Abstract
We identify a large coiled-coil protein, Sme4/PaMe4, that is highly conserved among the large group of Sordariales and plays central roles in two temporally and functionally distinct aspects of the fungal sexual cycle: first as a component of the meiotic synaptonemal complex (SC) and then, after disappearing and reappearing, as a component of the spindle pole body (SPB). In both cases, the protein mediates spatial juxtaposition of two major structures: linkage of homolog axes through the SC and a change in the SPB from a planar to a bent conformation. Corresponding mutants exhibit defects, respectively, in SC and SPB morphogenesis, with downstream consequences for recombination and astral-microtubule nucleation plus postmeiotic nuclear migration. Sme4 is also required for reorganization of recombination complexes in which Rad51, Mer3, and Msh4 foci relocalize from an on-axis position to a between-axis (on-SC) position concomitant with SC installation. Because involved recombinosome foci represent total recombinational interactions, these dynamics are irrespective of their designation for maturation into cross-overs or noncross-overs. The defined dual roles for Sme4 in two different structures that function at distinct phases of the sexual cycle also provide more functional links and evolutionary dynamics among the nuclear envelope, SPB, and SC.
Keywords: meiosis, pairing, coalignment, postmeiotic division, ascomycetes
The central event of meiotic prophase is recognition, pairing, and accompanying recombination of homologous maternal and paternal chromosomes (homologs). This complex program of chromosome dynamics occupies the entirety of a prolonged G2/prophase. Homologous chromosomes first become coaligned at a distance of 400 nm and then, at a distance of 200 nm (1, 2). Coalignment is mediated by axis-associated recombination complexes, each corresponding to the interaction of a single initiating double-strand break (DSB) with a partner chromosome (refs. 2 and 3 and references therein). Coalignment is followed by synapsis, where chromosome axes become linked all along their lengths at ~100-nm distance through a close-packed array of transverse filaments, plus other proteins, that together form the synaptonemal complex (SC). The SC is the most universally observed structure of meiosis. SC transverse filament proteins of budding yeast, mammals, Drosophila, Caenorhabditis elegans, fish, and plants share no sequence similarity but strong structural homology: all comprise long internal coiled-coil domains (with sizes correlated to the width of the SC) flanked by globular N- and C-terminal domains, one or both of which are predicted to have DNA binding capacity (reviewed in refs. 4 and 5) (Table S1). After completion of recombination, SC is lost, and these events are followed by a transient diffuse stage—reindividualization of chromosomes at diplotene where homologs are now only connected at sites of cross-overs (chiasmata) and finally, the two meiotic divisions (6).
The work presented here follows on the earlier identification of a Podospora anserina mutation, mei4-1, that conferred defects in both meiotic recombination and postmeiotic spindle pole body (SPB) morphogenesis (7). To understand how a single mutation might affect two such functionally and temporally disparate processes, we cloned the corresponding WT gene and its homolog in Sordaria macrospora, created corresponding deletion mutant derivatives in both organisms, and analyzed both protein localization and mutant phenotypes for meiotic prophase events (in Sordaria) and SPB roles (in both organisms).
Results
Molecular Identification of P. anserina PaMe4 and S. macrospora Sme4.
The PaMe4 gene corresponding to Podospora mutation mei4-1 and the homologous Sordaria gene, named SME4, were cloned, and WT and mei4-1 alleles were sequenced (SI Materials and Methods). Predicted Sme4/PaMe4 proteins, 1643/1503 amino acids, exhibit large globular and basic N- and C-terminal domains, with multiple Ser-Pro-X-X or Thr-Pro-X-X (S/TPXX) putative DNA binding motifs, linked by a pair of long coiled-coil domains with a central intervening hinge region (Table S1). The protein is well-conserved only among the Sordariales, with hints of rapid evolution (Fig. S1). PaMe4 and Sme4 play no role in the vegetative cycle (SI Results).
Sme4 Localization to Meiotic Prophase Chromosomes Matches the Pattern of Synapsis in Time, Space, and Functional Dependence.
Sme4 localization was examined throughout the life cycle of Sordaria in WT and selected isogenic mutants expressing fully active Sme4-GFP instead of Sme4 (SI Materials and Methods). Sme4 is visible on chromosomes only during pre-SC and SC stages of the sexual cycle.
