Abstract
Freshwater ecosystems are being heavily exploited and degraded by human activities all over the world, including in North America, where fishes and fisheries are strongly affected. Despite centuries of taxonomic inquiry, problems inherent to species identification continue to hamper the conservation of North American freshwater fishes. Indeed, nearly 10% of species diversity is thought to remain undescribed. To provide an independent calibration of taxonomic uncertainty and to establish a more accessible molecular identification key for its application, we generated a standard reference library of mtDNA sequences (DNA barcodes) derived from expert-identified museum specimens for 752 North American freshwater fish species. This study demonstrates that 90% of known species can be delineated using barcodes. Moreover, it reveals numerous genetic discontinuities indicative of independently evolving lineages within described species, which points to the presence of morphologically cryptic diversity. From the 752 species analyzed, our survey flagged 138 named species that represent as many as 347 candidate species, which suggests a 28% increase in species diversity. In contrast, several species of parasitic and nonparasitic lampreys lack such discontinuity and may represent alternative life history strategies within single species. Therefore, it appears that the current North American freshwater fish taxonomy at the species level significantly conceals diversity in some groups, although artificially creating diversity in others. In addition to providing an easily accessible digital identification system, this study identifies 151 fish species for which taxonomic revision is required.
Keywords: DNA barcoding, cytochrome c oxidase I, biodiversity, evolutionarily significant units, aquatic ecosystem
Assessing the state of life in a world that faces a sixth mass extinction (1, 2) represents one of the largest challenges of modern science. This assessment is particularly needed for North American freshwater fishes. Indeed, 40% of the fauna is in peril (3), and it continues to support a multibillion dollar commercial and recreational fishery (4). Despite more than 2 centuries of descriptive taxonomic inquiry, problems inherent to species identification have hampered the study, conservation, and management of the richest diversity of temperate freshwater fish (5). Indeed, the exact amount of diversity is still unknown, and it is thought that 10% of North American freshwater fish are still formally undescribed (6, 7). Part of this problem may stem from the fact that there is no universally accepted operational species concept (8, 9). Most biologists agree that species are independently evolving lineages of populations or metapopulations (8, 9), but a debate surrounds the choice of an exact cutoff in the divergence continuum. Morphological differentiation has long been the criterion of choice for taxonomists because of the relative ease with which those characters are assessed. Unfortunately, such differences are not always caused by independent evolutionary history, and reproductively isolated taxa can sometimes be morphologically indistinguishable.
Generating rapid and accurate molecular identifications using standardized tools can help to resolve distorted views of biodiversity. Indeed, DNA barcoding (10) surveys using partial cytochrome c oxidase subunit I (COI) sequences have revealed cryptic diversity across the animal kingdom. For instance, previous DNA barcoding studies in other taxonomic groups have found as many as nine undescribed species embedded within a single known species of skipper butterfly (11). These numbers are even larger in less studied groups, such as parasitic hymenopterans (12). Whereas many species may need to be split into distinct evolutionary lineages, others may need to be combined, given that not all morphological differences are the result of cladogenesis. In an outstanding case documented in fishes, individuals that belonged to two different species were found to be the female and male of a single species (13).
In this study, we established a barcode reference library for more than 80% of the named freshwater fish species of North America. We used this survey of standing genetic diversity as an independent calibration of current taxonomic resolution within the North American fish fauna to reveal key areas of uncertainty where discrepancies between genetic data and morphologically based taxonomy arise. In contrast, where DNA sequences and traditional taxonomy exhibit congruence, our data serve as an accessible key for the molecular identification of North American freshwater fishes.
Results
We obtained mitochondrial barcodes for 5,674 fish specimens belonging to 50 families, 178 genera, and 752 species (Table 1). This coverage includes more than 80% of the 902 Canadian and American species listed by Nelson et al. (14). In accordance with the Fish Barcode of Life Campaign (15), we deposited all sequences and collateral specimen information within the Barcode of Life Data System (BOLD) (16), where this information can be queried by users, annotated, and curated in light of new information. This list of 752 species also includes 22 exotic invasive species (non-native species that adversely affect ecosystems), 4 species closely related to North American species, and 30 worldwide species of lampreys (17) because those species are notoriously difficult to identify and can have important ecological and economic impacts. For most species, multiple specimens (mean = 7.5 specimens per species) from distant localities (mean = 3.2 localities per species) were analyzed to document intraspecific variability. Only 62 species were represented by a single specimen, and 1 species (Etheostoma radiosum) was represented by 108 specimens. We observed a hierarchical increase in mean divergence from within species (mean = 0.73%, SE = 0.053) to within congeners (mean = 13.67%, SE = 0.004), within families (mean = 15.91%, SE = 0.002), and within orders (mean = 21.24%, SE = 0.002) (Table S1). Within a genus, the mean distance to the most closely related species (nearest neighbor) was 5.71% (SE = 0.012); therefore, the mean distance to the nearest neighbor was 7.82 times higher than the mean intraspecific divergence.
Table 1.
