Abstract
The prevalent eye disease age-onset cataract is associated with aggregation of human γD-crystallins, one of the longest-lived proteins. Identification of the γ-crystallin precursors to aggregates is crucial for developing strategies to prevent and reverse cataract. Our microseconds of atomistic molecular dynamics simulations uncover the molecular structure of the experimentally detected aggregation-prone folding intermediate species of monomeric native γD-crystallin with a largely folded C-terminal domain and a mostly unfolded N-terminal domain. About 30 residues including a, b, and c strands from the Greek Key motif 4 of the C-terminal domain experience strong solvent exposure of hydrophobic residues as well as partial unstructuring upon N-terminal domain unfolding. Those strands comprise the domain–domain interface crucial for unusually high stability of γD-crystallin. We further simulate the intermolecular linkage of these monomeric aggregation precursors, which reveals domain-swapped dimeric structures. In the simulated dimeric structures, the N-terminal domain of one monomer is frequently found in contact with residues 135–164 encompassing the a, b, and c strands of the Greek Key motif 4 of the second molecule. The present results suggest that γD-crystallin may polymerize through successive domain swapping of those three C-terminal β-strands leading to age-onset cataract, as an evolutionary cost of its very high stability. Alanine substitutions of the hydrophobic residues in those aggregation-prone β-strands, such as L145 and M147, hinder domain swapping as a pathway toward dimerization. These findings thus provide critical molecular insights onto the initial stages of age-onset cataract, which is important for understanding protein aggregation diseases.
Keywords: beta-sheet protein folding, eye cataracts, nonnative interaction
Exploring the pathways of protein aggregation is crucial for preventing and/or treating a wide number of human degenerative diseases, such as Alzheimer’s disease, Huntington disease, type II diabetes, and cataract, which is a growing concern in today’s aging world population. Age-related cataract resulting from aggregation of lens crystallins (1) is responsible for 48% of world blindness (http://www.who.int/blindness/causes/priority/en/index1.html) and affects 20.5 million Americans age 40 and over (http://www.cdc.gov/visionhealth/basic_information/eye_disorders.htm). The α-, β- and γ-crystallins are structural proteins of the vertebrate eye lens, which must remain soluble and stable throughout lifetime in order to maintain lens transparency. Currently proposed models for cataract include protein unfolding as a result of oxidative or UV-induced damage (2, 3). Such partially unfolded protein conformations can participate in aberrant intermolecular interactions leading to crystallin aggregation. The remarkable stability of crystallins and chaperone function in the lens is presumed to prevent aggregation for a long period of time (i.e., approximately 40 years).
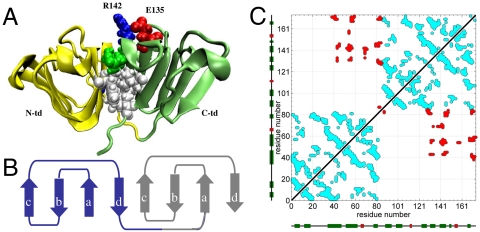
Human gamma D crystallin (γD-crys) is the third most abundant γ-crystallin in the lens and a significant component of the age-onset cataract. It is primarily located in the central lens nucleus, the oldest region of the body, and is therefore one of the longest-lived proteins in the human body. As shown in Fig. 1A, γD-crys is a monomeric protein composed of two structurally homologous domains (4). Each domain is composed of intercalated double β-sheet Greek key motifs (see Fig. 1B), a characteristic structural feature of the βγ-crystallin superfamily. The duplicated domains connected by a linker peptide form a highly conserved hydrophobic interface that plays a crucial role in determining long-term stability (see Fig. 1A). To further illustrate the complex topology of γD-crys protein, we plot the residue–residue contact map calculated based on its crystal structure in Fig. 1C. The contacts are colored differently, if they are intradomain or interdomain, to highlight the set of residues that comprise the domain–domain interface.
Fig. 1.
(A) A cartoon representation of human γD-crystallin. The N-terminal domain (N-td) and the C-terminal domain (C-td) are shown in yellow and green, respectively. The heavy side chain of the residues at the interdomain surface is shown in ball-stick representation. The E135–R142 residue pair is also shown that forms a stabilizing salt-bridge interaction, as predicted in earlier simulations. White color is used for nonpolar residues, while polar residues are shown in green. Acidic residues are colored in red and basic residues are colored in blue. (B) The complex topology of a crystallin domain consisted of two intercalated antiparallel β-sheet Greek Key motifs. Each motif is colored differently and the naming of the strands is illustrated. (C) The residue–residue contact map of the crystal structure of human γD-crystallin. A contact between residue i and j has been considered if any heavy atom of residue i is within 6.5 A of residue j in the crystal structure. The intradomain contacts are colored in cyan, whereas the interdomain contacts are colored in red. The secondary elements of the protein are also shown along the axes, with β-strands in green and helices in red.
Folding/unfolding experiments of γD-crys have indicated the existence of a partially unfolded intermediate with C-terminal domain (C-td) mostly folded and N-terminal domain (N-td) unfolded (5). Substitutions at the above-mentioned domain interface residues resulted in a sharp destabilization of the N-td and enhanced this folding intermediate population in experiments (5–7). The isolated N-td is able to fold on its own into the native configuration in experiments; however, the stability of the isolated N-td is much lower compared to that in the full monomer (8). These results suggest that (i) the C-td stabilizes the N-td in the context of full γD-crys monomer, and (ii) the C-td interface serves as a template for the N-td folding in the full protein.
