Abstract
The molecular mechanisms that activate expression of zygote genes after fertilization are obscure. In animals, receptor-ligand interactions during sperm–egg membrane fusion as well as delivery of putative regulatory molecules by the sperm into the egg cytoplasm are proposed to activate zygote development and subsequent transcription of zygote genes. The mechanisms of activation of zygote development in higher plants also are mysterious, in part because of the difficulty of isolating female gametes of higher plants. In the unicellular, biflagellated green alga Chlamydomonas, the early steps in zygote development are much more accessible to investigation. Within minutes after mating type plus (mt+) and mating type minus (mt−) gametes fuse, expression of several zygote-specific transcripts is induced independently of protein synthesis. Here, we show that ectopic expression in mt− gametes of an mt+ gamete-specific, homeodomain protein, GSP1, induces a zygote-like phenotype and activates expression of zygote genes. One of the genes, zsp2, expressed in these “haploid zygotes” encodes a zygote cell surface adhesion molecule that promotes formation of multicellular aggregates. In total, expression of six out of seven zygote genes examined was induced by ectopic expression of GSP1. Our experiments show that in addition to contributing their genomes to the zygote cytoplasm, gametes also deliver proteins that can activate gene transcription.
Keywords: Chlamydomonas, homeodomain protein, fertilization, zygote-specific genes, gametes, green alga
Because the cytoplasms from two cells with distinct developmental histories become rapidly mixed as a consequence, gamete fusion during fertilization offers a unique opportunity to regulate gene expression. Little is known, however, about if or how organisms have taken advantage of this possibility. Not surprisingly, examples of zygote gene activation without gamete fusion are rare. Endosperm development without fertilization occurs in some Arabidopsis maternal-effect mutants (Kiyosue et al. 1999), and a few plant species form seeds in the wild without fertilization (apomixis) (Chaudhury et al. 1998). The specific signals that initiate plant embryogenesis after gamete fusion, though, have not been identified (Harada 1999). In animals, gene transcription and zygote development can be initiated by introduction of sperm into intact eggs or somatic nuclei into enucleated eggs (Latham 1999; Tesarik and Mendoza 1999), but animal cloning is extremely inefficient (Wakayama and Yanagimachi 2001) and is associated with widespread gene dysregulation (Jaenisch and Wilmut 2001). Moreover, a full understanding of the molecular mechanisms essential for activation of animal zygote genes has remained elusive. In recent studies on fertilization in the green alga Chlamydomonas reinhardtii, we identified a homeodomain protein GSP1 that is expressed only in gametes of a single sex (mating type) and we speculated that GSP1 might be involved in regulation of zygote-specific gene expression (Kurvari et al. 1998). Here, we investigate the role of GSP1 in zygote development in Chlamydomonas.
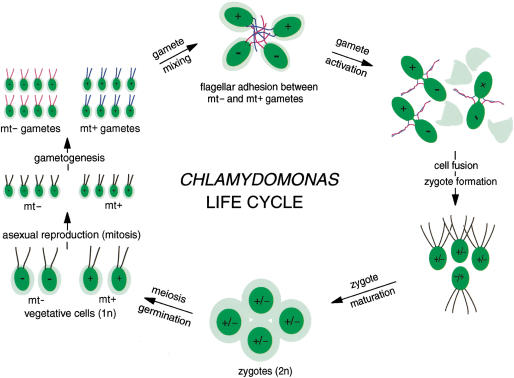
The biflagellated, unicellular alga Chlamydomonas is a highly motile, genetically tractable organism that has proved useful for studying cellular and molecular mechanisms of fertilization (Pan and Snell 2000b). Upon nutrient (nitrogen) deprivation, vegetatively growing haploid mt+ and mt− cells undergo gametogenesis to yield mt+ and mt− gametes (Fig. 1). Mating type is under the control of the mid (minus dominance) gene located in the mt− mating type locus (Ferris and Goodenough 1997). Flagella on gametes display mating type-specific adhesion molecules (agglutinins) that allow gametes of opposite mating types to adhere to each other via their flagella and undergo cAMP-dependent cell activation leading to gamete fusion. Formation of the zygote immediately (1–5 min) triggers loss of gametic properties and activation of the zygote developmental pathway, including degradation of mt− chloroplast DNA and synthesis of a new, zygote extracellular matrix (Goodenough et al. 1995). This tough, highly impervious matrix contains proteins whose properties allow the organism to survive environmental extremes incompatible with continued existence of vegetative cells or unfused gametes (Ferris and Goodenough 1987; Woessner and Goodenough 1989; Wegener and Beck 1991; Uchida et al. 1993, 1999). After an obligatory period in the dark followed by a return to nutrient-rich conditions, zygotes undergo meiosis and germination to yield two mt+ and two mt− haploid vegetative cells (Fig. 1).
Figure 1.