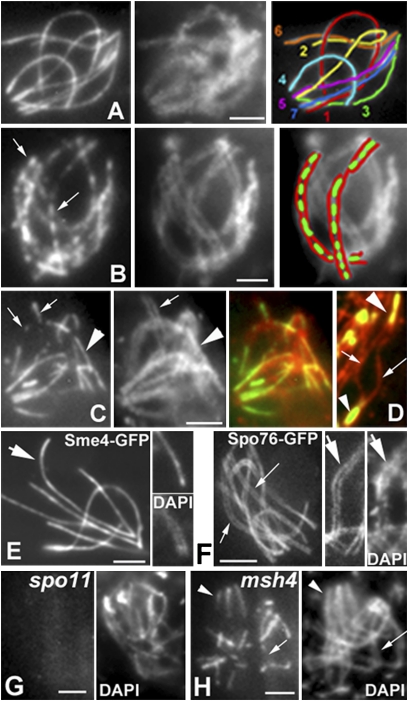
First, Sme4 localizes between chromosome axes in stage-specific correlation with synapsis. (i) During pachytene, Sme4-GFP delineates a bright line that runs in between the closely juxtaposed chromatin paths of fully synapsed homologs defined by DAPI staining (Fig. 1A). (ii) During zygotene, Sme4 signals follow the developing synapsis pattern: individual foci or very short lines throughout the entire nucleus at early zygotene (Fig. 1B) and short plus longer lines at midzygotene (Fig. 1C). (iii) In costaining with axis component Rec8-RFP, Sme4-GFP forms bright lines in regions where homolog axes are conjoined at the ~100-nm distance diagnostic of synapsis, and it is absent where homolog axes are farther apart (Fig. 1D). (iv) Sme4 lines disappear at the time that SC disassembles (diffuse stage), and they are never seen on chromosomes thereafter. Second, Sme4 is not a component of chromosome axes. Axes begin to develop during S-phase and form complete lines along chromosomes by early leptotene (Figs. 1 D and F and 2), whereas Sme4 is not detectable on the chromosomes until zygotene (Fig. 1B). At pachytene, bright and single Sme4-GFP lines occur between the chromatin of homologs (Fig. 1 A and E), whereas axis component Spo76/Pds5-GFP presents a double signal that delineates two tightly juxtaposed but clearly individualized homolog axes (Fig. 1F). Third, Sme4 loading exhibits the same functional dependence on recombination as synapsis. spo11Δ or ski8Δ mutants do not form DSBs and thus, do not exhibit synapsis (8, 9); neither mutant shows Sme4-GFP foci or lines (Fig. 1G). msh4Δ and mer3Δ show only partial synapsis (2); in both mutants, Sme4 appears in linear stretches of variable length at positions where homolog axes lie in close proximity but never form continuous lines (compare Fig. 1A with Fig. 1H) (n = 200).
Fig. 1.
Sme4-GFP localization during meiotic prophase. (A–F) WT. (A) Pachytene. (Left) Sme4-GFP. (Center) DAPI. (Right) Drawing of the seven bivalents. (B) Early zygotene. (Left) Short Sme4 lines (arrows) along all bivalents. (Center) DAPI. (Right) Drawing of two bivalents (green, Sme4; red, DAPI). (C) Midzygotene. (Left) Mixture of short (arrows) and longer Sme4 lines (arrowhead). Correlated differences in pairing seen in DAPI (Center) and merge (Right). (D) Sme4 localization in relation to Rec8-RFP axis staining (red). Sme4-GFP is only seen in synapsed regions (arrowheads) and not in nonsynapsed region (arrows). (E and F) Sme4 and Spo76 localization at pachytene. Sme4 forms single bright lines (E Left, arrow) between the homolog chromatin (E Upper Right compared with DAPI in E Lower Right). Spo76-GFP illuminates double lines of chromosome axes (F Left and Center, arrows) corresponding to homolog chromatin (DAPI in F Right). (G Left) No Sme4-GFP staining in spo11 (DAPI in Right). (H Left) In the absence of Msh4, Sme4 is seen where homologs synapse (arrowheads) and not in synapsing regions (arrow; DAPI in Right). (Scale bars: 2 μm.)
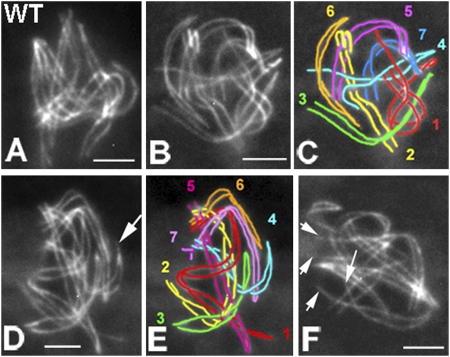
Fig. 2.
Meiotic prophase phenotypes of sme4. (A–F) Chromosome axes are visualized by Spo76-GFP. (A) WT late leptotene with all homologs aligned at 200 nm. (B–F) sme4. (B) Late leptotene homologs aligned at ~400 nm. (C) Drawing of B. (D) Nucleus with homologs aligned at ~200 nm; homologs are closer in a few telomere regions (arrow). (E) Drawing of D. (F) Nucleus with two open telomere regions (arrows). (Scale bars: 2 μm.)
Sme4 Is Not Required for Homolog Recognition or Coalignment but Is Essential for Synapsis.
In WT, during leptotene, homolog axes first align at a distance of 400 nm and then, progress rapidly and synchronously from 400 to 200 nm (Fig. 2A) before synapsing at a distance of 100 nm (Fig. 1 A and F). In sme4Δ, all seven chromosome pairs successfully complete presynaptic coalignment at 400 (Fig. 2 B and C) and 200 nm (Fig. 2 D and E), except that a few nuclei (9%, n = 300) show a mixture of fully and partially coaligned homologs (Fig. 2F). Absence of DSBs (by spo11Δ) eliminates coalignment in sme4Δ like in WT, which is in accord with recombination dependence of this process (9, 10). In WT meiosis, coalignment is a relatively brief stage: only 10–20% of prophase nuclei show homologs fully aligned at 400 or 200 nm (n = 300). Thus, after it is achieved, coalignment is rapidly followed by synapsis. In contrast, in sme4Δ, synapsis is never observed, and the coalignment configuration persists. In Sordaria, asci (meiocytes) increase progressively in size from leptotene to metaphase I independently of chromosomal defects, thus giving a mutation-independent marker for meiotic progression. In all sme4Δ prophase nuclei, homologs remain stiffly coaligned through what would be pachytene by ascus size without alteration of the interactions that hold them at a constant distance of ~200 nm (n = 500) (Fig. 2 B–F). Thus, Sme4 is not required for recombination-mediated recognition and coalignment of homologs but is essential for even the earliest stages of synapsis.