Characterization of cataractogenesis in the native environment has been difficult due to the physical integrity of the lens. The direct identification of the state of aggregation precursors within the lens fiber cells or the intact lens has not been achieved due to experimental complications. However, the experimentally found partially unfolded intermediate conformation of γD-crys undergoes aggregation that competes with productive refolding, as observed during equilibrium unfolding/refolding experiments (9, 10). This in vitro off-pathway aggregation in GdmCl at pH 7 provides a unique model for studying crystallin aggregation in vivo. Atomic force microscopy indicated that the aggregated state of γD-crys is ordered filament-like (10). The bis-ANS binding of γD-crys aggregate species suggested presence of exposed hydrophobic pockets (10). More recent experiments reveal that at low pH, γD-crys polymerize into amyloid fibrils (11), similar to those observed in neurodegenerative diseases.
Taken together, the detailed mechanism of crystallin polymerization is still unknown due to a lack of structural information of the monomeric and oligomeric aggregation-prone species. Mapping the initial pathways of crystallin aggregation can provide a route toward targeted searches for therapeutic agents inhibiting pathological deposition for a number of protein deposition diseases including cataract. Molecular dynamics simulations (12, 13) of protein models at different resolutions, from simple models (lattice and off-lattice) (14–17) to continuum solvent models (18, 19) to all-atom explicit solvent models (20, 21), have served as a powerful tool to complement existing experimental techniques to advance our fundamental understanding of protein folding and aggregation. In this study, using extensive atomistic molecular dynamics simulations performed on the IBM Blue Gene/L supercomputer we have characterized unfolding of human gamma D crystallin followed by oligomerization, which is consistent with the current models of cataract. A partially unfolded intermediate conformation strikingly similar to the experimentally observed aggregation-prone folding intermediate species is detected in our ≥2 μs denaturation simulations. This unfolded intermediate species has its C-td more native-like and the N-td largely unfolded. To explore the aberrant protein–protein interactions promoting γD-crys aggregation, we further simulated two partially unfolded monomeric conformations together. Our ≥1 μs simulated annealing molecular dynamics simulations of the modeled γD-crys dimeric system reveal successive domain swapping at the C-terminal β-strands as a molecular mechanism for γ-crystallin aggregation. Thus, this simulation study uncovers a molecular picture of the unfolding and polymerization reactions associated with the current models for cataract formation. Because crystallins are model proteins for understanding β-sheet folding, these molecular pictures obtained from simulated unfolding and oligomerization of human γD-crystallin can also offer important clues to solve the so-called protein folding as well as to understand the misfolding and aggregation of β-sheets implicated in protein conformational diseases.
Results and Discussion
An Intermediate State Is Populated During Unfolding of γD-crys Monomer.
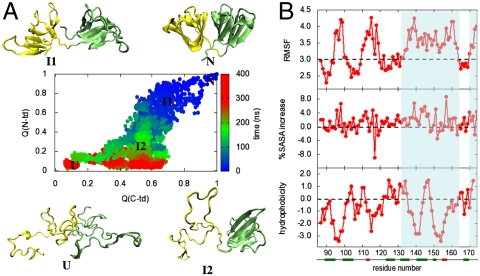
Fig. 2A summarizes the primary events of the simulated unfolding reaction of the native monomeric γD-crys protein performed at 425 K in 8 M aqueous urea. In this figure the time evolution of the fraction of native contacts formed for the two domains, Q(N-td) and Q(C-td) are plotted against each other, as obtained from different unfolding trajectories with a total simulation time of approximately 2 μs. A native contact is considered to be formed between residues i and j if a heavy atom of residue i is within 6.5 Å of a heavy atom of residue j in the crystal structure. For the unfolded protein, Q(N-td) and Q(C-td) are close to 0, whereas for the folded state, both Q(N-td) and Q(C-td) are ≈1. The time evolution of Q(N-td) and Q(C-td) clearly shows sequential unfolding of two domains, consistent with experimental results (5–7, 10). The unfolding of N-td always preceded that of C-td, as indicated by the rapid loss of Q(N-td) compared to Q(C-td). This higher stability of the C-td agrees well with experimental data (22). We also show characteristic conformations populated at different stages of unfolding in Fig. 2A. The structural details of these conformations suggest that the interdomain contacts are broken before denaturation of the N-td starts [at Q(C-td) = ∼ 0.6, Q(N-td) = ∼ 0.6, I1 configuration]. Next, a partially unfolded conformation (I2) with its C-td folded (Q(C-td) = ∼ 0.4) and N-td largely denatured [Q(N-td) < 0.3] is populated, in agreement with the experimental finding of a γD-crys folding intermediate (10). The C-td finally unfolds (to U conformation) to complete γD-crys denaturation.
Fig. 2.
