Abstract
Half a century has passed since the discovery of operons (groups of genes that are transcribed together as a single mRNA). Despite the importance of operons in bacterial gene networks, the relationship between their organization and gene expression remains poorly understood. Here we show using synthetic operons in Escherichia coli that the expression of a given gene increases with the length of the operon and as its position moves farther from the end of the operon. These findings can be explained by a common mechanism; increasing the distance from the start of a gene to the end of the operon (termed the “transcription distance”) provides more time for translation to occur during transcription, resulting in increased expression. We confirmed experimentally that the increased expression is indeed due to increased translation. Furthermore our analysis indicates the translation initiation rate for an mRNA is sixfold greater during transcription than after its release, which amplifies the impact of the transcription distance on gene expression. As a result of these mechanisms, gene expression increases by ∼40% for each 1,000 nucleotides of transcription distance. In summary, we demonstrate that a fundamental relationship exists between gene expression and the number, length, and order of the genes in an operon. This relationship has important implications for understanding the functional basis of genome organization and practical applications for synthetic biology.
Keywords: systems biology, mathematical model, polycistronic
Operons are a central feature of bacterial gene regulation (1). Each operon consists of a group of adjacent genes that are cotranscribed as a single mRNA. It is estimated that 50% of the genes in Escherichia coli are transcribed at least some of the time as part of an operon (2). The organization of genes in operons can alter gene expression when specific regulatory mechanisms are present such as translational coupling (3), polarity (4), and/or feedback (5, 6). However, it is unclear whether operon organization has any impact on gene expression in the absence of these specific mechanisms.
In this study we used synthetic operons to systematically examine whether the number, length, or order of the genes in an operon modulates gene expression. These synthetic operons lack the mRNA-specific, regulatory mechanisms commonly found in native operons. We found that gene expression increases with the distance between the start of a gene and the end of the operon (“transcription distance”) due to increased translation. These findings reveal that operon organization can modulate levels and patterns of gene expression via a general mechanism.
Results
Gene Expression Increases with Operon Length.
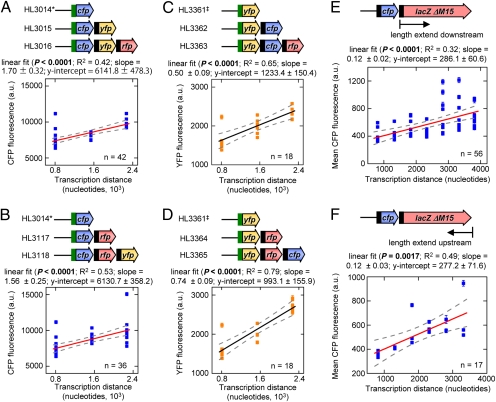
The relationship between gene expression and operon length was examined in four sets of operons constructed with the genes encoding the cyan (cfp), yellow (yfp), and/or mCherry (rfp) fluorescent proteins (Fig. 1 A–D and Fig. S1). The operons were constitutively transcribed from the pLlacO-1 promoter in the absence of the Lac repressor. The translation of the first gene in each operon was controlled by the highly efficient T710RBS7 ribosome binding sequence (RBS) and the other genes were translated from the moderately efficient st7 RBS (7) (Fig. S2). RFP was not used as a reporter because fluorescence varied with cell density and growth rate.
Fig. 1.
Linear relationship between transcription distance and gene expression in operons of different length. Each data point is the mean fluorescence of the first gene in each operon ± SEM from a separate culture. In the schematics of the operons, the green box indicates T710RBS7 and the black box indicates st7. (A and B) Two sets of operons with cfp in the first position of the operon. *, the monocistronic CFP fluorescence values are the same in both A and B. (C and D) Plots are the same as above except the two sets of operons have yfp in the first position. ‡, the monocistronic YFP fluorescence values are the same in both C and D. (E and F) Gene expression at different transcription distances in the lacZ1 (E) and lacZ2 (F) panels.
In the four sets of operons, cfp or yfp was placed in the first position (closest to the promoter) and operon length was increased by adding downstream genes. The mean expression of cfp and yfp in these operons was plotted as a function of the distance from the start codon of the reporter gene to the end of the operon (the transcription distance, λ) (Fig. 1 A–D). The transcription distance varies with operon length as well as with gene position in an operon (the latter is examined below). We found that gene expression increased linearly with the transcription distance in all four sets of operons.
