Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24:411–451 [Google Scholar]
- 4. Bernier L., Cooper R. M., Charnley A. K., Clarkson J. M. 1989. Transformation of the entomopathogenic fungus Metarhizium anisopliae to benomyl resistance. FEMS Microbiol. Lett. 60:261–265 [Google Scholar]
- 5. Braga G. U. L., Flint S. D., Miller C. D., Anderson A. J., Roberts D. W. 2001. Both solar UVA and UVB radiation impair conidial culturability and delay germination in the entomopathogenic fungus Metarhizium anisopliae. Photochem. Photobiol. 74:734–739 [DOI] [PubMed] [Google Scholar]
- 6. Braga G. U. L., Flint S. D., Messias C. L., Anderson A. J., Roberts D. W. 2001. Effect of UV-B on conidia and germlings of the entomopathogenic hyphomycete Metarhizium anisopliae. Mycol. Res. 105:874–882 [DOI] [PubMed] [Google Scholar]
- 7. Braga G. U., Rangel D. E., Flint S. D., Anderson A. J., Roberts D. W. 2006. Conidial pigmentation is important to tolerance against solar-simulated radiation in the entomopathogenic fungus Metarhizium anisopliae. Photochem. Photobiol. 82:418–422 [DOI] [PubMed] [Google Scholar]
- 8. Butler M. J., Day A. W. 1998. Fungal melanins: a review. Can. J. Microbiol. 44:1115–1136 [Google Scholar]
- 9. Butt T. M., Jackson C., Magan N. 2001. Introduction-fungal biological control agents: progress, problems and potential, p. 1–8 In Butt T. M., Jackson C., Magan N. (ed.), Fungi as biocontrol agents. CAB International, Wallingford, Oxon, United Kingdom [Google Scholar]
- 10. Cambareri E., Singer M. J., Selker E. U. 1991. Recurrence of repeat-induced point mutation (RIP) in Neurospora crassa. Genetics 127:699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Citovsky V., et al. 2007. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 9:9–20 [DOI] [PubMed] [Google Scholar]
- 12. Covert S. F., Kapoor P., Lee M.-H., Briley A., Nairn C. J. 2001. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol. Res. 105:259–264 [Google Scholar]
- 13. Dadachova E., et al. 2007. Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi. PLoS One 2:e457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Groot M. J., Bundock P., Hooykaas P. J. J., Beijersbergen A. G. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839–842 [DOI] [PubMed] [Google Scholar]
- 15. de Jong J. C., McCormack B. J., Smirnoff N., Talbot N. J. 1997. Glycerol generates turgor in rice blast. Nature 389:244–245 [Google Scholar]
- 16. Duarte R. T. D., et al. 2007. Development of a simple and rapid Agrobacterium tumefaciens-mediated transformation system for the entomopathogenic fungus Metarhizium anisopliae var. acridum. Lett. Appl. Microbiol. 44:248–254 [DOI] [PubMed] [Google Scholar]
- 17. Ellis D. H., Griffiths D. A. 1974. The location and analysis of melanins in the cell walls of some soil fungi. Can. J. Microbiol. 20:1379–1386 [Google Scholar]
- 18. Fang W., Bidochka M. J. 2006. Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can. J. Microbiol. 52:623–626 [DOI] [PubMed] [Google Scholar]
- 19. Fang W., Fernandes E. K., Roberts D. W., Bidochka M. J., St. Leger R. J. 2010. A laccase exclusively expressed by Metarhizium anisopliae during isotropic growth is involved in pigmentation, tolerance to abiotic stresses and virulence. Fungal Genet. Biol. 47:602–607 [DOI] [PubMed] [Google Scholar]
- 20. Goettel M. S., St. Leger R. J., et al. 1990. Pathogenicity and growth of Metarhizium anisopliae stably transformed to benomyl resistance. Curr. Genet. 17:129–132 [Google Scholar]
- 21. Hamann A., Feller F., Osiewacz H. D. 2000. The degenerate DNA transposon Pat and repeat-induced point mutation (RIP) in Podospora anserina. Mol. Gen. Genet. 263:1061–1069 [DOI] [PubMed] [Google Scholar]
- 22. Harki E., Talou T., Dargent R. 1997. Purification, characterisation and analysis of melanin extracted from Tuber melanosporum Vitt. Food Chem. 58:69–73 [Google Scholar]