(A) Simulated unfolding of human γD-crystallin at 425 K in 8 M aqueous urea. The fraction of native contacts formed, Q(?), for the two domains are plotted against each other, as obtained from an aggregate of approximately 2 μs of unfolding simulations. Each point on this plot is colored from blue to red according to its time sequence during unfolding. Typical conformations populated at different stages of unfolding are also shown. (B) Structural changes of the C-td in the I2 ensemble. The RMSF in Å from the native structure, the percentage of solvent-exposed surface area (%SASA) change, and the Kyte–Doolittle hydrophobicity are plotted for each residue of the C-td within the I2 ensemble consisting > 1,000 conformations with QC-td≥0.4 and QN-td ≤ 0.3. The regions that undergo strong conformational fluctuation and/or solvent exposure, as well as contain hydrophobic residues, are highlighted.
Exposure of Hydrophobic Patches Within the C-td upon N-td Unfolding.
Because the structure of the I2 ensemble populated in the simulated unfolding is highly similar to the experimentally detected folding intermediate with a more native-like C-td and an unfolded N-td that serves as the aggregating precursor in vitro, we further analyze the structure of the I2 ensemble to identify crucial structural changes. Although the C-td remains more native-like compared to the N-td in the I2 ensemble, the C-td also experiences partial unfolding, as indicated by an approximately 60% reduction of the Q(C-td) from the crystal structure. We also find that, upon N-td unfolding the C-td undergoes a net approximatley 23% increase in solvent-exposed surface area (probed using a sphere of 1.4 Å) from the crystal structure. Recent experiments also suggested that a partial unfolding of the C-td is required for aggregation, further demonstrating the structural similarity between the simulated folding intermediate and the experimentally detected aggregation-prone species (23). To map this structural change of the C-td at the residue level, the root-mean-square fluctuation (RMSF) and the %SASA increase in the I2 ensemble as well as the hydrophobicity of each residue within the C-td were calculated (Fig. 2B). The RMSF per residue plot clearly indicates that only the loop regions from Greek key motif 3 of the C-td become more flexible upon N-td unfolding. On the other hand, residues 132–164 from the Greek Key motif 4 that include a, b, and c strands suffer large structural fluctuation (> 3 Å). Those residues encompassing the interdomain interface also experience strongest solvent exposure within the C-td with a net contribution of > 50% to the total increase in SASA. The hydrophobicity analysis reveals presence of eight hydrophobic residues in the 132–164 region, as delineated by the widely used Kyte–Doolittle hydrophobicity scale, in which regions with values above 0 are defined as hydrophobic. Taken together, these analyses suggest that the a, b, and c strands from the Greek key motif 4 that contain a large hydrophobic region experience large structural perturbation as well as strong solvent exposure upon unfolding of the N-td.
Simulations of Oligomerization of Aggregation-Prone Monomeric Species.
Because of its high structural resemblance with the experimentally detected aggregation-prone folding intermediates, the partially unfolded intermediate with its C-td more native-like and N-td largely unstructured detected in our unfolding simulations provides us an excellent candidate to study the mechanism of intermolecular association during the competing off-pathway polymerization reaction in vitro. To characterize the intermolecular interactions between those aggregation-prone species of γD-crys we designed an in silico experiment. In this experiment, the conformational dynamics of one partially unfolded monomer in water is studied in presence of a second partially unfolded molecule by using extensive atomistic molecular dynamics simulations combined with a thermal annealing protocol (see System and Methods for details). The initial monomeric conformations for this experiment are selected from the I2 ensemble (see Fig. 1C). The simulations of the dimeric γD-crys are then performed in three different steps: (i) The N-tds of the monomers are first fully unfolded by thermal denaturation; (ii) the conformational space of the denatured N-tds is then explored by simulated annealing molecular dynamics; (iii) the whole system is allowed to relax at 350 K. During step i and step ii, the C-tds of the monomers remain constrained and only N-tds are allowed to move. Nine different sets of dimeric systems were simulated starting from different starting monomeric conformations representing the I2 ensemble. In the dimeric system, the distance between the C-tds was varied from 30 Å to 56 Å. In addition, the relative orientation of the monomers was also varied by changing the monomer–monomer angle from 70° to 160°. The initial (at the end of stage i) and final conformations (at the end of stage ii and iii) for these trajectories are shown in Fig. S1. For comparison, the individual monomers alone were also simulated using the same protocol stated above. During this in silico experiment with modeled monomers and dimers, we consider the N-td to be unfolded, if radius of gyration, Rg, is > 20 Å, and solvent accessible surface area (SASA) is > 10,000 Å2. A collapsed state of the N-td is considered if Rg is ≤ 15 Å and SASA is < 7,500 Å2 (the native N-td has a Rg of 11.7 Å and a SASA of 4,850 Å2). Thus, a > 5 Å lowering in Rg and a > 2,500 Å2 decrease in SASA define the collapse of the N-td in this study. We should also emphasize that the simulated annealing molecular dynamics simulations performed in this study are used to solely investigate the nonspecific collapse, but not the folding, of the N-td polypeptide chain; as such, study is impossible with current computational resources.
Collapsed N-td Forming Interdomain Contacts in the Dimeric Structure.