Diagrammatic representation of the Chlamydomonas life cycle. Gametes are formed when vegetative mt+ and mt− cells undergo gametogenesis induced by nitrogen deprivation (middle left). When gametes of opposite mating type (or sex) are mixed together, they adhere to each other via mating type specific adhesion molecules (agglutinins) on their flagella to form large collections of agglutinating cells (upper center). Flagellar adhesion induces gamete activation that leads to release of cell walls, erection of mating structures at the apical ends of the gamete cell bodies (upper right), and several other cellular and biochemical changes that prepare the gametes for cell fusion. All of the events associated with gamete activation can be induced in gametes of a single mating type by incubating them in dibutyryl cAMP (Pan and Snell 2000b). The tips of the mating structures fuse to establish cytoplasmic continuity, and soon thereafter the two gametes merge completely to become a zygote (lower right). Zygote formation can be extremely rapid, and within 5–10 min after gametes are mixed together, 70%–90% of the cells fuse. Immediately after cell fusion, the zygote developmental pathway is initiated and at 2–4 h after fusion, the zygotes form large aggregates of tightly adherent cells (lower center) as a consequence of synthesis of zygote-specific extracellular matrix molecules and cell body adhesion molecules. At about the same time, the mt− chloroplast DNA is degraded. After an obligatory several days in the dark and upon return to nutrient-rich medium, meiosis and germination occur, and each zygote forms four progeny, two mt+ and two mt− vegetative cells (lower right). Vegetative cells undergo mitotic divisions until the nitrogen is depleted from their medium and the cycle begins again. Diagram modified and adapted from Figure 1 of Pan and Snell 2000b.
In Chlamydomonas, zygote proteins are translated from newly made mRNAs whose transcription is activated within minutes after gamete fusion. Several years ago, Ferris and Goodenough (1987) reported identification of six genes whose transcription was induced following Chlamydomonas gamete fusion. Five of the transcripts appeared within 5–10 min after gametes of opposite mating types were mixed together and appearance of the transcripts was independent of protein synthesis. Transcripts for the sixth (the Class V molecule) appeared after 90 min and inhibitors of protein synthesis blocked their appearance. Since then, other zygote-specific molecules have been described (Woessner and Goodenough 1989; Wegener and Beck 1991; Matters and Goodenough 1992; Armbrust et al. 1993; Uchida et al. 1993, 1999; Suzuki et al. 2000). Many of the known zygote-specific transcripts encode molecules demonstrated or proposed to be extracellular matrix molecules. Zygote Ser Pro 1 (ZSP1), a molecule with a Ser Pro-rich domain (Woessner and Goodenough 1989; Suzuki et al. 2000) and the Class V molecule, a cysteine-rich protein (Ferris and Goodenough 1987; Matters and Goodenough 1992), have properties that implicate them as extracellular matrix molecules. Based on sequence analysis, immunolocalization analysis, and functional studies with antibodies, one of the early molecules, ZSP2, is a zygote cell surface, Ser Pro-rich adhesion molecule (Suzuki et al. 2000). ZSP2 is essential for adhesion of zygotes to one another to form the large multicellular aggregates and mats that typify suspensions of zygotes. The zygote-specific ZYS3 protein is of unknown function and likely a cytoplasmic molecule (Kuriyama et al. 1999).
Two other zygote-specific molecules, ZYS1A (Uchida et al. 1999) and EZY1 (Armbrust et al. 1993), are implicated in uniparental inheritance of chloroplasts. As is true for most plants (Birky 1995), chloroplasts from only one parent (the mt+ parent in Chlamydomonas) appear in zygotic progeny (Sager and Ramanis 1973); the mt− chloroplast DNA is degraded via as-yet-uncharacterized mechanisms. The expression of both zys1a and ezy1 transcripts is selectively inhibited when mt+, but not mt−, gametes are briefly ultraviolet (UV) irradiated just prior to mating, a treatment known to interfere with uniparental inheritance of chloroplasts (Armbrust et al. 1993; Uchida et al. 1999). Also strongly suggesting a role for EZY1 in chloroplast inheritance, Armbrust et al. (1993) reported that the EZY1 protein is transported rapidly to both mt− and mt+ chloroplasts about 30 min after gamete fusion, just before the mt− chloroplast DNA is degraded (Armbrust et al. 1993).
Although various models have been proposed for mechanisms of fusion-regulated activation of zygote gene transcription (Goodenough and Ferris 1987; Armbrust et al. 1993), only recently has a potential zygote regulatory molecule emerged. We showed that mt+ gametes, but not mt− gametes or mt+ or mt− vegetative cells express a homeodomain protein, GSP1, which we proposed to be involved in regulation of gamete and zygote genes (Kurvari et al. 1998; Wilson et al. 1999). Homeodomain proteins are transcription factors widely used in plants, fungi, and animals to regulate key steps in development (Gehring 1987; Vollbrecht et al. 1991; Madhani and Fink 1998). GSP1, the first homeodomain protein to be discovered in a green alga, was identified in a screen for molecules whose transcripts were up-regulated under conditions in which gametes underwent prolonged flagellar adhesion but were unable to fuse to form zygotes. One implication of its transcriptional up-regulation during gamete activation was that GSP1 itself was involved in regulation of other mt+-specific molecules such as the mt+ flagellar agglutinins. Consistent with a possible regulatory role during gamete activation, we determined that GSP1 was phosphorylated during gamete activation, even in an impotent mutant, imp1, that undergoes gamete activation but is unable to fuse (Wilson et al. 1999).