Summary of the fish taxa analyzed
| Species |
|||
| Family | Barcoded | Indistinguishable using barcodes | With UCS (no. of UCS) |
| Cyprinidae | 221 | 10 | 52 (121) |
| Percidae | 197 | 18 | 45 (120) |
| Catostomidae | 52 | 10 | 5 (10) |
| Centrarchidae | 32 | 6 | 6 (18) |
| Cottidae | 31 | 5 | 6 (16) |
| Ictaluridae | 30 | 0 | 11 (29) |
| Salmonidae | 30 | 7 | 0 |
| Petromyzontidae | 27 | 13 | 3 (8) |
| Fundulidae | 26 | 2 | 3 (6) |
| Cichlidae | 11 | 0 | 0 |
| Poeciliidae | 9 | 0 | 0 |
| Acipenseridae | 6 | 0 | 0 |
| Esocidae | 5 | 2 | 1 (2) |
| Gasterosteidae | 5 | 0 | 1 (2) |
| Lepisosteidae | 5 | 0 | 0 |
| Clupeidae | 4 | 2 | 0 |
| Elassomatidae | 4 | 0 | 2 (6) |
| Embiotocidae | 4 | 0 | 0 |
| Percichthyidae | 4 | 0 | 0 |
| Umbridae | 4 | 0 | 0 |
| Amblyopsidae | 3 | 0 | 1 (3) |
| Cyprinodontidae | 3 | 0 | 0 |
| Geotriidae | 3 | 0 | 0 |
| Gobiidae | 3 | 0 | 0 |
| Osmeridae | 3 | 0 | 0 |
| Atherinopsidae | 2 | 0 | 1 (2) |
| Characidae | 2 | 0 | 0 |
| Hiodontidae | 2 | 0 | 0 |
| Loricariidae | 2 | 0 | 0 |
| Sciaenidae | 2 | 0 | 0 |
| Achiridae | 1 | 0 | 1 (2) |
| Amiidae | 1 | 0 | 0 |
| Anguillidae | 1 | 0 | 0 |
| Aphredoderidae | 1 | 0 | 1 (2) |
| Ariidae | 1 | 0 | 0 |
| Belonidae | 1 | 0 | 0 |
| Channidae | 1 | 0 | 0 |
| Cobitidae | 1 | 0 | 0 |
| Dasyatidae | 1 | 0 | 0 |
| Doradidae | 1 | 0 | 0 |
| Elopidae | 1 | 0 | 0 |
| Fistulariidae | 1 | 0 | 0 |
| Gadidae | 1 | 0 | 0 |
| Lotidae | 1 | 0 | 0 |
| Myxinidae | 1 | 0 | 0 |
| Percopsidae | 1 | 0 | 0 |
| Pleuronectidae | 1 | 0 | 0 |
| Polyodontidae | 1 | 0 | 0 |
| Profundulidae | 1 | 0 | 0 |
| Sparidae | 1 | 0 | 0 |
| Total | 752 | 75 | 138 (347) |
This list includes the number of indistinguishable species and the number of species with UCS (represented by lineages that diverge by over 2%), along with the total number of UCS (SI Text).
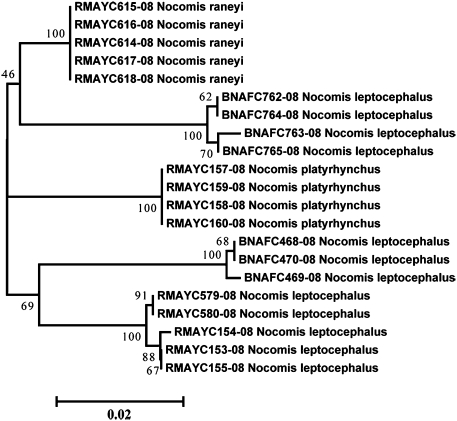
The resolution of named species using barcodes approached 90% (676 of 752) (Table 1, Table S1, and SI Text). Those delineated species could be identified using a diagnostic nucleotide approach (18) and were represented by a unique haplotype, a single tight cluster of haplotypes, or distinct clusters of haplotypes. Using the more traditional distance-based identification approach, we obtained a success rate of 81%. However, the inflated proportion of indistinguishable species identified using the distance-based identification approach was mainly caused by the presence of cryptic diversity within “known” species. Thus, approximately half (72 of 141) of the 19% of indistinguishable species identified using the distance-based approach were those also considered as problematic using the diagnostic nucleotide approach. Over half of the remaining problematic cases (37 of 69) involved species diverging by over 2% from any other species but were considered as indistinguishable using the distance-based approach because of an even deeper intraspecific divergence (e.g., Nocomis leptocephalus; Fig. 1 and Fig. S1). For N. leptocephalus, some of the taxa exhibiting deep intraspecific divergence values were recovered as poly/paraphyletic in phylogenetic trees; nevertheless, all lineages remained clearly distinct from any other lineages using the diagnostic nucleotide approach and always differed by over 10 characters (e.g., Nocomis spp.; Fig. S1). The lineages of the remaining 32 species were less differentiated (range: 0.15–1.99%) but could still be identified using diagnostic nucleotides (e.g., Fig. S2). Identification at the genus level was completely accurate (100%) using both the diagnostic nucleotide and distance-based approaches.