To gather further support for the linear relationship between expression and transcription distance, two additional panels of operons were constructed with varying sized fragments of the lacZ gene [with codons 11–41 deleted (ΔM15), resulting in an inactive β-galactosidase protein (8)] downstream of cfp. In these panels we also altered the RBS for cfp (resulting in a large decrease in CFP production) to determine whether the translational efficiency has an impact on the observed linear relationship (Fig. 1 E and F). The length of lacZ was extended from a fixed point at the 5′ end of the gene (“lacZ1”, Fig. 1E) or it was extended from the 3′ end of the gene (“lacZ2”, Fig. 1F). In both panels a significant linear relationship exists between gene expression and transcription distance (Fig. 1 E and F).
A quadratic regression was performed on the data from the lacZ1 (R2 = 0.36, df = 39, P = 0.0002) and lacZ2 panels (R2 = 0.51, df = 14, P = 0.007). Although these fits were significant, they were not significantly better than the linear regression (P = 0.83 and 0.62, respectively). Therefore, there is no evidence to indicate loss of the linear relationship between gene expression and transcription distance at longer distances.
A substantial amount of variation was observed in the fluorescence values at each transcription distance in the experiments (Fig. 1 A–F). Most of this variation reflects differences between cultures of the same strain measured on the same or different days. This variation was substantially reduced in operons that were integrated into the chromosome (see below), which suggests that it primarily arises from fluctuations in plasmid numbers (9).
Expression of a Gene Increases As Its Position Moves Proximally Within an Operon.
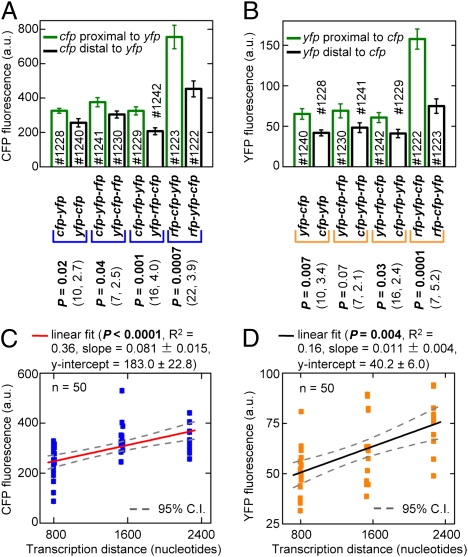
Within an operon, the transcription distance of a gene increases as its position moves away from the end of the operon. Therefore, we would expect the expression of a gene at the first position to be higher than that of an identical gene at the second position, which should be higher than that of an identical gene at the third position (assuming the same RBS at the different positions). To test this prediction, two and three gene operons were constructed with cfp and yfp in the first, second, or third position. All genes in the operons had the same RBS (st7).
The expression of CFP and YFP was compared in pairs of two and three gene operons that were identical except the positions of cfp and yfp were swapped (Fig. 2 A and B). As predicted, expression was always greater for the gene farthest from the end of the operon (i.e., the more proximal gene). The presence of rfp in the first position substantially increased the expression of all of the genes in an operon via a mechanism that is unclear (rfp also had an effect on gene expression that depended on whether it was in the second or the third position). Nonetheless, operons with rfp in the first position still showed significantly higher expression in the more proximal gene. This result indicates that sequence differences that alter absolute levels of transcription and translation do not alter the relationship between the transcription distance and gene expression. The CFP and YFP fluorescence values at each position were plotted as a function of transcription distance (excluding operons with rfp in the first position because the higher fluorescence would disproportionately affect the regression). This analysis showed a significant linear relationship between gene expression and transcription distance within an operon (Fig. 2 C and D).
Fig. 2.
Expression of a gene depends on its position within an operon. Error bars are ± SEM. # indicates the HL strain number. (A and B) Mean CFP and YFP fluorescence in operon pairs. In each pair, the mean was compared using the two-tailed t test with a significance threshold of 0.05. The values in parentheses are the degrees of freedom and the t value, respectively. (C and D) Mean CFP and YFP fluorescence as a function of the transcription distance within operons. Each data point is from a separate culture.
General Model of Operon Translation.