Sme4 and PaMe4 Are Essential for Cross-Over and Chiasma Formation.
Cross-over (CO) formation was analyzed genetically in Podospora WT and pame4Δ (Fig. S2 and SI Materials and Methods). In the mutant, CO levels are reduced 15- to 30-fold all along the chromosomes: from ~3 in WT to ~0.1 along metacentric chromosome 1 and from ~2 to ~0.2 along acrocentric chromosome 6 (Fig. 3A and Fig. S3). The frequency of diplotene chiasmata is reduced similarly in both organisms: 19–21 in WT Podospora (n = 50), 1–3 (n = 30) in pame4Δ (Fig. 3 B and C), and 21 ± 3 in WT Sordaria (n = 200) to 1–4 (n = 50) in sme4Δ (Fig. 3 D and E).
Fig. 3.
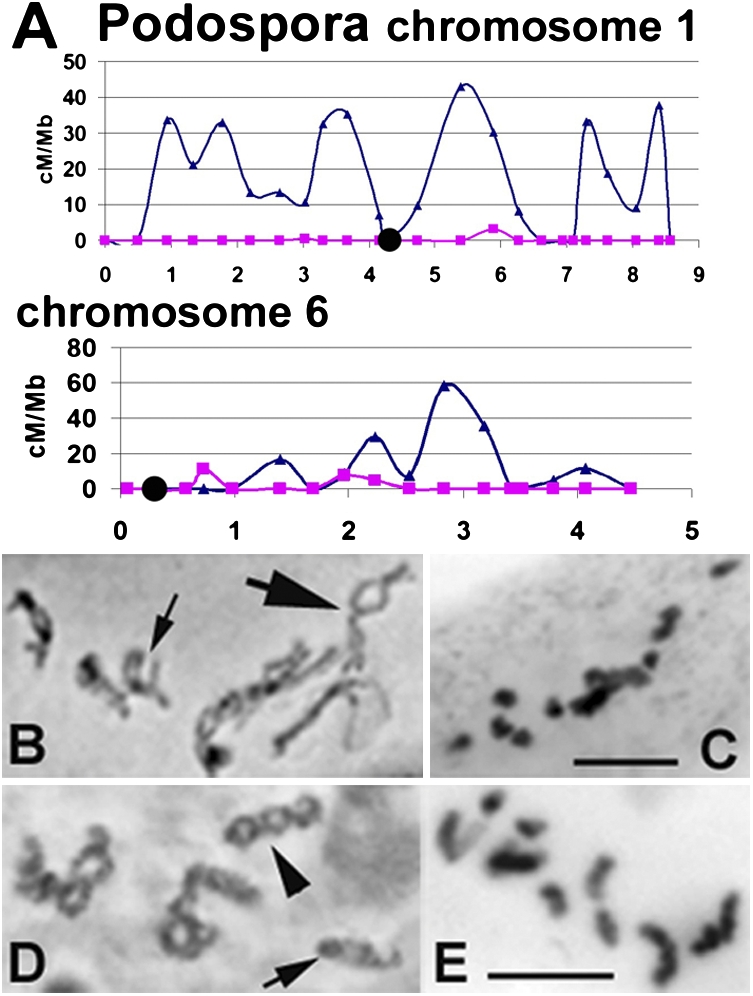
CO and chiasma distributions. (A–C) Podospora. (A) WT (blue) and pame4 (magenta) CO frequencies along long and short chromosomes 1 and 6. Black circles indicate centromeres. (B) WT diplotene with three to four and one to two chiasmata in chromosomes 1 (large arrow) and 6 (small arrow), respectively. (C) pame4 late diplotene with 14 univalents. (D) WT Sordaria diplotene long chromosomes show three to four (arrowhead) and short chromosomes show one to two chiasmata (arrow). (E) sme4 late diplotene with 14 univalents. (B–E) Hematoxylin staining. (Scale bars: 5 μm.)
Sme4 Mediates Synapsis-Correlated Relocalization of Recombination Complexes.
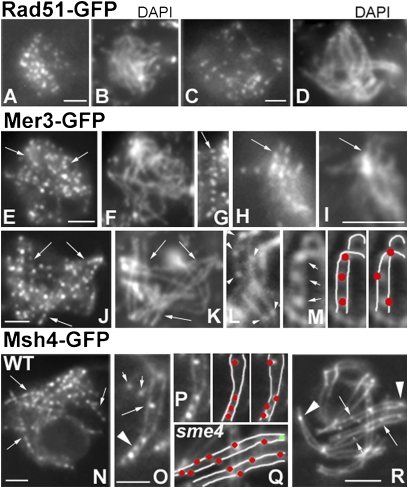
In WT meiosis, functional coupling of recombination and homolog coalignment plus synapsis is reflected in axis-related morphogenesis of foci corresponding to recombination complexes (2, 9). After DSBs at early leptotene (8), post-DSB proteins Rad51 and Mer3 appear as detectable foci on chromosome axes. When homologs are coaligned at ~200 nm, Mer3 foci occur as matching pairs of axis-associated foci and tend to be evenly spaced along the chromosomes (2). The numbers of Rad51 and Mer3 foci at leptotene correspond to the number of total recombinational interactions and not to the specific subset of interactions that ultimately mature into COs (chiasmata) (details in ref. 2).