Several properties of GSP1 also recommend it as a candidate regulatory molecule in zygotes. The observations that gene transcription is induced so quickly after zygote formation and that transcription of several (early) zygote-specific genes does not require protein synthesis, strongly suggest that preexisting transcription factors in one or both gametes are essential for activation of zygote-specific genes. Moreover, gamete activation alone is not sufficient to induce zygote gene expression. Chlamydomonas exhibits an absolute requirement for cell fusion and the consequent combining of cytoplasmic contents from both parents to activate zygote genes (Ferris and Goodenough 1987). Finally, our previous studies have shown that GSP1 persists in the zygote cytoplasm for at least 1 h after gamete fusion (Wilson et al. 1999). Here, we test the idea that ectopic expression of GSP1 in mt− gametes induces transcription of zygote-specific genes. We expressed GSP1 in mt− cells under the control of a non-gamete-specific, light-regulated promoter. While the mt− transformants did not display a detectable phenotype when cultured as vegetative cells, once they underwent gametogenesis, the resulting mt− gametes formed the multicellular aggregates and mats characteristic of wild-type zygotes. Moreover, the transformed mt− gametes synthesized transcripts for several, but not all, of the zygote-specific genes described above.
Results
GSP1-expressing mt− gametes, but not vegetative cells, form zygote-like aggregates
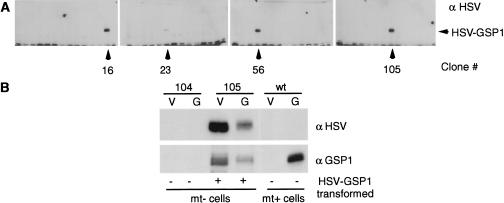
To test the idea that the rapid, protein synthesis-independent appearance of zygote transcripts after gamete fusion in Chlamydomonas is directed by GSP1, we ectopically expressed this mt+ gamete-specific protein in mt− cells. Nitrate reductase-deficient mt− cells were cotransformed with a plasmid containing the nitrate reductase gene (Kindle et al. 1989; Tam and Lefebvre 1993) and a separate plasmid containing an epitope (Herpes simplex virus; HSV)-tagged form of the gsp1 gene under control of a non-gamete-specific promoter—a fused ribulose bisphosphate carboxylase/oxygenase 2/heat shock promoter (Schroda et al. 2000). Screening by immunoblotting with an anti-HSV antibody (Fig. 2A) yielded clones 16, 23, 56, and 105 out of 120 vegetatively growing clones of the nit+ transformants that expressed HSV-tagged GSP1. This proportion of cotransformants is within the range normally obtained using conventional transformation methods with Chlamydomonas (Kindle et al. 1989; Tam and Lefebvre 1993). Clone 104 cells and three other non-HSV–GSP1-expressing clones originally in the culture wells adjacent to the HSV-positive clones were used as controls. Except for small variations in extent of expression of HSV–GSP1, the four HSV–gsp1 transformants were indistinguishable from one another in the properties to be described below and the control nit+ transformants not expressing HSV–GSP1 were indistinguishable from wild-type cells. Immunoblot analysis with anti-HSV and anti-GSP1 antibodies demonstrated that, as expected, wild-type mt+ vegetative cells did not express HSV-tagged or endogenous GSP1, nor did vegetative or gametic clone 104 mt− cells (Fig. 2B). Also as expected, wild-type mt+ gametes expressed endogenous GSP1, but not HSV–GSP1, and both vegetative and gametic clone 105 cells expressed HSV–GSP1 (Fig. 2B). The slightly decreased migration on SDS-PAGE of HSV–GSP1 from gametic 105 cells compared to that from vegetative 105 cells suggests that GSP1 might be unphosphorylated in vegetative cells. In addition to its gamete activation-induced phosphorylation (see Fig. 7A, below), we have shown that GSP1 is basally phosphorylated in wild-type mt+ gametes (Wilson et al. 1999). Examination of the protein levels of GSP1 detected by the anti-GSP1 antibody in the various cell types (Fig. 2B) indicated that the amount of GSP1 in the transformed gametes typically was less than the amount in wild-type mt+ gametes. Thus, any effects of the HSV–GSP1 on mt− gametes were not likely to be the result of overexpression of the molecule.
Figure 2.