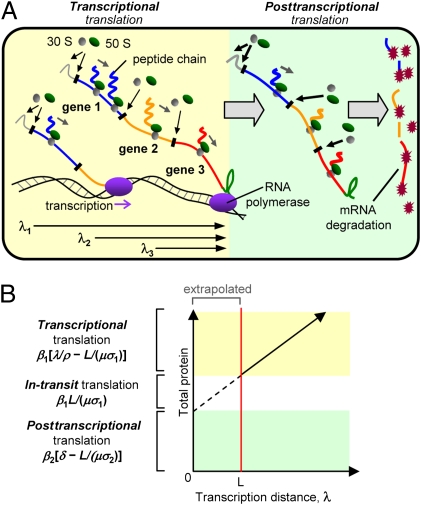
We created a general model to understand the relationship between gene expression and transcription distance. A detailed derivation of the model, with accompanying notes on the assumptions and simplifications, is in SI Materials and Methods. In the model, translation was separated into three phases; “transcriptional translation” (translation during mRNA transcription), “in-transit translation” (translation initiated during transcription and completed after mRNA release), and “posttranscriptional translation” (translation initiated and completed after mRNA release) (Fig. 3A).
Fig. 3.
Model of operon transcription and translation. (A) Translation during transcription (transcriptional translation) and following mRNA release (posttranscriptional translation). λ1, λ2, and λ3 are the transcription distances for genes 1, 2, and 3, respectively. (B) The contributions of transcriptional, in-transit, and posttranscriptional translation to the total protein in the cell. The estimated in-transit translation assumes the translation and transcription rates in units of codons per second are approximately equal.
Transcriptional translation can commence once the start codon of a gene is transcribed and continues until the RNA polymerase encounters the terminator and releases the mRNA. The amount of time available for transcriptional translation is therefore determined by the transcription distance (λ) divided by the transcription rate (ρ) minus the lag time to create the first protein. The lag time is calculated by dividing the gene length in nucleotides (L) by the number of nucleotides per codon (μ) and the translation rate (σ1; units of codons per second). Multiplying this time period by the rate of protein production (β1; units are proteins per mRNA per second) gives the amount of transcriptional translation per mRNA, which is
 |
By definition L ≤ λ and it has been shown that μσ1 ∼ ρ (10, 11). Both β1 and β2 (defined below) depend on the rate of translation initiation (ω1 and ω2, respectively) and the fractions of these initiations that result in a complete protein (θ1 and θ2, respectively) (SI Materials and Methods).
In-transit translation is typically determined by the number of ribosomes spanning the length of the gene before the mRNA is released (SI Materials and Methods). This number can be calculated by dividing the gene length (measured in codons) by the average spacing between each ribosome. The spacing is determined by the translation rate during transcription (σ1) divided by the average time between each successful translation initiation event (1/β1). Therefore, the amount of in-transit translation per mRNA is
 |
Posttranscriptional translation occurs after the mRNA is released until it is degraded. Therefore, the time available is determined by the mRNA lifetime (δ) minus the lag time to create the first protein. The lag time is the gene length in codons (L/μ) divided by the translation rate after mRNA release (σ2; units are codons per second). Therefore, the amount of posttranscriptional translation per mRNA is
 |
where β2 is the protein production rate for free mRNA.
The total protein per mRNA is the sum of the protein produced by transcriptional, in-transit, and posttranscriptional translation. To obtain the total amount of protein in the cell at steady state, which is experimentally measured, the total protein per mRNA must be multiplied by the number of mRNAs transcribed per second (m), which is governed by the promoter's strength and divided by the protein degradation rate constant (γ in units of s−1). That is,
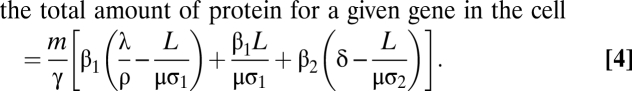 |
The values for transcriptional, in-transit, and posttranscriptional translation can be obtained from plots of gene expression as a function of transcription distance (Fig. 3B).
Normalizing the total protein by the amount of protein from posttranscriptional translation gives
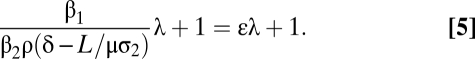 |
The advantage of this normalization is that the slope, termed the translation coefficient (ε), is independent of mRNA production, protein degradation, and the fluorescent reporter. Therefore, the translation coefficient can be compared across different datasets. It can also be used to determine the ratio of transcriptional and posttranscriptional protein production by
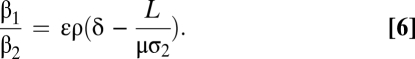 |
In summary, the model provides a simple explanation for the linear increase in gene expression with the transcription distance. It occurs because the transcription distance determines the length of time an mRNA is attached to the DNA and therefore the amount of transcriptional translation.