In the absence of Sme4 (Fig. 4 A–K), normal establishment of ~200 nm coalignment (above) is accompanied by normal morphogenesis of Rad51 (Fig. 4 A and B) and Mer3 foci (Fig. 4 E–I) through that stage with respect to (i) time of appearance at early leptotene (Fig. 4 A and E), (ii) number of foci (in WT and mutant, respectively: 58 ± 6 and 52 ± 5 for Rad51 and 113 ± 14 and 130 ± 10 for Mer3; n = 100 in each case), and (iii) for Mer3 foci, even spacing (Fig. 4 E, G, H, and J, arrows) and localization to matching sites (Fig. 4 H and I). Thus, Sme4 is not required for efficient DSB formation or early post-DSB events correlated with homolog coalignment (reviewed in ref. 11). Importantly, throughout this period, in WT and sme4Δ, all foci are tightly and symmetrically localized in straight rows along the center line of the axes as defined (e.g., by Spo76-GFP) (Fig. 4L for WT and Fig. 4 G–J for sme4Δ).
Fig. 4.
Rad51, Mer3, and Msh4 localization. Stages are assigned by ascus size (in the text). (A–D) Rad51-GFP in sme4. (A) Early leptotene. (B) Corresponding DAPI. (C) Pachytene. (D) Corresponding DAPI. (E–K) Mer3-GFP in sme4. (E) Early leptotene foci (arrows) along chromosomes seen by DAPI in F. (G) Late leptotene/zygotene foci are regularly spaced (arrow) and (H) occur on matching sites (arrow) along aligned homolog paths visualized by DAPI (I, arrow). (J) At midpachytene, foci (arrows) persist as evenly spaced entities along chromosomes seen by DAPI (K). (L, M, and O–R) Axes visualized by Spo76-GFP. (L) WT Mer3 foci first appear on axes (arrowheads). (M Left) At zygotene/pachytene, Mer3 foci are located between homolog axes (arrows). (M Center) Corresponding drawing (foci in red and axes in white). (M Right) Predicted pattern if foci remained on axes instead of relocalizing. (N–R) Msh4-GFP. (N) WT pachytene with lines of regularly spaced foci as predicted for association of foci with SC (M). (O) WT zygotene. In nonsynapsed regions, Msh4 foci are located either on axis (small arrows) or one side of the axis (long arrow); in synapsed regions (arrowhead), foci are in between axes. (P Left) Region from O with corresponding drawing of foci (red) between axes (white) as observed (Center) or predicted (Right) if foci remained on axes. (Q and R) Pachytene nucleus of sme4. (Q) Drawing of the middle region of R (red, on-axis foci along arms; green, telomere focus). (R) sme4 homologs never synapse, and Msh4 foci (arrows) remain located on axes except in a few telomere regions, where they occur between axes (arrowheads). The small arrow indicates the focus on one side of the axis. (Scale bars: 2 μm.)
In WT, two major changes then occur in local correlation with synapsis. (i) Mer3 foci become single, reflecting loss of one focus or fusion of focus pairs (Fig. 4M), and (ii) Mer3 plus Msh4 foci become localized between the homolog axes rather than along the center lines of axes (Fig. 4 M Center, O, and P Center, respectively). Thus, recombinosome foci change from an on-axis to a between-axis disposition. Notably, if foci retained their on-axis coalignment disposition along now synapsed axes, they would have remained centered on their respective axis midlines and would have exhibited a zigzag pattern along the bivalents (Fig. 4 M Right and P Right), which is not observed for Mer3 (Fig. 4M) or Msh4 (Fig. 4 N and O) foci. However, a slight tendency for some foci to be positioned closer to one axis than the other suggests that they could lie at the border of one axis and the interaxis SC central region. In accord with relocalization of foci, about to synapse regions of zygotene nuclei occasionally exhibit foci positioned off the axis in the direction of the partner homolog (Fig. 4O, long arrow), consistent with release of foci from the on-axis position in anticipation of assuming a between-axis state. Interestingly, despite the fact that differentiation of recombinational interactions into CO- and non cross-over (NCO)-fated types likely occurs immediately before synapsis, all interactions undergo the same spatial reorganization in local correlation with synapsis, regardless of their ultimate CO/NCO fate.
In contrast, sme4Δ is specifically defective in these synapsis-correlated recombinosome dynamics. (i) Msh4 foci first appear on axes (Fig. 4 Q and R) like in WT, but their number is strongly reduced [5–20 (n = 50) vs. 81 ± 8 in WT (n = 200)]. Thus, Sme4 plays a role in this presynapsis on-axis event. (ii) Rad51, Mer3, and Msh4 foci do not move to a between-axis position but instead, mostly remain in the late leptotene on-axis configuration along the persistently coaligned homologs (Fig. 4 J and R). Only occasional foci (13%, 22/171) do exhibit repositioning to the interhomolog region (Fig. 4R) such as at zygotene in WT (Fig. 4O) at chromosome ends, where there is also a tendency for homolog axes to be closer together (Fig. 4R, arrowheads). Thus, relocalization of recombinosome foci from the on-axis state to the between-axis state is not absolutely dependent on but is strongly promoted by Sme4. In sme4Δ as well, all three types of recombinosome foci, rather than disappearing at early pachytene (Rad51) or midpachytene (Mer3 and Msh4) like in WT, remain visible at what should be late pachytene by ascus size (Fig. 4 C, D, J, and K) and concomitantly, decrease in number (e.g., Rad51 in Fig. 4C).