Ectopic expression of HSV–GSP1 in mt− gametes. (A) Anti-HSV antibody screening of HSV–gsp1 transformed vegetative cells. Clones 16, 23, 56, and 105 out of 120 vegetatively growing mt− nit+ clones expressed a 120 kD HSV-reactive antigen detected by immunoblotting. (B) HSV–gsp1 transformed vegetative cells and gametes produced HSV–GSP1 protein that reacted with anti-GSP1 antibody. Immunoblotting showed that clone 105 cells expressed HSV–GSP1, which was reactive with both anti-GSP1 (lower panel) and anti-HSV (upper panel) antibodies, when grown as vegetative cells and gametes; endogenous GSP1 was expressed only by mt+ gametes. Clone 104 mt− cells, which were nit+ but did not express HSV–GSP1, were unreactive with both antibodies as vegetative cells and gametes.
Figure 7.
Regulation of transcripts for zys1A, but not ezy1, by gamete activation. (A) HSV–GSP1 undergoes posttranslational modification during gamete activation. Immunoblotting with an anti-HSV antibody showed that HSV–GSP1 underwent posttranslational modification during gamete activation induced either by adhesion with nonfusing mt+ imp1 gametes or during zygote formation after mixing with wild-type mt+ gametes. Because clone 104 is an mt− gamete not expressing HSV–GSP1, it did not react with the anti-HSV antibody. In addition, samples containing only wild-type mt+ gametes or only imp1 mt+ gametes did not react with the anti-HSV antibody, even though these cells contain endogenous GSP1 (data not shown). (B) Transcript levels for zys1A are up-regulated during gamete activation. Northern blotting showed that transcripts for the zygote-specific molecule zys1A were up-regulated several fold during activation of HSV–gsp1 transformed gametes that had been mixed with imp1 mt+ gametes, whereas levels of zsp2 and Class VIII transcripts were unaffected by gamete activation. Wild-type 30-min and 2-h zygotes, clone 104 gametes, and clone 104 and 105 vegetative cells are shown as controls. The total RNA loaded in each lane was identical, but only one half of the RNA in the four right-hand lanes is from mt− cells.
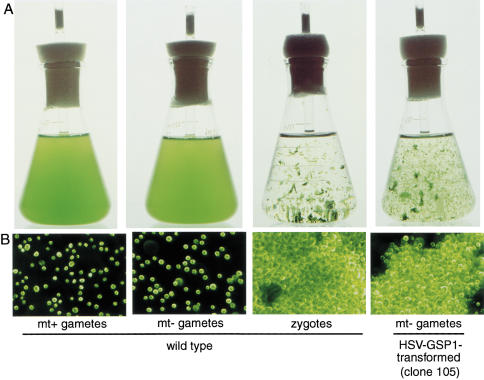
Under standard liquid culture conditions, Chlamydomonas cells are highly motile and distribute themselves homogeneously throughout their culture medium (Fig. 3A,B, left two panels). While none of the vegetatively growing HSV–gsp1 transformants displayed a phenotype different from wild-type cells, a striking phenotype appeared after gametogenesis: The cultures of HSV–gsp1 transformed mt− gametic cells formed large mats and clumps of cells that are characteristic of zygotes formed after fusion of wild-type mt+ and mt− gametes (Fig. 3A,B, right two panels). Gametes of the parental cells, of non-HSV–GSP1-expressing, nitrate reductase-transformed clones from the cotransformation experiment, and of wild-type mt+ and mt− cells did not exhibit this phenotype (Fig. 4). In similar experiments in which the HSV–gsp1 construct was expressed in mt+ vegetative and gametic cells, the mt+ transformants also failed to show a zygote-like phenotype (Fig. 4).
Figure 3.
HSV–gsp1 transformed mt− gametes exhibit a zygote-like phenotype. (A) Wild-type zygotes and the mt− transformants formed macroscopic aggregates visible to the unaided eye. The two left panels show photographs of 125-mL Erlenmeyer flasks containing wild-type mt+ and mt− gametes, which distributed themselves homogeneously in the medium. Wild-type zygotes and clone 105 gametes formed extensive aggregates as seen in the left two panels. (B) Microscopic, dark-field, pseudocolor images of wild-type and mt− transformants. Cells in the mt+ and mt− gamete cultures existed as individuals (left two panels), whereas cells in the cultures of wild-type zygotes and in cultures of the HSV–GSP1 transformed clone 105 gametes (two right panels) formed large, multicellular aggregates of closely adherent cells. Gametes are about 10 μm in diameter.
Figure 4.
The zygote-like phenotype was detected only in HSV–gsp1 transformed mt− cells. Photographs of cell cultures in 24-well plates show that only the HSV–gsp1 transformed mt− gametes (clones 16, 23, 56, and 105) and wild-type zygotes displayed the zygote-like phenotype. Nontransformed mt− parental cells (B215) and mt+ parental cells (G1), HSV–GSP1 transformed mt− vegetative cells and HSV–GSP1 transformed mt+ vegetative cells and gametes did not form aggregates and were homogeneously distributed in the wells.