Fig. 1.
Neighbor-joining tree shows the relationships among 615-bp COI sequences from three species of the genus Nocomis, producing five divergent clusters. The numbers at the nodes are bootstrap values based on 1,000 replications.
DNA barcode species identification success rates varied from nearly 50% in Clupeidae and Petromyzontidae to 100% in 40 families (Table 1). A literature review of data from all 75 species that cannot be delineated using a DNA barcode showed that hybridization, ancestral polymorphism sharing, and inadequate taxonomy can explain those cases of haplotype sharing. The lack of divergence observed in lampreys is particularly intriguing because it involves 13 species included in only five clusters (Fig. 2 and Table 1). Each of those clusters included at least 1 parasitic and 1 nonparasitic species of lamprey that are morphologically quite distinct.
Fig. 2.
Neighbor-joining tree shows the relationships among 620-bp COI sequences from six parasitic and seven nonparasitic species of lampreys, producing five clusters. The numbers at the nodes are bootstrap values based on 1,000 replications. The life history characteristic [parasitic (+) or nonparasitic (−)] and origin of the samples (states/provinces) are given for each individual.
We found deeply divergent intraspecific clusters (>2%) within 138 of the 752 analyzed species (Fig. 1, Table 1, Fig. S1, and SI Text). Those divergent intraspecific clusters, which correspond to divergent evolutionary lineages, were restricted to 15 of the 50 analyzed fish families (Table 1 and SI Text). The number of lineages by species varied from 2 to 7, for a total of 347 divergent lineages among 138 named species. Deeply divergent intraspecific lineages (>2%) were almost always (88%) found in different geographical locations.
Discussion
This continent-wide genetic survey of North American freshwater fishes offers a new perspective on their diversity. Indeed, results reveal that current species-level taxonomy significantly conceals diversity in some groups, although artificially creating diversity in others (Figs. 1 and 2 and Table 1). Furthermore, DNA barcodes provide a straightforward identification system when a perfect match exists between morphology-based taxonomy and genetic divergence.
Comparison Between Taxonomy and DNA Barcodes.
The DNA barcodes library provides an identification system with many applications, including the identification of fish parts or remnants, such as fish filet, sushi, smoked fish, caviar, eggs, and larvae, that are not recognizable using morphological characters (19, 20). It may facilitate tracking exotic invasive species through water samples (21) and aid food web reconstruction through gut content or fecal sample analyses (e.g., ref. 22). Barcodes are therefore highly valuable for enhancing wildlife protection from poaching and illegal trade by easing the application of different laws (e.g., Convention on International Trade in Endangered Species) and also help to protect consumers from market fraud (19).
Within North American freshwater fishes, the combined use of the distance-based identification approach with, when necessary, the diagnostic nucleotide approach appears to deliver the most reliable identifications using COI. For some of the 10% of indistinguishable species, the additional use of nuclear DNA markers (e.g., 28S, ITS1, microsatellites, amplified fragment length polymorphisms) may facilitate species-level identification. For some other indistinguishable species, it is quite possible that no genetic marker would ever allow delineation because they might not represent an isolated gene pool, and thus might not represent separate species.
With a correct species identification rate of 90%, freshwater fishes are among the groups of animals harboring the most frequent cases of interspecific haplotype sharing (Table S1): 8% in Canadian freshwater fishes, 4% in Cuban freshwater fishes, 2% in Australian marine fishes, 6% in North American birds, and 1% in Lepidoptera (23–27). The relatively elevated proportion of species sharing haplotypes in this study (10%) has four possible nonexclusive explanations: hybridization, incomplete lineage sorting, inadequate taxonomy, and erroneous identification (15). Hybridization is problematic for DNA barcode identification because mtDNA is maternally inherited; therefore, a hybrid will inevitably be diagnosed as its maternal species. Because of incomplete lineage sorting, some haplotypes can remain identical in two isolated gene pools. This situation mainly occurs when divergence is recent. Inadequate taxonomy can give the impression that two different species share barcodes if, in fact, the two “named” species are part of the same gene pool, and are therefore from the same actual species. Finally, erroneous identifications within the reference library could produce an apparent case of haplotype sharing if, in fact, specimens from the same species are wrongly given two different names by the identifier.