Aberrant focus morphogenesis in sme4Δ implies defective recombination progression, thus explaining the deficit of COs/chiasmata in sme4Δ and pame4Δ (above). Additionally, prophase chromosome morphologies persist for ~18 h longer in both mutants than in WT (leptotene to pachytene exit), and this delay is absent in sme4Δ spo11Δ, implying a recombination-dependent global delay in the chromosomal program. Nonetheless, there is no evidence of chromosome fragmentation at diplotene in sme4Δ (Fig. 3E), implying that all DSBs are repaired through interactions between sister chromatids or interhomolog NCOs.
Sme4 Is also a Component of the SPB.
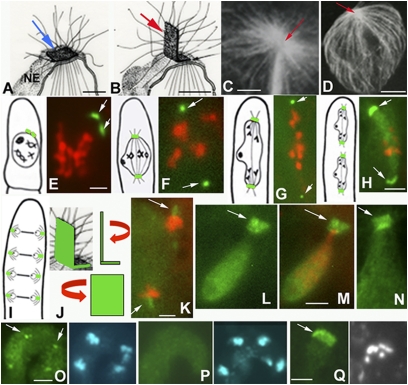
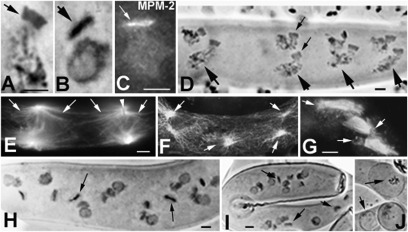
Like budding and fission yeasts, the filamentous fungi Sordaria, Neurospora, and Podospora undergo closed mitosis and meiosis, with the nuclear envelope (NE) remaining intact and a spindle extending within the nucleus between two SPBs (the fungal equivalent of the centrosome). SPBs are embedded in NE in a flattened shape (Fig. 5A) and are used for spindle microtubule (MT) assembly during mitotic and meiotic divisions (12, 13). SPBs also play roles after meiosis during ascospore formation (12, 14). In yeasts, SPBs remain flat on the NE during this process (12). In contrast, in Sordariales, meiosis is followed by a postmeiotic mitotic division (PMM), during which time SPBs undergo a dramatic morphological change; a major portion becomes perpendicular to the NE, whereas a minor portion remains embedded in NE (Fig. 5B) (7, 13). These modified SPBs mediate assembly of long cytoplasmic MTs on both sides (Fig. 5C), which become even longer after telophase when they form an umbrella-like aster that surrounds the nucleus (Fig. 5D) and delimits a spatial territory in the ascus before ascospore enclosure (13, 14).
Fig. 5.
Sme4 localization at the SPB. (A and B) Drawings from reconstruction of serial sections of SPBs in WT (28). (A) During meiosis, SPB (blue arrow) lies flat on the nuclear envelope (NE). (B) During PMM, most of the SPB (red arrow) protrudes perpendicular to the NE and nucleates MTs on both sides (gray lines). (C and D) Antitubulin staining in WT. (C) Pole of PMM anaphase spindle with large astral MTs nucleated by SPB (red arrow). (D) Post-PMM prespore nucleus covered by the MT umbrella nucleated by the SPB (red arrow). (E–M) Merged images of Sme4-GFP (arrows) and chromosomes (red, DAPI) with drawings (E–H Left) of corresponding stages (green, SPBs). (E) Sme4 reappears as double lines (arrows) at diplotene and becomes two foci at the two spindle poles of metaphase I (F), anaphase I (G), and metaphase/anaphase II (H). (I) The four PMM spindles lie across the long axis of the ascus. (J) Drawing of SPB rotations. (Upper) Profile and (Lower) frontal views. (K) Profile view of anaphase PMM (arrows point to SPBs). (L) Frontal view of SPB (arrow), and (M) SPB merged with DAPI. (N) MPM-2 staining confirms SPB localization and the shape of Sme4-GFP. (O) MPM-2 (Left) stains the four SPBs of the two synchronous anaphase spindles of the premeiotic croziers (arrows indicate upper poles) and corresponding DAPI (Right). (P) At the same stage as in O, no Sme4-GFP signal is seen at the poles (Left) or chromosomes (Right; DAPI). (Q) In spo11, Sme4 localizes to all postdiplotene SPBs (e.g., PMM SPB and corresponding DAPI). (Scale bars: A, B, and K–Q, 2 μm; C–J, 5 μm.)