Operon Length and Gene Position Have Quantitatively Similar Effects on Gene Expression.
We examined the translation coefficient (ε) across the different experiments by normalizing the fluorescence values (i.e., total protein) by the extrapolated fluorescence values at λ = 0 (i.e., posttranscriptional translation). The translation coefficients were as follows (data source in parentheses): 2.77 × 10−4 (Fig. 1A), 2.54 × 10−4 (Fig. 1B), 4.05 × 10−4 (Fig. 1C), 7.45 × 10−4 (Fig. 1D), 4.19 × 10−4 (Fig. 1E), 4.33 × 10−4 (Fig. 1F), 4.43 × 10−4 (Fig. 2C), and 2.74 × 10−4 (Fig. 2D). The translation coefficients from the different datasets are remarkably similar given the error in the fits; that the measurements are in vivo; and that manipulating the order, length, and sequence of the genes will have had confounding effects on expression (e.g., via altered mRNA folding). The differences most likely reflect errors in the data fits but they may also indicate true variation in the constants (β1, β2, δ, ρ, and σ2).
The average translation coefficient was 4.06 × 10−4 ± 0.60 × 10−4 (range = 7.45 × 10−4–2.54 × 10−4 and SD = 1.58 × 10−4). That is, gene expression increases by 40% for each 1,000 nucleotides of transcription distance. The differences in the coding sequence, 5′-UTR, and RBS between the datasets do not appear to have had a major effect on the translation coefficient.
Dynamics of Gene Expression Within an Operon.
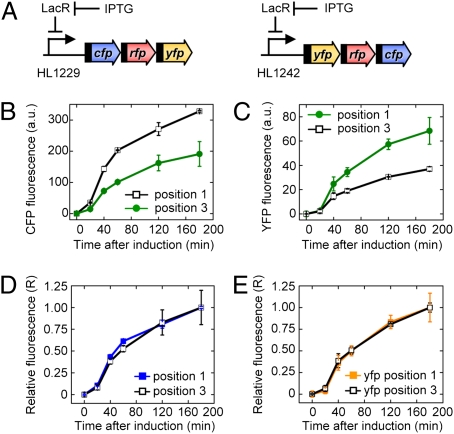
The dynamics of expression were examined to confirm that gene expression increases with the transcription distance due to increased protein production (as predicted by the model) rather than decreased protein degradation or a delay in the expression of distal genes (12, 13). Gene expression was measured in two operons (5′ cfp-rfp-yfp 3′ and 5′ yfp-rfp-cfp 3′) after induction of transcription with isopropyl-β-d-thiogalactopyranoside (IPTG) (Fig. 4 A–C). We found that CFP and YFP expression at the first position in the operons was higher than at the third position for each time point. The protein concentration at each time point was normalized by the final concentration to determine their relative expression (R). R is independent of protein production and depends only on the protein degradation rate and any delay in the induction (SI Materials and Methods). When R was calculated, it was the same for positions 1 and 3 at each time point (Fig. 4 D and E). Therefore, protein degradation and any delay after induction are the same at both positions, indicating the differences in expression must be due to greater protein production in the more proximal position of the operons.
Fig. 4.
The effect of gene position on the dynamics of expression. Data are the mean fluorescence ± SEM. (A) The experimental system used to measure the dynamics of gene expression at different positions within the operon following the induction of transcription with IPTG. All genes had the st7 RBS. (B and C) Mean CFP and YFP fluorescence in the first and third positions of a three-gene operon. (D and E) Relative CFP and YFP fluorescence in the first and third positions of the operon (see main text).
Transcription Distance Primarily Modulates Translation.
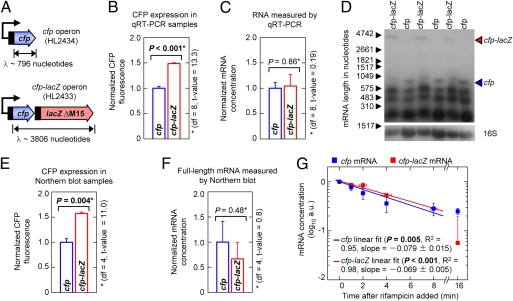
Protein production may increase for genes with longer transcription distances due to increased translation from each mRNA or a higher mRNA concentration. To distinguish between these possibilities, gene expression and the mRNA concentration were compared between a monocistronic cfp gene and the cfp-lacZ operon (Fig. 5A). We found that CFP expression was 1.5-fold (±0.1) greater for cfp-lacZ compared with cfp (Fig. 5B) but there was no significant difference in their mRNA concentrations as measured by quantitative RT-PCR (the 5′ end of cfp was amplified in both mRNAs; Fig. 5C). Therefore, the increased CFP expression with the longer transcription distance is due to greater translation from each mRNA.