The time evolution of the radius of gyration of the N-td (see Fig. S2A) was monitored during the in silico experiment to follow the conformational change of the monomers. We find that the denatured N-tds of the partially folded monomers, both in isolation and in presence of a second monomer, undergo a collapse at 350 K, as indicated by the sharp decrease in both radius of gyration and solvent accessible surface area. The time scale of this simulated collapse (the first time N-td experiences a nonspecific collapse starting from the completely denatured state generated at the end of step i) of the N-td is approximately 20–100 ns. Fig, S2A, Top, illustrates the characteristic behavior of the radius of gyration of the N-td during one such simulation of the dimeric system, suggesting formation of a collapsed state (Rg < 15 Å) from an extended state (Rg > 25 Å) of the N-td. Table S1 summarizes the initial intermonomer orientation and distance as well as the radius of gyration of the N-td in the final dimeric ensemble for all nine runs performed in this study. We found that the collapse of the N-td is favored in presence of a second monomer, if the two monomers are oriented in an antiparallel manner (the angle between two monomers approaching 180°) and the distance between two monomers is lowered (see Table S1). This observation suggests possible formation of intermolecular interactions facilitating the N-td collapse.
Next, the number of contacts between the N-td and the C-td, NQN-C, was estimated as a function of simulation time (Fig. S2A, Bottom). An interdomain contact between the N-td and the C-td is considered, if the Cα atom of any residue i from the N-td is within 10 Å of the Cα atom of any residue from the C-td. An increase in the total number of interdomain contacts, NQN-C, was observed during the N-td collapse for the monomers as well as for the dimers, as shown in Fig. S2A. Such an increase in NQN-C advocates the essential interaction of the N-td with certain regions of the C-td during its collapse.
To investigate the time order of the collapse of the N-td and its interaction with the C-td, we have estimated the cross-correlation function between Rg and NQN-C time series. Fig. S2B shows the typical cross-correlation found between Rg of the N-td and NQN-C as a function of time lags for one I2 conformation during simulations of the isolated molecule and in presence of a second conformation. The cross-correlation reaches its peak at time lag approximately 0 for all simulations, suggesting that the collapse of the N-td is consistently accompanied by formation of contacts with the C-td for simulations of both monomers and dimers.
Mechanism for γD-protein Oligomerization.
The final collapsed conformations at the end of simulations were always found to form interdomain interactions. Those interactions were both intramonomer and intermonomer for the dimeric system—i.e., the N-td of a monomer can interact with its own C-td or with the C-td of the neighboring monomer, which is strongly determined by the initial conformations. For example, if the two monomers are aligned to each other in an antiparallel manner and they are closer to each other, more intermonomer interactions are formed between the N-td and the C-td (see Table S1). A few representative conformations from the ensemble of dimeric γD-crys with collapsed N-tds, which is populated at the end of our simulations, are shown in Fig. 3A (see also Fig. S1). Such conformations represent stable endpoints of the simulation and are stable for tens of nanoseconds. In the first example, we show a final conformation in which both N-tds interact with the C-tds in intra- as well as in intermolecular fashion, forming a structure similar to a close-ended dimer. In the second example, one N-td forms contacts with the C-td from the same monomer, whereas the other N-td interacts with the C-d in both intra- and intermolecular fashion. We also find conformations in which the N-td of one monomer remains denatured, whereas the N-td of the second monomer undergoes collapse by forming interchain contacts with the C-td, thus forming conformation similar to a N-terminal open-ended dimer (example 3 and 4 in Fig. 3A). These simulations thus reveal that the oligomerization of the partially unfolded intermediates can occur through interactions at the domain interface. The final dimeric ensemble obtained from those trajectories contains structurally different domain-swapped conformations, which are either close-ended or open-ended. However, estimating the relative abundance and stability of those dimeric species require simulations, which is beyond the limit of the current study. From the current simulations, it appears that the close-ended domain-swapped dimer formation is favorable, when the two monomers are oriented in an almost antiparallel manner. As the relative orientation starts deviating from antiparallel, formation of more open-ended domain-swapped dimers is noticed.
Fig. 3.
Domain-swapped dimers of γD-crys. (A) Final domain-swapped dimer configurations are shown from three different runs. The folded C-td and the unfolded N-td of one monomer are colored in green and yellow, respectively. For the second monomer, the folded C-td and the unfolded N-td are colored in blue and red, respectively. (B) The average number of contacts with the collapsed N-td is plotted for each residue within the C-td, as obtained from an ensemble of > 1,000 conformations, for both monomeric and dimeric systems. Those conformations are populated during the last 50 ns of the simulations at approximately 350 K, in which the N-tds are collapsed. The results obtained from isolated monomer simulations are colored in black. The probabilities of intra-, inter-, and total (intra- + inter-) interdomain contact formation per residue in dimers are plotted in cyan, red, and gray, respectively. (C) The number of interchain contacts formed between the N-td and every residue of the Greek key motif 4 during the last 25 ns of MD at 350 K, for (left) the wild-type dimer and for (right) the double mutant dimer. The number of interchain interdomain contacts is colored according to the color scale shown. The alanine substitution sites are indicated with an red arrow, where a decrease in intermolecular interaction is noticed in the double mutant system. Results for one representative trajectory are shown.