Sme4-GFP is not seen on the SPBs of vegetative nuclei or nuclei undergoing early steps of the sexual cycle (below) through the prophase diffuse stage. However, after Sme4 is shed from between the homologs at the end of pachytene (above), it reemerges at diplotene on the SPB as a bright double-bar signal (Fig. 5E) that matches the underlying SPB structure (13, 14). The two halves of this double bar then become two well-separated single foci on the two poles of the metaphase I (Fig. 5F), and anaphase I spindles (Fig. 5G) as well as on both poles in metaphase II and anaphase II nuclei (Fig. 5H). Foci of second division are larger and brighter, implying recruitment of more Sme4 in anticipation of later roles (below). Finally, during PMM, when spindles are positioned perpendicular to the long axis of the ascus (Fig. 5I), Sme4 decorates the modified SPB (Fig. 5 J–M) as especially evident in a frontal view at telophase (Fig. 5 L and M). This pattern is confirmed by staining with anti-phospho-Ser/thr-Pro mitotic protein monoclonal 2 (MPM-2) antibody, which colocalizes with γ-tubulin at the SPB (15). In WT, at all stages where Sme4 is present at the SPB, Sme4 localization corresponds to that of MPM-2 signals (Fig. 5N, telophase). In contrast, whereas MPM-2 stains the SPBs of all vegetative nuclei, including crozier nuclei (Fig. 5O), Sme4-GFP is not seen at these stages (Fig. 5P). Sme4 localization to SPBs is independent of both initiation of recombination and pairing/synapsis in spo11Δ (Fig. 5Q) and mer3 plus msh4 null mutants, implying functional independence of SC roles (above).
Sme4 and Pame4 Are Required Specifically for PMM SPB Morphogenesis and Function.
Both sme4Δ and pame4Δ exhibit normal MPM-2 staining of SPBs during vegetative growth and fruiting-body development, and they also exhibit normal spindle formation during both meiotic divisions, although at these latter stages, Sme4 is already present on WT SPBs (Fig. 5 F–H). In contrast and in accord with Sme4 localization to the protruding portion of the structurally modified postmeiotic SPB (Fig. 5 B and K–N), Sme4 plays major roles during and after PMM. Prominently and contrary to WT SPBs (Fig. 6A), sme4Δ and pame4Δ SPBs do not develop the bent shape specific to this period; instead, the entire structure remains flat on the NE (compare Fig. 6A with Fig. 6B; for MPM-2, compare Fig. 5N with Fig. 6C) (EM illustrations are shown in ref. 7). This defect does not affect chromosome segregation or emergence of eight telophase nuclei after PMM. However, the entire unique constellation of events that would normally be mediated by the bent portion of the SPB and its long cytoplasmic astral MTs is now defective. (i) WT PMM spindles and nuclei are equally distributed across the ascus (Fig. 6D), with SPBs leading the movement (Fig. 6D, small arrows), and both sides of their modified SPBs nucleate long astral MTs (Fig. 6E). In sme4Δ and pame4Δ, PMM spindles are randomly oriented (Fig. 6 F and G), and their SPBs form only short astral MTs (Fig. 6 F and G, arrows). Consequently, the resulting eight nuclei are abnormally distributed within the asci in both Podospora (compare Fig. 6D with Fig. 6H) and Sordaria (Fig. 6I). (ii) The long umbrella of astral MTs that guides descent of the prospore double membrane in WT (Fig. 5D) is absent in mutants (Fig. 6F), resulting in abnormal enclosure of the ascospores (Fig. 6J). Thus, Sme4 and PaMe4 play indispensable roles in the morphological modifications of PMM SPBs that are needed to form orderly arrays of eight (Sordaria) or four (Podospora) ascospores in a single cell, the ascus. Furthermore, spo11Δ eliminates SC formation and Sme4 localization to chromosomes (above) but has no effect on PMM SPB morphogenesis or function (Fig. 5Q); conversely, these latter features are as defective in spo11Δ sme4Δ as in sme4Δ. Thus, the two roles of Sme4 are temporally and functionally distinct from and independent of their roles for recombination and chromosome synapsis.
Fig. 6.
Postmeiosis in WT and mutants. (A, B, D, and H–J) Hematoxylin staining. (A–C) Frontal views of SPBs at PMM telophase. (A) WT SPB with typical rectangle shape. (B) sme4 SPB is flat and dense and is confirmed by MPM-2 staining (C). (D) WT Podospora ascus with the eight post-PMM nuclei arranged in four rows of two nuclei (large arrows); small arrows show one pair of SPBs. (E–G) Antitubulin staining. (E) Two WT PMM anaphase spindles. Large arrays of astral MTs (arrows) delimit clear areas along the ascus; arrowhead points to SPB (unstained rectangle). (F) Same stage as E in sme4. Flat SPBs nucleate only short astral MTs (arrows) that are intermingled with persisting cortical MTs (absent in WT). (G) pame4 spindles grouped in the center of the ascus because of defective astral MT-mediated (arrows) nuclear movement. In pame4 (H) and sme4 (I), post-PMM nuclei are arranged randomly and often degenerate (compare with WT chromatin in D) before spore enclosure. (J) Abortive ascospores (arrows). (Scale bars: 5 μm.)
Discussion
Sme4/Pame4 is a large coiled-coil protein that is highly conserved in the Sordariale group of fungi, which includes over 500 known species. It plays central roles in two temporally and functionally distinct aspects of the sexual cycle, first as a component of the meiotic SC and then, after disappearing and reappearing, as a component of the SPB. This protein, with its two important dual roles, is a molecular component of either the SC or SPB in this large group of filamentous fungi. Analysis of corresponding mutants also gives insights into the functional interactions between SC formation and recombinosome localization.