Fig. 5.
Increasing transcription distance increases translation. Error bars are ±SEM. *, the means were compared using a two-tailed t test with a significance threshold of 0.05. (A) Schematics of the cfp and cfp-lacZ operons. (B) Normalized CFP fluorescence in the cfp and cfp-lacZ operons. The difference in the expression of the short and long operons was slightly less than previously observed; this difference may be due to the higher cell density required for RNA extractions. (C) The relative concentration of cfp and cfp-lacZ mRNA normalized to the 16S RNA control as determined by quantitative RT-PCR. (D) Northern blots showing full-length cfp and cfp-lacZ mRNA. The contrast of the whole image was altered solely to enable visualization of the full-length cfp and cfp-lacZ mRNA transcripts; it played no role in the analysis. (E) CFP expression for the samples shown in D. (F) Relative concentration of full-length cfp and cfp-lacZ mRNA for the samples shown in D. (G) Decay of full-length cfp and cfp-lacZ mRNA measured by Northern blots following treatment with rifampicin (t = 0). The fits did not include the final time points due to the inaccuracy of the measurements at such low concentrations.
We also performed Northern blots to measure the mRNA concentrations and their decay (Fig. 5 D–G). The probe was located at the 5′ end of cfp (a probe using the entire cfp sequence gave similar results; Fig. S3). The Northern blots showed the full-length cfp-lacZ mRNA transcript had slightly lower abundance than full-length cfp mRNA (Fig. 5 E and F). Again there was no increase in the cfp-lacZ mRNA concentration that would account for the increased CFP expression. It was further demonstrated that the cfp and cfp-lacZ mRNAs have similar half-lives; both are ∼4 min (dividing the half-life by loge2 yields the mRNA lifetime = 352 s; Fig. 5G). It should be noted that because the mRNA lifetime is calculated from the degradation rate, it does not include the time taken to transcribe the mRNA.
The mRNA lifetime (δ = 352 ± 34 s), the translation coefficient (ε = 4.06 × 10−4 ± 0.60 × 10−4), gene length (L = 720 nucleotides), and the transcription rate measured under similar conditions (ρ = 42 ± 2 nucleotides/s) (14) were substituted into Eq. 6. Furthermore, we assume that μσ2 ∼ ρ. This calculation yielded a β1/β2 ratio of 5.7 ± 1.2. That is, the protein production rate is approximately six times greater during mRNA transcription than after its release.
Chromosomal Gene Expression Increases with Transcription Distance.
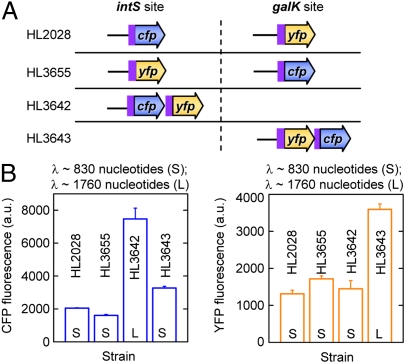
We tested whether the transcription distance also modulates the expression of genes in the chromosome. Strains were created with cfp and yfp at separate locations (intS and galK) (9) or at the same location as an operon (Fig. 6A). Unfortunately, we were unable to insert operons with three fluorescent genes into the chromosome. We found that expression was greater at the first position of an operon compared with a monocistronic gene with a shorter transcription distance (Fig. 6B). In contrast, the last gene in an operon and a monocistronic gene, which have similar transcription distances, have approximately the same expression levels.
Fig. 6.
Chromosomal gene expression increases with the transcription distance. (A) The arrangement of the genes at the intS and galK sites in the chromosome. The pLlacO-1 promoter is used for all genes. The 5′-UTR, RBS, and the first 11 codons of the T7 10 gene (purple box) are fused to cfp and yfp. (B) The mean CFP and YFP fluorescence for each strain. Strains were measured in triplicate and error bars indicate the SEM.