To examine the propensity of forming inter- or intramolecular contacts within the C-td, we analyze the average number of inter- and intramolecular contact formation per residue of the C-td in an ensemble of > 1,000 final dimeric conformations that are collected from trajectories of runs 1–4 (Table S1) showing domain swapping, in which N-tds are collapsed (Fig. 3B). For comparison, we also plot the propensity of interdomain contact formation in the final conformational ensemble of isolated monomers. The regions from the C-td that were identified with high number of contact formation with the collapsed N-td, both in dimers and monomers, resemble the native interdomain interface. One exception is the participation of the 110–120 loop in the interaction with the N-td in the isolated monomers, which becomes partially unstructured in the I2 ensemble (see Fig. 3B). In the dimeric structures, the C-td residues are found to interact with the N-td in both intra- and intermonomer fashion. Particularly, residues 135–164 comprising a, b, and c strands from the Greek Key motif 4 are frequently found to interact with the N-td in an intermolecular manner (see Fig. 3B). Remarkably, the same region containing six hydrophobic residues experience partial unfolding and strong solvent exposure in the monomeric partially unfolded folding intermediate species. Thus, these findings suggest that, about 30 amino acids including three antiparallel β-strands from the C-td, which also comprise the native domain-domain interface, are swapped between adjacent monomers to facilitate the intermolecular association of partially unfolded aggregation-prone conformations of γD-crys. As a result, a “native-like” interdomain surface is formed in the dimers. In the current context, presence of a native-like interdomain surface in the dimeric system suggests participation of residues from the C-td, which also comprise the native interdomain surface (see Fig. 1C), in the formation of intermonomer linkage. As seen in the crystal structure, the interdomain contacts primarily involve a, b, c, and d strands of the Greek Key motif 4 (Fig. 1C). From the simulated dimric ensemble, it is clear that a, b, and c strands of the Greek Key motif 4 significantly contribute to the formation of the intermonomer surface, thus resulting into a native-like interdomain surface in the dimers (Fig. 3B).
As mentioned earlier, six of those residues are strongly hydrophobic. To test if those hydrophobic residues indeed play a key role in promoting dimerization, we have substituted in silico two of those six residues, L145 and M147, with alanine and simulated the dimerization of the double mutant protein using similar protocol as used for the wild-type protein. Strikingly, the final ensemble for the double mutant shows less inter- as well as intrachain interdomain contact formation compared to the wild-type protein. For direct comparison, simulations of the double mutant and wild-type dimers were started from identical initial conformations, except that the residues L145 amd M147 were substituted with alanines. Fig. 3C shows the residues from the Greek Key motif 4 of the C-td of one monomer that form contact with the N-td of the second molecule during the last 25 ns of one representative MD trajectory, for the wild-type (left) and for the double mutant (right). Clearly, the intermolecular swap of the ab hairpin from the Greek Key motif 4 takes place to a much lower extent in the double mutant compared to the wild-type protein. The residues L145 and M147 are major components of the hydrophobic interdomain interface in the monomeric γD-crys, which significantly contributes to the overall native stability. Our simulations suggest that substituting those two residues with less hydrophobic ones may hinder domain swapping as a pathway toward dimerization in γD-crystallin.
Role of Interdomain Surface in Crystallin Stability.
All known βγ-crystallins from extant vertebrate species have duplicated Greek key domains that are presumably evolved by gene duplication and gene fusion from an ancestral single domain crystallin (24, 25), as suggested by the high sequence and structural similarity of the two domains. In fact, such single domain crystallins have been identified—for example, in the Sea squirt Ciona (26). These organisms may represent descendants of lineages that are candidates for the origin of the vertebrates. It is also noteworthy that most βγ-crtstallins exhibit differential domain stability (27), suggesting that addition of a second domain and the interdomain interface to the ancestral single domain protein contribute to the overall stability of the full-length protein. In fact, stability comparisons of the isolated domains with the full γD-crys monomer indicated that the domain interface contributes a ΔGH2O of approximately 4.2 kcal/mol to the stability of the full monomer (22). This additional stability conferred by the interfacial interactions between the duplicated domains is likely very important for maintaining the long life time of proteins of the lens nucleus, and thus explains the selection for duplicated forms.
The crucial role of the interdomain interface in the folding, stability, and aggregation of βγ-crystallins is supported by experiments (5–7, 28). In a previous unfolding simulation study of the isolated domains of γD-crys, we have shown that the a and b strands from the Greek Key motif 4 comprising the interdomain interface are the most stable structure within full γD-crys (29). We also uncovered that a Glu-Arg salt-bridge at the topologically equivalent positions of residues E135 and R142 (see Fig. 1A and ref. 29) plays a significant role in determining the stability of a Greek Key motif. Disrupting the E135-R142 salt-bridge in silico resulted in destabilizing the interdomain interface and facilitated the N-td unfolding (29). Similarly, an Arg14 to Cys (R14C) mutation on γD-crys breaking the E7-R14 salt-bridge in the Greek Key motif 1, which is associated with a juvenile-onset hereditary cataract (30), triggers formation of intermolecular aggregates in physiological pH.
Domain Swapping as a Molecular Mechanism for Crystallin Polymerization.
Domain swapping (31, 32) has been recognized as an aggregation mechanism for a number of proteins—for example, human prion protein (33) and β2-microglobulin (34). Aggregation by domain swapping in an open-ended fashion has been observed for cystatin C (35). Successive domain swapping of a single domain has been previously suggested as a mechanism for polymerization on the basis of the dimeric and/or trimeric structures of Serpin (36), RNAse A (37), and cytochrome C (38). Domain swapping of an α-helix has been reported in the minor dimeric structure of RNase A (39) and in staphylococcal nuclease (40), as well as in the dimeric and trimeric structures of cytochrome c (38). The major dimeric component of RNase A is formed by swapping of the C-terminal β-strands (41). The crystal structure of a stable dimer of serpin, a protein family that forms large stable multimers leading to intracellular accretion and disease, revealed a domain-swapped structure of two long antiparallel β-strands (36). Domain-swapped trimeric structures have been also reported for Barnase (42) and an antibody fragment (43), but the mechanism of polymerization was not elucidated.