In this study, most cases of haplotype sharing appear to result from hybridization and inadequate taxonomy. A literature review involving all species lacking barcode divergence shows that 47% of the cases reported in this survey are corroborated by other studies that also report a lack of genetic divergence or hybridization between the same species (SI Text). According to available literature, hybridization is the most common explanation, consistent with the status of freshwater fishes as the group of vertebrates with the highest occurrence of hybridization (28). Here, the proportion of species sharing haplotypes with another species (10%) was identical to the classic, and still widely accepted, estimated proportion of hybridizing freshwater fishes (10%) (28). An example of a suspected case of hybridization that was determined to be of unexpectedly large scale involves two pike species: Esox americanus and Esox niger (23, 29). Nuclear markers and morphology clearly distinguish these species and also the two subspecies of E. americanus (29). Grande et al. (29) found a lack of mtDNA divergence between one specimen of E. americanus americanus and one specimen of E. niger but a clear differentiation between E. americanus vermiculatus and E. niger/E. americanus americanus Here, our analyses of 54 specimens from 10 different states/provinces showed that the DNA barcodes from the chain pickerel (E. niger) and red fin pickerel (E. americanus americanus) are all part of the same cluster, whereas all sequences from the grass pickerel (E. americanus vermiculatus) belong to a distinct cluster that diverges by over 2% from any other Esocidae. The most likely explanation for this pattern is that the mitochondrial genome of E. niger has been completely replaced over its entire distribution range (i.e., over thousands of square kilometers) by the mtDNA from red fin pickerel (E. americanus americanus) through introgressive hybridization. A literature review indicates that many other nondistinguishable species are part of a “species complex” for which taxonomic boundaries are still debated, and phylogenetic studies have also revealed a lack of monophyly among recognized species. These include North American ciscoes [Coregonus spp., Salmonidae (23, 30)], sunfishes [Lepomis spp., Centrarchidae (31)] and sculpins [Cottus bairdi species complex, Cottidae (32)] (SI Text). It remains impossible to completely rule out the possibility that misidentifications explain some of the uncorroborated 5% of cases of haplotype sharing. However, this possibility is unlikely, because morphological identifications were performed by specialized taxonomists and most specimens are from museum collections. Furthermore, if there are some cases of misidentification, it is indicative of a pressing need for complementary identification tools, such as DNA barcodes. Overall, this survey highlights the failure of many recognized North American “species” to meet all the criteria of biological (33), phylogenetic (34), and phenetic (35) species concepts.
This study is among the first to show that DNA barcodes can differentiate recently radiated species. Indeed, we found that DNA barcodes allowed the identification of 94% of the species within the two most diversified and recently radiated genera of the North American freshwater fauna, Notropis (71 species) and Etheostoma (139 species). This identification success rate is even higher than the overall mean for North American freshwater fishes. It has been suggested that such an evolutionary history of adaptive radiations would illustrate inherent pitfalls on DNA barcoding, as for some recently diverged African cichlids (15) and the Coregonus species complex (23, 30). On the contrary, our results clearly show that DNA barcodes can correctly distinguish these often morphologically similar species in many circumstances.
Deep Intraspecific Divergence and New Candidate Species.
The mean level of intraspecific divergence of 0.73% observed in North American freshwater fishes was approximately two to three times higher than for any other animal groups thoroughly surveyed with DNA barcodes (Table S1), including the following: 0.39% in Australian marine fishes, 0.23% in North American birds, and 0.43% in Lepidoptera (25–27). Such a high level of intraspecific divergence may be explained by the effect of highly restricted gene flow attributable to the fragmented nature of freshwater ecosystems. The limited dispersal capabilities of freshwater fishes relative to flying or marine organisms (36) can, in turn, promote lineage divergence and enhanced speciation rates (37, 38). Indeed, nearly 90% of the deep intraspecific lineages recovered through this survey are allopatric. This finding reinforces the fact that such lineages have independent evolutionary histories (39) and do not reflect patterns of genetic variation suggestive of a single large population. This intraspecific diversification among lineages from different geographical regions demonstrates that individuals from some taxa can be identified not only according to species but linked to a particular watershed.
The proportion of species that possess deep intraspecific lineages (>2%) among North American freshwater fishes is high relative to the other animal groups surveyed (19%) (Table S1). The proportion of species that exceed this divergence threshold is only 5.1% for North American Lepidoptera (27), 3.2% for North American birds (26), and 2.1% for Australian marine fishes (25). This proportion of cryptic diversity within North American freshwater fishes is surprising, considering that they represent one of the best taxonomically studied groups of organisms. Perhaps, unsurprisingly, most cryptic diversity was found within taxa of minimal direct economic value.
The highest proportion of cryptic diversity was found among pygmy sunfish [Elassomatidae, an increase of 100% (4 of 4)], cave fish [Amblyopsidae, an increase of 67% (2 of 3)], and catfish [Ictaluridae, an increase of 60% (18 of 30)] (Table 1 and SI Text). Nonetheless, 70% of all cryptic diversity occurs in the two most diversified families, Percidae and Cyprinidae. Indeed, Etheostoma (family Percidae) and Notropis (family Cyprinidae), already the most species-rich genera (with species often distinguished by only a few subtle morphological characters), still harbor a large proportion of cryptic diversity (Table 1 and SI Text). For three species (Etheostoma brevirostrum, Etheostoma artesiae, and Aphredoderus sayanus), the maximum intraspecific divergence was over 15%, and thus closer to the level of divergence observed among genera (13.5%) and families (15.9%) than between sister species (5.7%). Those extreme cases of intraspecific divergence are unlikely to be the result of misidentification. Indeed, it is highly implausible that the highlighted diverging clades corresponded to misidentified specimens from another described species not included in this analysis, creating a false excess of intraspecific divergence. For the genus Etheostoma, for example, we have already included nearly all described species. Thus, we included specimens from 139 Etheostoma species, whereas for the same genus, Nelson et al. (14) listed a total of 131 described Canadian and American species. More recently, 142 species (98.6%) were listed in FishBase (17). For A. sayanus, the extreme level of intraspecific divergence is unlikely the result of misidentification because it is the sole member of the Aphredoderidae family, which is quite distinct from any other fish family. Overall, it appears that, just like for other components of biodiversity, the distribution of cryptic diversity is not uniform. Although little is known about factors that have an impact on the distribution of cryptic diversity both taxonomically and regionally, some answers might be gained from the ever-growing coverage of DNA sequence data. Because we detected a significantly higher intraspecific divergence for fishes found in the United States (mean = 0.73, SE = 0.05) compared with those from Canada (mean = 0.27, SE = 0.01), as already suggested (37), a promising avenue may be to investigate further a possible relationship between latitude and cryptic diversity.