Discussion
In this study we found that gene expression increases linearly with the distance from the start of a gene to the end of the operon (transcription distance). This relationship was observed in multiple sets of operons of different lengths, at different gene positions in multiple operons, with multiple coding sequences, and with different 5′-UTR sequences. Furthermore, the relative increase in translation per nucleotide of transcription distance (i.e., the translation coefficient) was similar across the different experiments and operons. Together these findings provide compelling support for a relationship between gene expression and the transcription distance and they indicate a common mechanism.
We proposed a general model that shows that genes with longer transcription distances have increased expression because they have a longer period for translation during transcription. There are three points in support of the model. First, it correctly predicts from first principles that the relationship between gene expression and the transcription distance is linear. Second, increasing the transcription distance was found to increase translation as predicted. Third, it provides a single explanation for why varying operon length and varying gene position have the same effect on gene expression (i.e., the same translation coefficient).
The most intriguing prediction of the model is that the production rate of proteins during transcription (β1) is sixfold greater than after mRNA release (β2). This result means that increasing the transcription time by 24 s (resulting from a 1-kb increase in transcription distance) has an equivalent effect on expression to increasing the mRNA lifetime by 144 s. The difference in transcriptional and posttranscriptional protein production rates could be due to local differences in ribosome concentrations and/or due to a different mRNA structure during transcription (15). In support of the former, ribosomes have been shown to be preferentially located at sites of active transcription in Bacillus subtilus (16) and there is mounting evidence that spatial localization is important for bacterial translation (17, 18). Both mechanisms would increase translation initiation.
The transcription distance had a measurable impact on expression in a wide variety of operons and there is evidence to suggest an association between gene expression and transcription distance in some native operons (19, 20). Therefore, although the transcription distance has only a moderate effect on gene expression, its role should not be ignored. In addition, altering gene expression by varying the transcription distance is fundamentally different from changing the RBS (21) or the transcription rate (22, 23); consequently it may have unique effects on gene noise and for coordinating expression from multiple genes. Furthermore, varying the transcription distance has a predictable effect on gene expression and this could be exploited to tune patterns and levels of gene expression in synthetic and native operons (24). In particular, it could be used to optimize gene order in operons to increase the output of a pathway (25) and to generate specific stoichiometries for protein complexes.
In conclusion, we show that operon organization can modulate levels and patterns of gene expression. It was known that the proximal genes in an operon can influence the expression of distal genes (e.g., polar mutations). Here we demonstrated that the converse also occurs; distal genes can regulate proximal genes in an operon via their effect on the transcription distance. These findings also provide an example of how synthetic biology can help deconstruct complex biological processes. In this case, synthetic operons enabled the effect of operon organization on gene expression to be decoupled from the regulatory mechanisms that exist in native operons, thereby making it easier to identify. The next and more difficult task will be to investigate the role of the transcription distance in modulating the expression of nonfluorescent genes in native operons.
Materials and Methods
Plasmids and Strains.
Details of the strains and plasmids are in Fig. S1 A and B, Tables S1–S3, and SI Materials and Methods.
Measurements of Gene Expression.
For steady-state and dynamics experiments, cells during early exponential growth (OD600 = 0.1) were harvested and placed on ice followed by centrifugation at 16,100 × g for 1 min to concentrate the cells. The cells were resuspended in LB media and gene expression was measured by fluorescence microscopy using a TE2000E microscope (Nikon) with an X-cite 120PC lamp (Exfo) and 100× objective with phase 3 contrast. Images were captured with a Pixus 1024 pixel CCD camera (Princeton Instruments). Image capture and analysis were automated with Metamorph 7.0 (Molecular Devices). Only objects ±2 SD from the mean were included in the analysis to eliminate cell debris and rare, exceptionally bright fluorescent cells. The mean fluorescence was determined by subtracting the background autofluorescence in cells without the plasmid. Further details are in SI Materials and Methods.
Quantitative RT-PCR Measurements.
Total RNA was extracted from five separate exponentially growing cell cultures using TRIzol (Invitrogen). The RNA was treated with DNase I (New England Biolabs) and cDNA was synthesized using the iScript select cDNA synthesis kit with random primers (Bio-Rad). Quantitative PCR was performed with oligonucleotides located at the 5′ end of cfp or at the 3′ end of rrsB (control for extraction and cDNA synthesis). Full details are in SI Materials and Methods.