Sme4 Is an SC Transverse Filament Component.
Eleven previously identified coiled-coil SC components lack significant primary sequence identity outside of immediate family members (4, 5). Sme4 family proteins fit this same pattern. Where examined, SC transverse filament components form parallel-oriented coiled-coil homodimers projecting from each of the two axes and meeting in the center of the SC (Table S1 and Fig. S4). Sme4 family proteins could have this same disposition (Fig. S4). However, the large size of these proteins plus other unique features (Table S1) raise the possibility of other organizations (Fig. S4). Specifically, single Sme4 coiled-coil homodimers (parallel or antiparallel) might extend from one axis to the other, which is seen for transverse filaments in tomography pictures of beetle SCs (16).
Sme4 Mediates Synapsis-Correlated Recombinosome Relocalization.
In WT meiosis, synapsis is immediately preceded by on-axis appearance of Msh4 foci and is then accompanied, on a region by region basis, by relocalization of Mer3, Rad51, and Msh4 foci from an on-axis to a between-axis disposition. Foci may be positioned at the axis/SC central region border. Sme4 is required for all of these events (i.e., efficient on-axis appearance of Msh4 foci before synapsis and ensuing on-axis to between-axis recombinosome relocalization). The role of Sme4 could be direct: Sme4-mediated changes that bring homolog axes together and install SC between them could concomitantly mediate transfer of recombinosome contacts from axis components to central region components, possibly Sme4 itself, at the axis/SC interface.
Studies of DNA events in budding yeast suggest that the recombinational interactions between homologs that mediate coalignment involve nascent (prestable strand exchange) interactions that are not yet physically identified (11). During onset of strand exchange, probably for both CO- and NCO-fated interactions, one (first) DSB end undergoes extensive strand invasion at its 3′ single-stranded DNA tail, whereas the other (second) end remains quiescent (11, 17). Thus, synapsis-correlated between-axis recombinosome foci could represent first DSB ends, which are prominently associated with the SC central region as they execute invasion of 3′ tails.
Dual Roles of Sme4/Pame4 for the SC and SPB.
Sme4 is not only a component of the SC but also the first known SPB component of filamentous fungi. SPB components have so far only been identified in budding and fission yeasts, where in both species, core SPB components are also prevalently coiled-coil proteins (reviewed in refs. 12 and 18). Interestingly, in both of its roles, Sme4 directly mediates the juxtaposition of two prominent structures: linking the homolog chromosome axes through the SC and raising one part of the PMM SPB relative to the other to produce a bent configuration, albeit with very different length scales (100 nm in SC vs. microns in the SPB). A similar dual role of cohesin for chromosomes and centrioles has recently emerged (19).
Sme4 is the unique case in which a molecule first localizes to the SC and then later, localizes to the SPB. Meiosis-specific relocalization of SPB proteins has been observed for budding and fission yeasts Mps3 and Sad1 proteins, respectively, that relocate from SPB to the NE during meiotic prophase and return to the SPB at metaphase I (20, 21). Also, the mouse Death Inducer-Obliterator DIDO3 isoform moves from centrosome to the SC during meiosis, although it is likely not an intrinsic structural component of either structure (22).
Links Among the SC, NE, and SPB.
Sme4/Pame4 proteins are specifically found during the sexual cycle, implying coordinate evolution within this program. Their presence in Sordariales and not in other ascomycetes like budding and fission yeasts (Fig. S1) can be correlated with the fact that only Sordariales undergo PMM. In yeasts, the SPB is the site of the prospore wall initiation, and proteins involved in vesicle docking are associated with the SPB during this process (23). In contrast, in Sordaria, Podospora, and Neurospora species, the prospore wall is not formed on the SPB; it is laid down along the ascus wall as a peripheral cylinder during PMM (14). Therefore, the SPB per se is not directly involved in ascospore wall formation like in yeasts. Instead, it plays crucial roles through its bent configuration for regular spacing of both spindles and PMM nuclei (above), which are, in turn, necessary for the formation of eight or four ascospores in the ascus cell (14).
The three major nucleus-related structures of meiosis, the SC, NE, and SPB, are linked in diverse phenomena. The roles of Sme4 for the SC and NE-associated SPB provide an additional link. Pairing and CO formation between homologs could provide a single unifying motivation for these relationships. Early in evolution, chromosome pairing and SC formation might have occurred in subtelomeric and/or centromeric regions, with juxtaposition of pairing centers mediated by cytoskeletal forces (linked through the NE) to these specialized chromosomal regions in complexes related to centrosomes/SPBs (24). Later in evolution, these effects would be integrated with recombinational interactions evolved from mitotic DSB repair to give DSB-mediated homolog coalignment plus a tendency for SC formation at sites of CO-designated interactions (10). The meiotic process in C. elegans (24) could reflect dominance of the primordial process, albeit with DSB-related events now present (25). In other organisms, the DSB-mediated process predominates, whereas the primordial process is now used to regularize topological relationships among already-paired homologs (2, 6) and in budding yeast, underlying early transient SC formation at paired centromeres during a backup homolog pairing/segregation process (26).
Materials and Methods
Strains, Cloning, and Transformation.