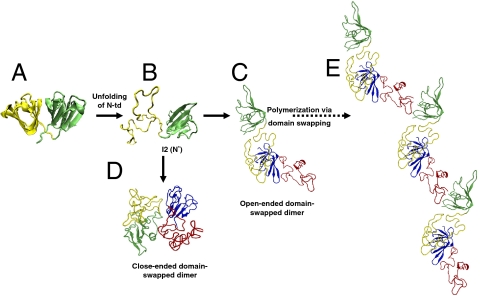
Domain-swapped polymers have been detected for a number of functional proteins as well, such as T7 helicase, RecA, and carbonic anhydrase (32). Solved structures of the βB2-crystallins show natural close-ended domain-swapped dimer, in which the N-td interacts with the C-td intermolecularly (44). Based on the presence of duplicated domains and the crucial role of native interdomain surface in γD-protein stability, together with the in vitro off-pathway aggregation reaction of the folding intermediate, domain swapping has been proposed as a plausible model for γD-crys polymerization. However, aggregation by a domain swap mechanism has not been experimentally observed for the crystallins. Our large-scale simulations provide direct evidence supporting this model, in which the aggregation-prone folding intermediate is found to form domain-swapped dimeric structures. Structures similar to both open-ended and close-ended domain-swapped dimers were detected in our simulations, which suggests successive domain swapping as a possible mechanism for crystallin aggregation in aged lens leading to cataract (Fig. 4). We identify structural components of γD-crystallin that are prone to domain swapping. This information can help us to design drug-like molecules that can prevent cataractogenesis in eyes. For example, small molecules that can attach to the native domain–domain interface and, therefore, hinder unfolding of the N-td can serve as drugs to avoid cataract. Those molecules can provide additional stability to the interdomain surface and prevent the C-terminal β-strands from solvent exposure. In addition, based on simulations we propose two potential mutation sites, L145 and M147, in which alanine substitution can retard dimerization via domain swapping.
Fig. 4.
Schematic summary of human γD-crys polymerization. (A) Crystal structure of human γD-crys. (B) Simulated monomeric aggregation precursor (I2), often referred as N* in the general mechanism of protein aggregation in literature. (C) Simulated structure of open-ended domain-swapped dimer. (D) Simulated structure of close-ended domain-swapped dimer. (E) Model of human γD-crys hexamer formed via domain swapping. The coloring scheme for the dimer system is same as in Fig. 3.
The findings of this simulation study, together with the previous experiments, shed important insights onto the pathways of crystallin aggregation, which can be connected to the more general theory of protein aggregation, suggesting the critical role of an aggregation-prone monomeric species (termed as I2 in this study, often referred as N* in literature; see refs. 45 and 46 and references therein). The role of N* state has been implicated in the aggregation pathways of transthyretin, prion proteins, and a variety of amyloid proteins including aβ protein. Any changes in the solution or environmental conditions (e.g., heat, UV rays) or mutations that destabilize the native γ-crystallin will trigger the formation of the aggregation-prone partially unfolded monomeric folding intermediates (N*). In such circumstances, successive domain swapping of the C-terminal strands may lead to polymerization in a highly crowded lens environment (Fig. 4). The emerging role of the native interdomain surface in triggering polymerization indicate that the crystallin aggregation in aged eye lens is an evolutionary cost of the long-term native state stability, which, in turn, is determined by its complex domain architecture. Because the successive intermolecular protein association by means of β-sheet expansion (47) is also the mechanism underlying a multitude of diseases including Alzheimer’s disease, Huntington disease, Parkinson disease, and the prion encephalopathies, the detailed picture of crystallin unfolding followed by polymerization revealed in this study may provide valuable information toward understanding those conformational diseases (48).
Conclusion
Large-scale atomistic simulations were used to reveal the molecular structures of the aggregation-prone intermediate species of human γD-crystallin, an eye lens protein implicated in cataracteogenesis. The partially unfolded conformation with a mostly folded C-td and a largely denatured N-td identified in the unfolding simulations has a structure strikingly similar to the aggregation-prone folding intermediate species observed in experiments. A detailed structural characterization of this pathogenic monomeric state N* reveals the presence of a solvent-exposed partially unfolded region within the more native-like C-td, which is approximately 30 amino acids long and contains eight hydrophobic residues. This region includes the first three antiparallel strands from the Greek Key motif 4, which also encompasses the interdomain interface crucial for native γD-crystallin stability. We further simulated the intermolecular association of these monomeric partially unfolded aggregation precursors (N*) to elucidate the mechanism of γD-crystallin aggregation. The denatured N-tds experience a distinct nonspecific collapse that is essentially accompanied by formation of specific interactions with the C-tds. As a result, domain-swapped dimeric conformations were populated in our simulations. The partially unfolded and solvent-exposed region encompassing a, b, and c strands from the Greek Key motif 4 in N* ensemble was found to be exchanged between adjacent monomers, forming a native-like interdomain interface in the dimeric structures. We also observed that alanine substitutions of the hydrophobic residues in those aggregation-prone β-strands, such as L145 and M147, located at the end of the ab hairpin of the Greek Key motif 4 hinders domain swapping as a pathway toward dimerization. In an older lens, the native proteins may aggregate by propagating domain swapping in an open-ended fashion along multiple partially unfolded monomers leading to cataract. This simulation study thus provides critical insights onto the molecular mechanism of initial stages of age-onset cataract formation, which is also important toward understanding other protein aggregation diseases.