This exhaustive genetic survey of North American freshwater fishes revealed a significant amount of previously unrecognized cryptic diversity. By comparing our results with studies that have previously reported cases of deep (>2%) intraspecies divergence, we conclude that our study highlights some 87 new taxa. This represents an increase of 42% relative to what has been previously reported over more than 20 y of phylogeographical research. This estimate is likely conservative, because for 36 species analyzed at two to three sites in the literature, it is likely that our study, which detected divergent lineages within those species, flagged different, and therefore new, lineages. Finally, we also found divergent lineages that have not been detected before in 8 species for which phylogeographical studies have been conducted, reflecting the failure of many published phylogeographical studies to cover the entire species range.
The genetically dissimilar taxa flagged in this study may represent new species. For the time being, however, we view those lineages as unconfirmed candidate species (UCS) (9). Our calibration suggests that taxonomic revisionary work is warranted for those UCS, with an emphasis on watersheds where genetic variants were detected. We first suggest a careful reexamination of the morphological variation within those described species that possibly harbor cryptic species, because, at this point, it remains possible that some of the newly identified lineages possess some slight morphological differences that have simply been overlooked. The reproductive biology and ecology of such cryptic lineages also require investigation. However, because species are lost at an alarming rate and looking for reproductive isolation is time-consuming, the precautionary principle suggests that the lineages highlighted here should be considered evolutionarily significant units that need to be taken into account in conservation strategies (40, 41). Despite uncertainties surrounding mtDNA molecular clock calibration, deeply divergent mtDNA clusters (e.g., >2%) are likely indicative of a million years or more of unique evolutionary legacy (39, 42). This represents a relatively long evolutionary time period compared with the mean time between origination and extinction, which is estimated to be 2.5 million years for mammals (43), the vertebrate group with the best estimates of species life span. Conserving as many “genetic building blocks” (sensu ref. 44) as possible will hopefully enhance the capacity of biodiversity to evolve and adapt to an ever-changing environment and reduce these species’ chances of extinction.
Lack of Interspecific Divergence and Distinct Species That May Be Single Species.
At the other end of the spectrum were the cases where different species were found to form a single genetic cluster, and probably a single evolutionary lineage. This situation seems particularly recurrent in lampreys. Indeed, there are 13 recognized species included in only five clusters, and each one of those clusters includes at least 1 parasitic and 1 nonparasitic species of lamprey (Fig. 2 and Table 1). Interestingly, it has been known for decades that most lamprey genera harbor paired species with morphologically similar larvae (45, 46). Nevertheless, following metamorphosis, adults differ both morphologically and ecologically and are either parasitic to other fish or nonparasitic (i.e., do not eat anything at all). Adults of nonparasitic species do not migrate, are smaller, and have less developed teeth than parasitic lampreys (46). Although the current taxonomy considers all parasitic and nonparasitic lampreys as distinct species, our results, along with those of several previous studies (46–53), stress the need to revisit lamprey taxonomy.
The lack of significant genetic divergence between parasitic and nonparasitic lampreys appears to be a ubiquitous worldwide pattern. Our analysis, based on 174 specimens representing 30 lamprey species that were characterized by ∼650 bp of mtDNA, documents new relationships among lampreys and further corroborates results of other studies. In the most exhaustive previous study, 46 specimens from 23 species had been characterized by 384 bp of mtDNA (48). Lack of divergence between two pairs of lamprey has also been observed at allozymes [Lampetra fluviatilis/Lampetra planeri (49) and Lampetra ayresii /Lampetra richardsoni (50)]. The relationship between L. fluviatilis and L. planeri was previously analyzed using mtDNA (47, 49). Okada et al. (51) and Yamazaki et al. (52) also investigated the relationships between the nonparasitic Lethenteron kessleri and two other nonparasitic species using mtDNA. Our analysis complements that study by showing that this clade also comprises two other nonparasitic species (Lampetra alaskense and Lampetra appendix) and the parasitic species Lampetra camtschaticum (Fig. 2).