HSV–gsp1 transformed mt− gametes express several zygote-specific genes
Having obtained morphological evidence that ectopic expression of HSV–GSP1 in mt− gametes led to a zygote-like phenotype, we tested transformants for expression of zygote-specific genes using Northern blotting analysis. Suzuki et al. (2000) recently showed that a Chlamydomonas cell body adhesion molecule, ZSP2, which is expressed only in zygotes, is involved in formation of zygote aggregates. Confirming their Northern blotting results, we also found that the zsp2 transcript appeared within 5 min after wild-type mt+ and mt− gametes were mixed together and had begun to form zygotes (Fig. 5A). As reported previously (Ferris and Goodenough 1987), expression was similar in the presence and absence of the protein-synthesis inhibitor cycloheximide (Fig. 5A). Northern blotting showed that HSV–gsp1 transformed clones 16, 23, 56, and 105 also expressed zsp2 transcripts, but only after the cells had undergone gametogenesis (Fig. 5B). Neither the non-HSV–GSP1-expressing nit+ transformed clones (17 and 104), the parental cells used for the transformation experiment (B215), nor HSV–gsp1 transformed mt+ vegetative cells or gametes contained zsp2 transcripts (Fig. 5B).
Figure 5.
Expression of the zygote-specific extracellular matrix adhesion molecule, zsp2, is restricted to zygotes and HSV–gsp1 mt− transformed gametes. (A) zsp2 expression in wild-type zygotes does not require protein synthesis. Northern blotting indicated that zsp2 transcripts were not expressed in unmixed mt+ (mt+ G) or mt− (mt− G) gametes. Within 5 min after wild-type mt+ and mt− gametes were mixed together to induce zygote formation, the zsp2 transcript appeared and continued to be present until at least 40 min after zygote formation began; expression was not inhibited if the cells were mixed together in the presence of the protein-synthesis inhibitor cycloheximide (10 μg/mL). cblp was used as a loading control. (B) HSV–gsp1 transformation induced expression of zsp2 transcripts, but only in mt− gametes. HSV–gsp1 transformed clones 16, 23, 56, and 105 expressed zsp2 transcripts if they had undergone gametogenesis (G), but failed to express zsp2 transcripts as vegetative cells (V). Transformed cells that were nit+, but did not express HSV–GSP1 (clones 16 and 104), also did not express zsp2 as vegetative cells or gametes. Neither the mt− parental cells (B215, G) nor the mt+ parental cells (G1, G) expressed zsp2, nor did HSV–gsp1 transformed mt+ gametes (HSV–GSP1, G). Wild-type zygotes at 1 h after mixing expressed substantial amounts of zsp2 transcripts (wt, 1 h, Z). (C) ZSP2 protein is expressed only in zygotes and HSV–GSP1–expressing mt− gametes. Vegetative cells (V) and gametes (G) of clones 104 and 105, wild-type mt+ gametes, and wild-type zygotes were analyzed for the presence of ZSP2 by use of immunoblotting. The arrowhead on the right indicates the ZSP2 antigen. The band above ZSP2 was present in all of the lanes except the zygote one, and presumably represents a cross-reactive, constitutively expressed cell-wall molecule, which would have been released by zygotes and discarded when the samples were harvested by centrifugation in preparation for SDS-PAGE and immunoblotting.
In their studies on ZSP2, Suzuki et al. (2000) investigated the appearance of the ZSP2 protein using immunoblotting. They showed that the antigen was not present in vegetative cells or gametes, but was present only in zygotes. We used an anti-ZSP2 antibody provided by Suzuki et al. to study expression of the ZSP2 protein in the HSV–gsp1 transformed mt− gametes. Immunoblotting confirmed that the ZSP2 protein was expressed in the HSV–gsp1 transformed mt− gametes and in wild-type zygotes, but not in wild-type mt+ gametes, in transformed mt− vegetative cells, or in non-HSV–GSP1-expressing clone 104 mt− cells (Fig. 5C).