System and Methods
The initial structure of the wild-type γD-crystallin protein (see Fig. 1A) containing 173 residues has been taken from the crystal structure deposited in the Protein Data Bank (PDB ID code 1HK0). The molecular system for unfolding studies was prepared by immersing the full monomer in 8M urea. The system contained approximately 7,650 water molecules and approximately 1,775 urea molecules—a total of approximately 40,000 atoms. The partially unfolded intermediate conformations were generated by unfolding the full γD-crys monomer at 425 K using 8 M aqueous urea as a denaturant. Typical length of such unfolding simulations was > 300 ns. Details of the simulation setup and analysis of γD-crys unfolding can be also found in SI Text and ref. 29.
To explore the oligomerization pathway(s) of γD-crys, two partially unfolded γD-crys molecules were placed together in a box of size ∼100 Å × 100 Å × 100 Å containing approximately 26,000 TIP3P water molecules. Simulations were performed with different relative orientation ranging from 70° to 160° and with distance between two C-tds ranging from 30 Å to 56 Å. The system containing a total of approximately 100,000 atoms then went through a sophisticated annealing process (details in SI Text). Finally, we allowed the complete system to relax at 350 K for > 50 ns. This in silico experiment on simulating dimeric structures was repeated nine times starting from different configurations, in which the monomer conformations as well as the intermonomeric distance and orientation were varied. The partially folded monomers alone were also simulated in water using the same protocol to compare the intramolecular interactions with the intermolecular interactions present in the dimeric system. L144 and M146 were mutated in silico to alanine to create the initial structure of the double mutant dimer, which was then simulated using a similar protocol stated above. The aggregate MD simulation time for both wild-type and the mutant oligomers is approximately 2 μs.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019152108/-/DCSupplemental.
References
- 1.Benedek GB. Cataract as a protein condensation disease: The Proctor lecture. Invest Ophthalmol Vis Sci. 1997;38:1911–1921. [PubMed] [Google Scholar]
- 2.Aarts HJM, Lubsen NH, Schoenmakers JGG. Crystallin gene expression during rat lens development. Eur J Biochem. 1989;183:31–36. doi: 10.1111/j.1432-1033.1989.tb14892.x. [DOI] [PubMed] [Google Scholar]
- 3.Lampi KJ, Shih M, Ueda Y, Shearer TR, David LL. Lens proteomics: Analysis of rat crystallin sequences and two-dimensional electrophoresis map. Invest Ophthalmol Vis Sci. 2002;43:216–224. [PubMed] [Google Scholar]
- 4.Basak A, et al. High-resolution X-ray crystal structures of human gamma D crystallin (1.25 angstrom) and the R58H mutant (1.15 angstrom) associated with aculeiform cataract. J Mol Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 5.Flaugh SL, Kosinski-Collins MS, King J. Interdomain side-chain interactions in human gamma D crystallin influencing folding and stability. Protein Sci. 2005;14:2030–2043. doi: 10.1110/ps.051460505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaugh SL, Kosinski-Collins MS, King J. Contributions of hydrophobic domain interface interactions to the folding and stability of human gamma D-crystallin. Protein Sci. 2005;14:569–581. doi: 10.1110/ps.041111405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaugh SL, Mills IA, King J. Glutamine deamidation destabilizes human gamma D-crystallin and lowers the kinetic barrier to unfolding. J Biol Chem. 2006;281:30782–30793. doi: 10.1074/jbc.M603882200. [DOI] [PubMed] [Google Scholar]
- 8.Iliopoulos I, et al. Evaluation of annotation strategies using an entire genome sequence. Bioinformatics. 2003;19:717–726. doi: 10.1093/bioinformatics/btg077. [DOI] [PubMed] [Google Scholar]
- 9.Kosinski-Collins MS, Flaugh SL, King J. Probing folding and fluorescence quenching in human gamma D crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 2004;13:2223–2235. doi: 10.1110/ps.04627004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosinski-Collins MS, King J. In vitro unfolding, refolding, and polymerization of human gammaD crystallin, a protein involved in cataract formation. Protein Sci. 2003;12:480–490. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papanikolopoulou K, et al. Formation of amyloid fibrils in vitro by human gamma D-crystallin and its isolated domains. Mol Vis. 2008;14:81–89. [PMC free article] [PubMed] [Google Scholar]
- 12.Warshel A. Molecular dynamics simulations of biological reactions. Acc Chem Res. 2002;35:385–395. doi: 10.1021/ar010033z. [DOI] [PubMed] [Google Scholar]
- 13.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Mol Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 14.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 15.Dill KA, et al. Principles of protein folding—a perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das P, et al. Characterization of the folding landscape of monomeric lactose repressor: Quantitative comparison of theory and experiment. Proc Natl Acad Sci USA. 2005;102:14569–14574. doi: 10.1073/pnas.0505844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das P, Matysiak S, Clementi C. Balancing energy and entropy: A minimalist model for the characterization of protein folding landscapes. Proc Natl Acad Sci USA. 2005;102:10141–10146. doi: 10.1073/pnas.0409471102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feig M, Brooks CL. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Zhou R, Berne BJ. Can a continuum solvent model reproduce the free energy landscape of a beta -hairpin folding in water? Proc Natl Acad Sci USA. 2002;99:12777–12782. doi: 10.1073/pnas.142430099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmerling C, Strockbine B, Roitberg AE. All-atom structure prediction and folding simulations of a stable protein. J Am Chem Soc. 2002;124:11258–11259. doi: 10.1021/ja0273851. [DOI] [PubMed] [Google Scholar]