- 23. Hu G., St. Leger R. J. 2002. Field studies using a recombinant mycoinsecticide (Metarhizium anisopliae) reveal that it is rhizosphere competent. Appl. Environ. Microbiol. 68:6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ikeda K.-I., et al. 2002. Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Mol. Microbiol. 45:1335–1364 [DOI] [PubMed] [Google Scholar]
- 25. Jacobson E. S. 2000. Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13:708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Javitt J. C., Hugh R. T. 1995. Cataract and latitude. Doc. Ophthalmol. 88:307–325 [DOI] [PubMed] [Google Scholar]
- 27. Kawamura C., et al. 1997. The melanin biosynthesis genes of Alternaria alternata can restore pathogenicity of the melanin-deficient mutants of Magnaporthe grisea. Mol. Plant Microbe Interact. 10:446–453 [DOI] [PubMed] [Google Scholar]
- 28. Kawamura C., Tsujimoto T., Tsuge T. 1999. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 12:59–63 [DOI] [PubMed] [Google Scholar]
- 29. Kimura N., Tsuge T. 1993. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 175:4427–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langewald J. 1997. Green Muscle user handbook. Lubilosa, Cotonou, Benin [Google Scholar]
- 31. Lazarovits G., Stoessl A. 1988. Tricyclazole induces melanin shunt products and inhibits altersolanol A accumulation by Alternaria solani. Pestic. Biochem. Physiol. 31:36–45 [Google Scholar]
- 32. Liao D.-I., Thompson J. E., Fahnestock S., Valent B., Jordan D. B. 2001. A structural account of substrate and inhibitor specificity differences between two naphthol reductases. Biochemistry 40:8696–8704 [DOI] [PubMed] [Google Scholar]
- 33. Liu Z.-Y., et al. 2002. Molecular evidence for teleomorph-anamorph connections in Cordyceps based on ITS-5.8S rDNA sequences. Mycol. Res. 106:1100–1108 [Google Scholar]
- 34. Matewele P., Trinci A. P. J., Gillespie A. T. 1994. Mutants of entomopathogenic fungi that germinate and grow at reduced water activities and reduced relative humidities are more virulent to Nephotettix virescens (green leafhopper) than the parental strains. Mycol. Res. 98:1329–1333 [Google Scholar]
- 35. Michielse C. B., Hooykaas P. J., van den Hondel C. A., Ram A. F. 2005. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48:1–17 [DOI] [PubMed] [Google Scholar]
- 36. Morley-Davies J., Moore D., Prior C. 1996. Screening of Metarhizium and Beauveria spp. conidia with exposure to simulated sunlight and a range of temperatures. Mycol. Res. 100:31–38 [Google Scholar]
- 37. Nosanchuk J. D., Casadevall A. 2003. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 5:203–223 [DOI] [PubMed] [Google Scholar]
- 38. Nosanchuk J. D., Casadevall A. 2006. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 50:3519–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nuss D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632–642 [DOI] [PubMed] [Google Scholar]
- 40. Piattelli M., Fattorusso E., Nicolaus R. A., Magno S. 1965. The structure of melanins and melanogenesis-V: ustilagomelanin. Tetrahedron 21:3229–3236 [DOI] [PubMed] [Google Scholar]
- 41. Polar P., Kairo M. T. K., Moore D., Pegram R., John S. 2005. Comparison of water, oils and emulsifiable adjuvant oils as formulating agents for Metarhizium anisopliae for use in control of Boophilus microplus. Mycopathologia 160:151–157 [DOI] [PubMed] [Google Scholar]
- 42. Rangel D. E. N., Braga G. U. L., Anderson A. J., Roberts D. W. 2005. Variability in conidial thermotolerance of Metarhizium anisopliae isolates from different geographic origins. J. Invertebr. Pathol. 88:116–125 [DOI] [PubMed] [Google Scholar]
- 43. Rangel D. E., et al. 2006. Mutants and isolates of Metarhizium anisopliae are diverse in their relationships between conidial pigmentation and stress tolerance. J. Invertebr. Pathol. 93:170–182 [DOI] [PubMed] [Google Scholar]
- 44. Rosas Å. L., Casadevall A. 1997. Melanization affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol. Lett. 153:265–272 [DOI] [PubMed] [Google Scholar]
- 45. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 46. Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. 1987. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell 51:741–752 [DOI] [PubMed] [Google Scholar]