Recurrent lack of genetic differentiation suggests that many genera of lamprey possess an intrinsic capacity to develop two alternative morphotypes that correspond to parasitic and nonparasitic life history strategies. Therefore, it is plausible that when facing different environmental conditions, larvae from several taxa may adopt one of two trophic tactics, each with its own costs and benefits, in a similar fashion to the winged or nonwinged morphs in pea aphids (54). This hypothesis is also supported by rare observations of plasticity in feeding type for some populations of lampreys (53) and the distribution overlap between parasitic and nonparasitic species (46). If the large anadromous parasitic lampreys and the small toothless nonparasitic lampreys effectively represent alternative life history tactics within the same gene pool, this places lampreys among the most phenotypically plastic vertebrates known.
Conclusion
Overall, this study demonstrated the ability of DNA barcoding to help calibrate current taxonomic resolution and shed new light on the biodiversity of North American fishes. Indeed, although differently described species might, in fact, represent single evolutionary lineages, as in the case of lampreys, as much as 28% of the American and Canadian freshwater fish species could be waiting for a formal taxonomic description. This finding reinforces the status of North American fresh waters as harboring some of the world's greatest fish diversity and deserving more important conservation efforts (6). Further DNA barcoding surveys will reveal whether the extremely high proportion of cryptic evolutionary lineages detected in this study is a common characteristic of freshwater organisms. Finally, our results stress the need for more taxonomic research, because it appears that even for economically important vertebrates that have benefited from over a century of scientific inquiry, additional work is required to create a more accurate picture of species diversity.
Materials and Methods
For each of the 902 freshwater fish species occurring north of Mexico and recorded by Nelson et al. (14), our objective was to sample five individuals from each main watershed within the species range. More individuals were analyzed when many specimens were readily available from the museum collection. Despite our effort to get a representative sampling design, because of our somewhat opportunistic approach and the rarity of some species, it was not possible to achieve a randomly structured sampling design with respect to biogeographical regions and drainages. Samples and DNA sequence acquisition, as well as the analyses of genetic distance and phylogenetic trees, are described in SI Text. Specimen and sequence data are accessible in the BOLD projects “Freshwater fishes of North America and Barcoding of Canadian freshwater fishes,” and sequences have also been submitted to GenBank.
The diagnostic nucleotide identification method assumes that a described species can be correctly identified using DNA barcodes if all reference specimens morphologically identified as this species by taxonomists possess one or more unique and nonhomoplasic (i.e., diagnostic) nucleotides relative to the other species (18). The distance-based approach assumes that a species can be correctly identified when the mean distance to the most closely related species (nearest neighbor) is higher than the maximum intraspecific distance. The latter approach offers more stringent criteria because all analyzed specimens of the species of interest not only need to have private haplotypes with a diagnostic nucleotide (as in the former approach) but need to appear monophyletic in a phylogenetic tree, because the level of divergence between species needs to be higher than the intraspecific variability. Even if neither of the two approaches is absolutely impervious to the possibility that an unsampled population presents a case of haplotype sharing, other DNA barcoding studies (26, 27) and some of the species analyzed here using a nearly complete geographical coverage have shown that this situation is certainly rare. Furthermore, both the number of samples and the number of localities by species remain relatively high compared with other large-scale DNA barcoding projects (Table S1).
A threshold of 2% of maximum divergence was applied to refer to cryptic diversity. This level of divergence is commonly observed between distinct vertebrate species (55) and has also been used as a standard in recent DNA barcoding projects (27). It is noteworthy that a lower threshold would allow recovery of more cryptic diversity but could also erroneously inflate species diversity before further investigations are performed on the candidate species. In contrast, a higher threshold would give more conservative estimates of diversity but might also miss many lineages that should be described as species. The number of divergent lineages within recognized species was calculated as the number of haplotypes, or clusters of haplotypes, with a mean divergence of over 2% from any other haplotypes or clusters of haplotypes.
Supplementary Material
Acknowledgments
We thank the following people for their valuable help in collecting samples: David Ward from Arizona Game and Fish; Tom Near and Benjamin Keck from the Peabody Museum of Natural History; Christian Smith from the Abernathy Fish Technology Center; Paul Rister from the Kentucky Department of Fish and Wildlife Resources; John Lyons from the Wisconsin Department of Natural Resources and the University of Wisconsin Zoological Museum; and Larry M. Page, Rob Robins, and Molly Phillips from the Florida Museum of Natural History. We also thank Miranda G. Haskins for assistance in subsampling biological material and Heather Braid for assistance in generating DNA sequences. We are grateful to Paul Hebert for his support and interest throughout the present study. We also thank the editor and three anonymous referees for their very constructive and useful comments. This research is a contribution to the research program of Québec Océan and was supported through funding to the Canadian Barcode of Life Network from the Natural Sciences and Engineering Research Council of Canada and other sponsors (listed at http://www.BOLNET.ca).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.M. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (for accession nos. see SI Materials and Methods). They have also been deposited in Barcode of Life Data System (http://www.boldsystems.org) (projects Freshwater fishes of North America and Barcoding of Canadian freshwater fishes).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016437108/-/DCSupplemental.