- 21.Snow CD, Nguyen H, Pande VS, Gruebele M. Absolute comparison of simulated and experimental protein-folding dynamics. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- 22.Mills IA, Flaugh SL, Kosinski-Collins MS, King JA. Folding and stability of the isolated Greek key domains of the long-lived human lens proteins gammaD-crystallin and gammaS-crystallin. Protein Sci. 2007;16:2427–2444. doi: 10.1110/ps.072970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau KL, King J. Hydrophobic core mutations associated with cataract development in mice destabilize human gamma D-crystallin. J Biol Chem. 2009;284:33285–33295. doi: 10.1074/jbc.M109.031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubsen NH, Aarts HJM, Schoenmakers JGG. The evolution of lenticular proteins- the beta-crystallin and gamma-crystallin super gene family. Prog Biophys Mol Biol. 1988;51:47–76. doi: 10.1016/0079-6107(88)90010-7. [DOI] [PubMed] [Google Scholar]
- 25.Piatigorsky J. Crystallin genes: Specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics. 2003;3:131–137. [PubMed] [Google Scholar]
- 26.Shimeld SM, et al. Urochordate beta gamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr Biol. 2005;15:1684–1689. doi: 10.1016/j.cub.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 27.Mayr EM, Jaenicke R, Glockshuber R. The domains in gamma B-crystallin: Identical fold-different stabilities. J Mol Biol. 1997;269:260–269. doi: 10.1006/jmbi.1997.1033. [DOI] [PubMed] [Google Scholar]
- 28.Palme S, Slingsby C, Jaenicke R. Mutational analysis of hydrophobic domain interactions in gamma B-crystallin from bovine eye lens. Protein Sci. 1997;6:1529–1536. doi: 10.1002/pro.5560060717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das P, King JA, Zhou R. β-strand interactions at the domain interface critical for the stability of human lens γD-crystallin. Protein Sci. 2010;19:131–140. doi: 10.1002/pro.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pande A, Gillot D, Pande J. The cataract-associated R14C mutant of human gamma D-crystallin shows a variety of intermolecular disulfide cross-links: A Raman spectroscopic study. Biochemistry. 2009;48:4937–4945. doi: 10.1021/bi9004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melanie J, Bennett MPSDE. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knaus KJ, et al. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat Struct Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 34.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by beta-2 microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 35.Wahlbom M, et al. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J Biol Chem. 2007;282:18318–18326. doi: 10.1074/jbc.M611368200. [DOI] [PubMed] [Google Scholar]
- 36.Yamasaki M, Li W, Johnson DJD, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 37.Sambashivan S, Liu YS, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 38.Hirota S, et al. Cytochrome c polymerization by successive domain swapping at the C-terminal helix. Proc Natl Acad Sci USA. 2010;107:12854–12859. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Hart PJ, Schlunegger MP, Eisenberg D. The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1-Å resolution. Proc Natl Acad Sci USA. 1998;95:3437–3442. doi: 10.1073/pnas.95.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green SM, Gittis AG, Meeker AK, Lattman EE. One-step evolution of a dimer from a monomeric protein. Nat Struct Mol Biol. 1995;2:746–751. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 41.Liu YS, Gotte G, Libonati M, Eisenberg D. Structures of the two 3D domain-swapped RNase A trimers. Protein Sci. 2002;11:371–380. doi: 10.1110/ps.36602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zegers I, Deswarte J, Wyns L. Trimeric domain-swapped barnase. Proc Natl Acad Sci USA. 1999;96:818–822. doi: 10.1073/pnas.96.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei XY, Holliger P, Murzin AG, Williams RL. The 2.0-A resolution crystal structure of a trimeric antibody fragment with noncognate VH-VL domain pairs shows a rearrangement of VH CDR3. Proc Natl Acad Sci USA. 1997;94:9637–9642. doi: 10.1073/pnas.94.18.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith MA, Bateman OA, Jaenicke R, Slingsby C. Mutation of interfaces in domain-swapped human beta B2-crystallin. Protein Sci. 2007;16:615–625. doi: 10.1110/ps.062659107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Straub JE, Thirumalai D. Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu Rev Phys Chem. 62:437–463. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Straub JE, Thirumalai D. Principles governing oligomer formation in amyloidogenic peptides. Curr Opin Struct Biol. 20:187–195. doi: 10.1016/j.sbi.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison RS, Sharpe PC, Singh Y, Fairlie DP. In: Reviews of Physiology, Biochemistry and Pharmacology. Kramer K, Krayer O, Lehnartz E, Muralt Av, Weber HH, editors. Vol. 159. Berlin: Springer; 2007. pp. 1–77. [DOI] [PubMed] [Google Scholar]
- 48.Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.