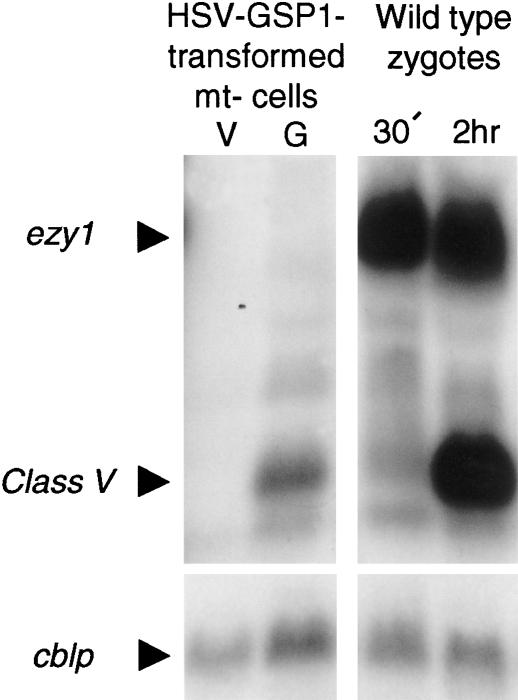
In addition to zsp2, we found that the HSV–gsp1 mt− gametes also expressed transcripts for several other zygote-specific genes including zsp1 (Woessner and Goodenough 1989), which encodes a protein with a Ser Pro-rich domain, and zys3 (Kuriyama et al. 1999), which encodes a protein with ankyrin-like repeats (data not shown). Like zsp2, these transcripts appeared within 5–10 min after fusion of wild-type gametes and their appearance did not require protein synthesis. We detected only slight induction of transcripts for another zygote-specific molecule, zys1A (Uchida et al. 1999), a cysteine- and glutamine-rich protein that accumulates in the nucleus and is highly expressed in wild-type zygotes (see below). Interestingly, the HSV–gsp1 transformed mt− gametes also expressed transcripts for the “late” Class V gene (Matters and Goodenough 1992) (Fig. 6), which encodes another cysteine-rich protein. In wild-type zygotes, the Class V transcript appears ∼2 h after gamete fusion (Fig. 6) and its expression is blocked by protein-synthesis inhibitors (Matters and Goodenough 1992; data not shown). Class V transcript expression in the HSV–gsp1 transformed mt− gametes suggests that one of the “early” genes whose expression is regulated by GSP1 also is involved in regulating transcription of late genes.
Figure 6.
Northern blotting analysis of transcripts for ezy1 and Class V genes. Transcripts for ezy1 were not induced in HSV–gsp1 transformed mt− gametes vegetative cells (V) or gametes (G). Transcripts for the late-appearing, zygote-specific Class V molecule, which were detected in 2-h zygotes but not 30-min zygotes, were also detected in the HSV–gsp1 transformed mt− gametes (G) but not vegetative cells (V). cblp was used as a loading control.
Genes implicated in uniparental inheritance of chloroplasts exhibit unique mechanisms of regulation
Interestingly, not all zygote-specific genes examined were activated in the HSV–gsp1 transformants. One notable exception was the ezy1 gene, whose protein product has been implicated in uniparental inheritance of the chloroplast genome. Armbrust et al. (1993) showed that transcripts for ezy1 appear within minutes after zygote formation, and by 30 min after fusion, the EZY1 protein translocates to both mt+ and mt− chloroplasts. Soon thereafter, the mt− chloroplast genome is destroyed in a process that occurs over a 15–30 min time (Nishimura et al. 1999). UV irradiation of mt+, but not mt−, gametes prior to cell fusion blocks appearance of EZY1 and destruction of mt− chloroplasts (Armbrust et al. 1993). Figure 6 shows that while the ezy1 transcript was strongly expressed in wild-type zygotes, it was not detected in the HSV–gsp1 transformants, even though the Class V transcript was detected by a Class V probe in the same blot. We also were unable to detect EZY1 protein in immunoblotting experiments in which the protein was strongly expressed in wild-type zygotes (not shown).
A central feature of zygote formation in most organisms is that it is the culmination of a complex series of events triggered by gamete interactions and accompanied by regulation of a network of intracellular signaling pathways (Wassarman et al. 2001). Chlamydomonas gametes also undergo dramatic changes in preparation for cell fusion when activated by adhesion with gametes of the opposite mating type (Pan and Snell 2000b). They recruit flagellar adhesion molecules (Goodenough 1989; Hunnicutt et al. 1990) and a protein kinase (Pan and Snell 2000a) from the cell body to the flagella, they erect cell fusion organelles (Friedmann et al. 1968; Wilson et al. 1997), and activated mt+ gametes phosphorylate GSP1 (Wilson et al. 1999). One surprising aspect of our current results was that expression of most of the zygote-specific genes we examined in the HSV–gsp1 mt− transformants could be detected simply by inducing gametogenesis without a requirement for gamete activation. It was possible, however, that gamete activation would induce ezy1 expression in the transformants or that expression of other zygote-specific transcripts would be modified.
To determine if expression of zygote-specific genes was regulated by gamete activation, we mixed HSV–gsp1 transformed mt− gametes with impotent, imp1 mt+ gametes, which can undergo flagellar adhesion with mt− gametes, but are unable to fuse (Ferris et al. 1996). Gamete activation was induced as evidenced by release of the gamete extracellular matrix (data not shown) as well as by the adhesion-induced shift in migration of the HSV-tagged GSP1 protein (Fig. 7A). This shift, which also occurred when the transformed mt− gametes were mixed with wild-type mt+ gametes, was similar to the gamete activation-induced shift that accompanies phosphorylation of endogenous GSP1 in wild-type gametes (Wilson et al. 1999). Gamete activation did not influence the levels of most of the zygote-specific genes we examined including zsp2, Class VIII (Fig. 7B), zsp1, zys3, or Class V (not shown). Notably, gamete activation did not induce ezy1 transcripts (not shown). On the other hand, transcripts for zys1A were increased nearly fourfold after gamete activation (Fig. 7B).