- 47. Selvakumar P., Rajasekar S., Periasamy K., Raaman N. 2008. Isolation and characterization of melanin pigment from Pleurotus cystidiosus (telomorph of Antromycopsis macrocarpa). World J. Microbiol. Biotechnol. 24:2125–2131 [Google Scholar]
- 48. Steenbergen J. N., Shuman H. A., Casadevall A. 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc. Natl. Acad. Sci. U. S. A. 98:15245–15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stipanovic R. D., Bell A. A. 1977. Pentaketide metabolites of Verticillium dahliae. II. Accumulation of naphthol derivatives by the aberrant-melanin mutant Brm-2. Mycology 69:164–172 [PubMed] [Google Scholar]
- 50. St. Leger R. J., Charnley A. K., Cooper R. M. 1988. Production of polyphenol pigments and phenoloxidase by the entomopathogen, Metarhizium anisopliae. J. Invertebr. Pathol. 52:215–220 [Google Scholar]
- 51. St. Leger R., Joshi L., Bidochka M., Rizzo N., Roberts D. 1996. Characterization and ultrastructural localization of chitinases from Metarhizium anisopliae, M. flavoviride, and Beauveria bassiana during fungal invasion of host (Manduca sexta) cuticle. Appl. Environ. Microbiol. 62:907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. St. Leger R. J., Joshi L., Bidochka M. J., Roberts D. W. 1996. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. U. S. A. 93:6349–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. St. Leger R. J. 2008. Studies on adaptations of Metarhizium anisopliae to life in the soil. J. Invertebr. Pathol. 98:271–276 [DOI] [PubMed] [Google Scholar]
- 54. Takano Y., Kubo Y., Kawamura C., Tsuge T., Furusawa I. 1997. The Alternaria alternata melanin biosynthesis gene restores appressorial melanization and penetration of cellulose membranes in the melanin-deficient albino mutant of Colletotrichum lagenarium. Fungal Genet. Biol. 21:131–140 [PubMed] [Google Scholar]
- 55. Tanaka C., Tajima S., Furusawa I., Tsuda M. 1992. The Pgr1 mutant of Cochliobolus heterostrophus lacks a p-diphenol oxidase involved in naphthalenediol melanin synthesis. Mycol. Res. 96:959–964 [Google Scholar]
- 56. Thompson J. E., et al. 2000. The second naphthol reductase of fungal melanin biosynthesis in Magnaporthe grisea. J. Biol. Chem. 275:34867–34872 [DOI] [PubMed] [Google Scholar]
- 57. Tokousbalides M. C., Sisler H. D. 1979. Site of inhibition by tricyclazole in the melanin biosynthetic pathway of Verticillium dahliae. Pestic. Biochem. Physiol. 11: 64:73 [Google Scholar]
- 58. Tsuji G., Takeda T., Furusawa I., Horino O., Kubo Y. 1997. Carpropamid, an anti-rice blast fungicide, inhibits scytalone dehydratase activity and appressorial penetration in Colletotrichum lagenarium. Pestic. Biochem. Physiol. 57:211–219 [Google Scholar]
- 59. Tsuji G., Fujikawa J., Ishida H., Horino O., Kubo Y. 2001. Laccase gene LAC1 of Colletotrichum lagenarium is not essential for melanin biosynthesis and pathogenicity. J. Gen. Plant Pathol. 67:182–190 [Google Scholar]
- 60. Wheeler M. H., Stipanovic R. D. 1985. Melanin biosynthesis and the metabolism of flaviolin and 2-hydroxyjuglone in Wangiella dermatitidis. Arch. Microbiol. 142:234–241 [DOI] [PubMed] [Google Scholar]
- 61. Wheeler M. H., Klich M. A. 1995. The effects of tricyclazole, pyroquilon, phthalide, and related fungicides on the production of conidial wall pigments by Penicillium and Aspergillus species. Pestic. Biochem. Physiol. 52:125–136 [Google Scholar]
- 62. White L. P. 1958. Melanin: a naturally occurring cation exchange material. Nature 182:1427–1428 [DOI] [PubMed] [Google Scholar]
- 63. Wilson R. A., Talbot N. J. 2009. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7:185–195 [DOI] [PubMed] [Google Scholar]
- 64. Zheng Y.-J., Basarab G. S., Jordan D. B. 2002. Roles of substrate distortion and intramolecular hydrogen bonding in enzymatic catalysis by scytalone dehydratase. Biochemistry 41:820–826 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.