References
- 1.Vitousek PM, Mooney HA, Lubchenco J, Melillo JM. Human domination of Earth's ecosystems. Science. 1997;277:494–499. [Google Scholar]
- 2.Sala OE, et al. Global biodiversity scenarios for the year 2100. Science. 2000;287:1770–1774. doi: 10.1126/science.287.5459.1770. [DOI] [PubMed] [Google Scholar]
- 3.Jelks HL, et al. Conservation status of imperiled North American freshwater and diadromous fishes. Fisheries (Bethesda, Md) 2008;33:372–407. [Google Scholar]
- 4.FAO . The State of World Fisheries and Aquaculture 2008. Rome: Fisheries and Aquaculture Department, Food and Agriculture Organization of the United Nations; 2009. [Google Scholar]
- 5.Abell RA, et al. Freshwater Ecoregions of North America: A Conservation Assessment. Washington, DC: Island Press; 2000. [Google Scholar]
- 6.Warren ML, et al. Diversity, distribution, and conservation status of the native freshwater fishes of the southern United States. Fisheries (Bethesda, Md) 2000;25:7–31. [Google Scholar]
- 7.Butler RS, Mayden RL. Cryptic biodiversity. Endangered Species Bulletin. 2003;28:24–26. [Google Scholar]
- 8.Mayden RL. In: Species, the Units of Biodiversity. Claridge MF, Dawah HA, Wilson MR, editors. London: Chapman & Hall; 1997. pp. 381–424. [Google Scholar]
- 9.Pedial JM, Miralles A, De la Riva I, Vences M. The integrative future of taxonomy. Front Zool. 2010;7:16. doi: 10.1186/1742-9994-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith MA, et al. Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc Natl Acad Sci USA. 2008;105:12359–12364. doi: 10.1073/pnas.0805319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrkjedal I, Rees DJ, Willassen E. Lumping lumpsuckers: Molecular and morphological insights into the taxonomic status of Eumicrotremus spinosus (Fabricius, 1776) and E. eggvinii Koefoed, 1956 (Teleostei: Cyclopteridae) J Fish Biol. 2007;71:111–131. [Google Scholar]
- 14.Nelson JS, et al. Common and Scientific Names of Fishes from the United States, Canada, and Mexico. Special Publication 29, 6th Ed. Bethesda, MD: American Fisheries Society; 2004. [Google Scholar]
- 15.Ward RD, Hanner R, Hebert PDN. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 2009;74:329–356. doi: 10.1111/j.1095-8649.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 16.Ratnasingham S, Hebert PDN. bold: The Barcode of Life Data System ( http://www.barcodinglife.org) Mol Ecol Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froese R, Pauly D. FishBase. 2010. Available at www.fishbase.org, version (07/2010). Accessed August 15, 2010.
- 18.Wong EHK, Shivji MS, Hanner RH. Identifying sharks with DNA barcodes: Assessing the utility of a nucleotide diagnostic approach. Mol Ecol Resour. 2009;9:243–256. doi: 10.1111/j.1755-0998.2009.02653.x. [DOI] [PubMed] [Google Scholar]
- 19.Wong EHK, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Res Int. 2008;41:828–837. [Google Scholar]
- 20.Victor BC, Hanner R, Shivji M, Hyde J, Caldow C. Identification of the larval and juvenile stages of the Cubera Snapper, Lutjanus cyanopterus, using DNA barcoding. Zootaxa. 2009;2215:24–36. [Google Scholar]
- 21.Ficetola GF, Miaud C, Pompanon F, Taberlet P. Species detection using environmental DNA from water samples. Biol Lett. 2008;4:423–425. doi: 10.1098/rsbl.2008.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaartinen R, Stone GN, Hearn J, Lohse K, Roslin T. Revealing secret liaisons: DNA barcoding changes our understanding of food webs. Ecol Entomol. 2010;35:623–638. [Google Scholar]
- 23.Hubert N, et al. Identifying Canadian Freshwater Fishes through DNA Barcodes. PLoS ONE. 2008;3:e2490. doi: 10.1371/journal.pone.0002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lara A, et al. DNA barcoding of Cuban freshwater fishes: Evidence for cryptic species and taxonomic conflicts. Mol Ecol Resour. 2010;10:421–430. doi: 10.1111/j.1755-0998.2009.02785.x. [DOI] [PubMed] [Google Scholar]
- 25.Ward RD, Zemlak TS, Innes BH, Last PR, et al. DNA barcoding Australia's fish species. Philos Trans R Soc Lond B Biol Sci. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr KCR, et al. Comprehensive DNA barcode coverage of North American birds. Mol Ecol Notes. 2007;7:535–543. doi: 10.1111/j.1471-8286.2007.01670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebert PDN, Dewaard JR, Landry JF. DNA barcodes for 1/1000 of the animal kingdom. Biol Lett. 2010;6:359–362. doi: 10.1098/rsbl.2009.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubbs CL. Hybridization between fish species in nature. Syst Zool. 1955;4:1–20. [Google Scholar]