Northern Blots.
For each strain, the total RNA was extracted from three separate cultures at OD600 = 0.2–0.5. Northern blots were performed using standard protocols and the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche). The membranes were visualized on radiographic film and quantified using Quantity One Analysis software (Bio-Rad). Further details are in SI Materials and Methods. The mRNA lifetime was determined by a linear fit to a semilog plot of the relative amount of mRNA as a function of time. The slope divided by log10e is the decay constant (k) and 1/k is the mRNA lifetime.
Supplementary Material
Acknowledgments
We thank Lusha Liang for constructing the chromosomal operons, Dena Block for constructing some of the plasmids, and David Adamson for helpful discussions and comments on the model and the manuscript. This work was supported by a University of California (Berkeley) Power Excellence Award (W5646).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105692108/-/DCSupplemental.
References
- 1.Rocha EP. The organization of the bacterial genome. Annu Rev Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 2.Okuda S, et al. Characterization of relationships between transcriptional units and operon structures in Bacillus subtilis and Escherichia coli. BMC Genomics. 2007;8:48. doi: 10.1186/1471-2164-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oppenheim DS, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wek RC, Sameshima JH, Hatfield GW. Rho-dependent transcriptional polarity in the ilvGMEDA operon of wild-type Escherichia coli K12. J Biol Chem. 1987;262:15256–15261. [PubMed] [Google Scholar]
- 5.Mattheakis LC, Nomura M. Feedback regulation of the spc operon in Escherichia coli: Translational coupling and mRNA processing. J Bacteriol. 1988;170:4484–4492. doi: 10.1128/jb.170.10.4484-4492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada M, Saier MH., Jr Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J Mol Biol. 1988;203:569–583. doi: 10.1016/0022-2836(88)90193-3. [DOI] [PubMed] [Google Scholar]
- 7.Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 8.Prentki P. Nucleotide sequence of the classical lacZ deletion delta M15. Gene. 1992;122:231–232. doi: 10.1016/0378-1119(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 9.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 10.Lacroute F, Stent GS. Peptide chain growth of -galactosidase in Escherichia coli. J Mol Biol. 1968;35:165–173. doi: 10.1016/s0022-2836(68)80044-0. [DOI] [PubMed] [Google Scholar]
- 11.Young R, Bremer H. Polypeptide-chain-elongation rate in Escherichia coli B/r as a function of growth rate. Biochem J. 1976;160:185–194. doi: 10.1042/bj1600185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berberich MA, Kovach JS, Goldberger RF. Chain initiation in a polycistronic message: Sequential versus simultaneous derepression of the enzymes for histidine biosynthesis in Salmonella typhimurium. Proc Natl Acad Sci USA. 1967;57:1857–1864. doi: 10.1073/pnas.57.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alpers DH, Tomkins GM. Sequential transcription of the genes of the lactose operon and its regulation by protein synthesis. J Biol Chem. 1966;241:4434–4443. [PubMed] [Google Scholar]
- 14.Gotta SL, Miller OL, Jr, French SL. rRNA transcription rate in Escherichia coli. J Bacteriol. 1991;173:6647–6649. doi: 10.1128/jb.173.20.6647-6649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: A quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascarenhas J, Weber MH, Graumann PL. Specific polar localization of ribosomes in Bacillus subtilis depends on active transcription. EMBO Rep. 2001;2:685–689. doi: 10.1093/embo-reports/kve160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montero Llopis P, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli. Science. 2011;331:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 19.Kovács K, Hurst LD, Papp B. Stochasticity in protein levels drives colinearity of gene order in metabolic operons of Escherichia coli. PLoS Biol. 2009;7:e1000115. doi: 10.1371/journal.pbio.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishizaki T, Tsuge K, Itaya M, Doi N, Yanagawa H. Metabolic engineering of carotenoid biosynthesis in Escherichia coli by ordered gene assembly in Bacillus subtilis. Appl Environ Microbiol. 2007;73:1355–1361. doi: 10.1128/AEM.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy KF, Balázsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox RS, 3rd, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil AS, Collins JJ. Synthetic biology: Applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klipp E, Heinrich R, Holzhütter HG. Prediction of temporal gene expression. Metabolic opimization by re-distribution of enzyme activities. Eur J Biochem. 2002;269:5406–5413. doi: 10.1046/j.1432-1033.2002.03223.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.