Culture conditions, media, and genetic methods for P. anserina are available at http://podospora.igmors.u-psud.fr. The PaMe4 gene was identified by positional cloning, and the SME4 gene (embl CABT01000092.1) was identified by sequence similarity (SI Materials and Methods). S. macrospora WT is St Ismier FGSC 4818. Null mutants were obtained by gene replacement where a hygromycin resistance cassette replaces the ORF (SI Materials and Methods).
SME4-GFP Fusion and Cytology.
The GFP coding sequence (p-EGFP-1; Clontech) was fused to SME4 just after the last C-terminal amino acid. The SME4-GFP gene, under control of its promoter, was ectopically integrated into a WT strain and by crosses to the different mutant strains (SI Materials and Methods). Immunofluorescence and hematoxylin staining are as described in ref. 27. Primary antibodies were MPM-2 (1:300) and anti–α-tubulin (1:600; Amersham). Secondary antibodies were Cy2 anti-mouse (1:100; Jackson). Observations were made on a Zeiss Axioplan microscope with a CCD Princeton camera.
Supplementary Material
Acknowledgments
We acknowledge the excellent technical assistance of Michel David, Sylvie Francois, and Philippe Le Flèche. E.E., C.V., and D.Z. were supported by grants from the Centre National de la Recherche Scientifique (Unité Mixte de Recherche 8621) and from the National Institutes of Health (GMO44794) (to N.E.K.). A.S. was supported by the Consiglio Nazionale delle Ricerche (Italy). N.E.K. was supported by National Institutes of Health Grant GM044794. P.S. and F.M. were supported by grants from Unité Mixte de Recherche 8621 and Agence Nationale de la Recherche Grant ANR-05-Blan-O385-02.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107272108/-/DCSupplemental.
References
- 1.Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
- 2.Storlazzi A, et al. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell. 2010;141:94–106. doi: 10.1016/j.cell.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH. Analysis of replication protein A (RPA) in human spermatogenesis. Mol Hum Reprod. 2007;13:837–844. doi: 10.1093/molehr/gam076. [DOI] [PubMed] [Google Scholar]
- 4.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanov YF, Grishaeva TM, Dadashev SY. Similarity of the domain structure of proteins as a basis for the conservation of meiosis. Int Rev Cytol. 2007;257:83–142. doi: 10.1016/S0074-7696(07)57003-8. [DOI] [PubMed] [Google Scholar]
- 6.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 7.Simonet JM, Zickler D. Mutations affecting meiosis in Podospora anserina. I. Cytological studies. Chromosoma. 1972;37:327–351. doi: 10.1007/BF00319874. [DOI] [PubMed] [Google Scholar]
- 8.Storlazzi A, et al. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003;17:2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci USA. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson KA, Keeney S. Synaptonemal complex formation: Where does it start? Bioessays. 2005;27:995–998. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- 11.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Heidelberg: Springer; 2006. pp. 381–442. [Google Scholar]
- 12.Jaspersen SL, Winey M. The budding yeast spindle pole body: Structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- 13.Thompson-Coffe C, Zickler D. Three microtubule-organizing centers are required for ascus growth and sporulation in the fungus Sordaria macrospora. Cell Motil Cytoskeleton. 1992;22:257–273. [Google Scholar]
- 14.Thompson-Coffe C, Zickler D. How the cytoskeleton recognizes and sorts nuclei of opposite mating type during the sexual cycle in filamentous ascomycetes. Dev Biol. 1994;165:257–271. doi: 10.1006/dbio.1994.1251. [DOI] [PubMed] [Google Scholar]
- 15.Horio T, et al. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- 16.Schmekel K, Daneholt B. The central region of the synaptonemal complex revealed in three dimensions. Trends Cell Biol. 1995;5:239–242. doi: 10.1016/s0962-8924(00)89017-0. [DOI] [PubMed] [Google Scholar]
- 17.Kim KP, et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zizlsperger N, Keating AE. Specific coiled-coil interactions contribute to a global model of the structure of the spindle pole body. J Struct Biol. 2010;170:246–256. doi: 10.1016/j.jsb.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura A, Arai H, Fujita N. Centrosomal Aki1 and cohesin function in separase-regulated centriole disengagement. J Cell Biol. 2009;187:607–614. doi: 10.1083/jcb.200906019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chikashige Y, et al. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 22.Prieto I, et al. Synaptonemal complex assembly and H3K4Me3 demethylation determine DIDO3 localization in meiosis. Chromosoma. 2009;118:617–632. doi: 10.1007/s00412-009-0223-7. [DOI] [PubMed] [Google Scholar]
- 23.Mathieson EM, Schwartz C, Neiman AM. Membrane assembly modulates the stability of the meiotic spindle-pole body. J Cell Sci. 2010;123:2481–2490. doi: 10.1242/jcs.062794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Smolikov S, Schild-Prüfert K, Colaiácovo MP. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 2008;4:e1000088. doi: 10.1371/journal.pgen.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsubouchi T, Macqueen AJ, Roeder GS. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008;22:3217–3226. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zickler D. Observing meiosis in filamentous fungi: Sordaria and Neurospora. Methods Mol Biol. 2009;558:91–114. doi: 10.1007/978-1-60761-103-5_7. [DOI] [PubMed] [Google Scholar]
- 28.Zickler D. Division spindle and centrosomal plaques during mitosis and meiosis in some ascomycetes. Chromosoma. 1977;30:287–304. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.