- 29.Grande T, Laten H, Lopez JA. Phylogenetic relationships of extant esocid species (Teleostei: Salmoniformes) based on mophological and molecular characters. Copeia. 2004;4:743–757. [Google Scholar]
- 30.Turgeon J, Estoup A, Bernatchez L. Species flock in the North American Great Lakes: Molecular ecology of Lake Nipigon Ciscoes (Teleostei: Coregonidae: Coregonus) Evolution. 1999;53:1857–1871. doi: 10.1111/j.1558-5646.1999.tb04568.x. [DOI] [PubMed] [Google Scholar]
- 31.Harris PM, Roe KJ, Mayden RL. A mitochondrial DNA perspective on the molecular systematics of the sunfish genus Lepomis (Actinopterygii: Centrarchidae) Copeia. 2005;2:340–346. [Google Scholar]
- 32.Kinziger AP, Raesly RL, Neely DA. New species of Cottus (Teleostei: Cottidae) from the middle Atlantic eastern United States. Copeia. 2000;4:1007–1018. [Google Scholar]
- 33.Mayr E. Systematics and the Origin of Species. New York: Columbia Univ Press; 1942. [Google Scholar]
- 34.Cracraft J. Species concepts and speciation analysis. Curr Ornithol. 1983;1:159–187. [Google Scholar]
- 35.Sokal RR, Crovello TJ. The biological species concept: A critical evaluation. Am Nat. 1970;104:127–153. [Google Scholar]
- 36.Ward RD, Woodwark M, Skibinski DOF. A comparison of genetic diversity levels in marine, freshwater and anadromous fishes. J Fish Biol. 1994;44:213–232. [Google Scholar]
- 37.Bernatchez L, Wilson CC. Comparative phylogeography of nearctic and palearctic fishes. Mol Ecol. 1998;7:431–452. [Google Scholar]
- 38.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 39.Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard Univ Press; 2000. [Google Scholar]
- 40.Waples RS. Pacific salmon, Oncorhynchus spp., and the definition of “species” under the Endangered Species Act. Mar Fish Rev. 1991;53:11–22. [Google Scholar]
- 41.Fraser DJ, Bernatchez L. Adaptive evolutionary conservation: Towards a unified concept for defining conservation units. Mol Ecol. 2001;10:2741–2752. [PubMed] [Google Scholar]
- 42.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alroy J. New methods for quantifying macroevolutionary patterns and processes. Paleobiology. 2000;26:707–733. [Google Scholar]
- 44.Waples RS. Evolutionarily significant units and the conservation of biological diversity under the Endangered Species Act. Evolution and the Aquatic Ecosystem: Defining Unique Units in Population Conservation. 1995;17:8–27. [Google Scholar]
- 45.Zanandrea G. Speciation among lampreys. Nature. 1959;184:380. [Google Scholar]
- 46.Docker MF. Biology, management, and conservation of lampreys in North America. Am Fish Soc Symp. 2009;72:71–114. [Google Scholar]
- 47.Espanhol R, Almeida PR, Alves MJ. Evolutionary history of lamprey paired species Lampetra fluviatilis (L.) and Lampetra planeri (Bloch) as inferred from mitochondrial DNA variation. Mol Ecol. 2007;16:1909–1924. doi: 10.1111/j.1365-294X.2007.03279.x. [DOI] [PubMed] [Google Scholar]
- 48.Blank M, Jurss K, Bastrop R. A mitochondrial multigene approach contributing to the systematics of the brook and river lampreys and the phylogenetic position of Eudontomyzon mariae. Can J Fish Aquat Sci. 2008;65:2780–2790. [Google Scholar]
- 49.Schreiber A, Engelhorn R. Population genetics of a cyclostome species pair, river lamprey (Lampetra fluviatilis L.) and brook lamprey (Lampetra planeri Bloch) J Zoological Syst Evol Res. 1998;36:85–99. [Google Scholar]
- 50.Beamish RJ, Withler RE. In: Indo-Pacific Fish Biology: Proceedings of the Second International Conference on Indo-Pacific Fishes. Uyeno T, Arai R, Taniuchi T, Matsuura K, editors. Tokyo, Japan: Ichthyological Society of Japan; 1986. pp. 31–49. [Google Scholar]
- 51.Okada K, Yamazaki Y, Yokobori S, Wada H. Repetitive sequences in the lamprey mitochondrial DNA control region and speciation of Lethenteron. Gene. 2010;465:45–52. doi: 10.1016/j.gene.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Yamazaki Y, Yokoyama R, Nishida M, Goto A. Taxonomy and molecular phylogeny of Lethenteron lampreys in eastern Eurasia. J Fish Biol. 2006;68:251–269. [Google Scholar]
- 53.Cochran PA. Observations on giant American brook lampreys (Lampetra appendix) J Freshwat Ecol. 2008;23:161–164. [Google Scholar]
- 54.Podjasek JO, Bosnjak LM, Brooker DJ, Mondor EB. Alarm pheromone induces a transgenerational wing polyphenism in the pea aphid, Acyrthosiphon pisum. Can J Zool. 2005;83:1138–1141. [Google Scholar]
- 55.Avise JC, Walker D, Johns GC. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc Biol Sci. 1998;265:1707–1712. doi: 10.1098/rspb.1998.0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.