Discussion
Our study identifies an mt+ gamete-specific homeodomain protein as an activator of zygote-specific genes in Chlamydomonas. The observation that GSP1 is expressed only in mt+ gametes, that it persists in zygotes for at least 1 h after cell fusion, and that transcription of several zygote genes does not require protein synthesis, suggested to us that GSP1 might regulate zygote-specific gene expression in Chlamydomonas. When we tested this idea by introducing the gsp1 gene into mt− cells under the control of a light-regulated promoter, we discovered that presence of the GSP1 protein in the cytoplasm of mt− gametes initiated the zygote developmental pathway.
GSP1 induces formation of zygote-like cell aggregates and expression of several zygote-specific genes
Within 2–4 h after wild-type gametes fuse to form zygotes, they begin to adhere tightly to each other via their cell bodies and form large aggregates or mats of cells. Our first evidence that ectopic expression of GSP1 in mt− gametes influenced their phenotype was the appearance of the zygote-like, macroscopic aggregates in the culture flasks, often involving 60%–90% of the cells in the culture (Fig. 3A). Examination by dark-field microscopy also confirmed the similarity between the gsp1-transformed mt− gametes and wild-type zygotes (Fig. 3B). The aggregates in both types of samples were composed of extensive clumps of cells that were tightly adherent to one another. We should note that the effects we observed probably were not the result of overexpression of GSP1, because the immunoblot analysis (Fig. 2B) showed that the amount of GSP1 detected by the anti-GSP1 antibody in the mt− transformed mt− gametes was less than that detected in wild-type mt+ gametes.
Two other results also argue against a nonspecific effect of the HSV–GSP1 in inducing zygote-like aggregates. The possibility that an unanticipated property of the epitope-tagged molecule induced the phenotype was ruled out by our results that mt+ vegetative cells and gametes expressing the same construct did not exhibit the zygote-like phenotype (Fig. 4). This indicated that, just as the presence of the wild-type form of GSP1 normally expressed in mt+ gametes does not initiate zygote development, neither does the presence of the HSV-tagged GSP1 in mt+ gametes. It also was possible that the morphological phenotype, while similar to that of zygotes, had a different explanation that was unrelated to the events that normally occur when wild-type mt+ and mt− gametes fuse. This possibility was ruled out by our results that only mt− gametes displayed the zygote-like phenotype. We could not detect any morphological or biochemical difference between wild type and HSV–gsp1 transformed mt− vegetative cells. In addition to arguing against nonspecific effects, the sex- and gamete-specificity of GSP1 suggests that molecules synthesized in mt− gametes during gametogenesis are required for ectopically expressed GSP1 to exert its effects.
Northern blot analysis of the transformants provided a direct demonstration that ectopically expressed GSP1 initiated the zygote developmental pathway. Of the seven zygote-specific genes we examined, transcripts for six were expressed in the transformed mt− gametes (Figs. 5,6,7; data not shown). As with the morphological phenotype, none of the zygote-specific transcripts were expressed in transformed mt− vegetative cells or in transformed vegetative or gametic mt+ cells. These Northern blotting data along with the immunoblotting results also provided an explanation for the large cell aggregates that we observed in the transformed mt− gametes. Not only did they express the zsp2 transcript, but they also synthesized ZSP2 protein (Fig. 5C), a cell body adhesion molecule that Suzuki et al. (2000) have demonstrated is essential for formation of zygote aggregates. The simplest explanation for these results is that GSP1, which is a homeodomain protein, indeed is a DNA-binding transcription factor that is involved in gene activation in the zygote. Moreover, GSP1 likely acts early in the gene induction pathway as it induced transcription of early zygote genes such as zsp2 and Class VIII, whose expression in zygotes does not require protein synthesis. Additionally, expression of the late gene Class V, whose transcript appearance in wild-type zygotes requires protein synthesis, suggested that GSP1 activates expression of a second wave of gene regulatory molecules.
Genes implicated in destruction of mt− chloroplasts have a more complex regulatory mechanism
Not unexpectedly, our results suggested that Chlamydomonas utilizes multiple mechanisms for regulating zygote genes. While levels of transcripts for five of the seven genes we examined were unchanged after gamete activation induced by mixing with nonfusing mt+ gametes, transcript levels for one gene, zys1A, were increased >fourfold by gamete activation (Fig. 7). Additionally, despite extensive efforts, we never detected transcripts for ezy1 in the transformants, even though it gave strong signals in wild-type zygote samples (Fig. 6). The property common to zys1A and ezy1 is that they both are implicated in uniparental inheritance of chloroplasts, the phenomenon that chloroplasts from only the mt+ parent are passed on to the next generation after germination. Expression of both proteins in wild-type zygotes is blocked if mt+, but not mt−, gametes undergo UV irradiation before being mixed with gametes of the opposite mating type. This treatment also blocks destruction of mt− chloroplast DNA and allows mt− chloroplasts to appear in the progeny of the cross (Armbrust et al. 1993; Uchida et al. 1999).
Nevertheless, that the two genes show different regulatory properties in the transformed mt− gametes (one being regulated by gamete activation and one not being expressed at all) is consistent with the observations that they have unique cellular and molecular properties (Armbrust et al. 1993; Uchida et al. 1999). In their studies of ZYS1A protein localization, Uchida et al. (1999) determined that the protein accumulated in both mt+ and mt− zygote nuclei. Based on this localization and the requirement of protein synthesis for preferential degradation of mt− chloroplast DNA, Uchida et al. speculated that ZYS1A might not be involved directly in degradation of mt− chloroplast DNA. Instead, they suggested that the protein might activate other genes required for uniparental chloroplast inheritance. We do not have an explanation for the regulation of zys1A transcripts by gamete activation in the transformed cells. The transcripts are not induced in activated wild-type mt− gametes (Fig. 7B), indicating that gamete activation, per se, does not activate the gene in either mt+ or mt− gametes. It is possible that gamete activation is accompanied by changes in other proteins that influence GSP1 activation of the zys1A gene. Or, the phosphorylation of GSP1 that occurs during gamete activation may regulate its ability to activate zys1A. The selective advantage, however, of using phosphorylation to influence regulation of some genes and not others is unclear, especially because gametes must undergo activation before zygote formation and thus, zygotes always receive a phosphorylated form of GSP1.
Unlike ZYS1A, the EZY1 protein does not localize to the nucleus, but accumulates in both mt+ and mt− chloroplasts (Armbrust et al. 1993), suggesting that it might be more directly involved in the selective degradation of mt− chloroplast DNA. The complete failure of its transcripts to be expressed in the transformed mt− gametes has several possible explanations. One is that an entirely different regulatory system is in place for this potentially lethal molecule, and that GSP1 is not required for its expression. Or, two molecules, GSP1 and another mt+ gamete-specific molecule might be required for ezy1 activation. An intriguing possibility would be that the ezy1 gene is imprinted in mt− gametes, and not capable of being expressed. Thus, because the mt+ genome is not present in the transformed mt− gametes, the ezy1 cannot be expressed. Obviously, the expression of at least six out of seven zygote genes examined in the transformed mt− gametes demonstrates that the mt− genome is competent for expression of several zygote-specific genes. On the other hand, the recent evidence indicating that there might be uniparental (paternal), global genome silencing in Arabidopsis early embryos (Vielle-Calzada et al. 2000) makes it important to learn if any zygote-specific genes from the mt+ parent are expressed in Chlamydomonas zygotes.
A model for GSP1 regulation of zygote-specific genes
Any model to explain the effects of ectopic expression of GSP1 in mt− gametes must incorporate both the sex (mt− cells) specificity and the cell-stage (gametic) specificity of the response. Only mt− gametes initiated zygote development after transformation. One model would be that the zygote-specific gene promoters are accessible to transcriptional regulatory machinery only in gametes, not vegetative cells, and that these genes are imprinted in mt+ gametes. The model we favor is that GSP1 interacts with an mt− gamete-specific molecule, and that the newly formed complex activates zygote-specific gene expression (Ferris and Goodenough 1987, 1997). Of course, we cannot rule out the possibility that GSP1 inactivates a gene repressor molecule in mt− gametes, but the rapid onset of gene expression after fusion hints at a direct rather than an indirect effect. According to this model, neither mt+ nor mt− vegetative cells nor mt+ gametes would express the putative GSP1 binding partner. Expression of such an mt−, gamete-specific molecule likely would be under the control of the MID protein, the master regulator of mating type, and the GSP1 partner could even be MID itself (Ferris and Goodenough 1997).
Cell–cell fusion-induced formation of a transcriptional regulatory complex from components contributed by each fusion partner is employed by some fungi (Johnson 1995; Kamper et al. 1995; Spit et al. 1998; Vershon and Pierce 2000). After formation of a diploid cell in Saccharomyces cerevisiae, the homeodomain protein MATα2 from the haploid α cell and the transcription factor a1 from the haploid a cell form a complex that represses α-specific and a-specific genes and eventually (and probably indirectly) brings about meiosis (Johnson 1995; Vershon and Pierce 2000). To our knowledge, this Chlamydomonas system offers the first evidence that plants use such a mechanism. Taken together with the fungal systems, our results suggest that use of ancient, atypical homeodomain proteins present in haploid gametes to regulate zygote molecules after cell–cell fusion arose in evolution before the divergence of plants and animals. Similar mechanisms that require protein contributions from two different gametes might underlie regulation of zygote genes during early development in multicellular organisms of both evolutionary groups.
Materials and methods
Cells
Chlamydomonas reinhardtii strains 21gr (mt+) (CC-1690), 6145C (mt−) (CC-1691), and imp1-15 (mt+) (CC-462) are available from the Chlamydomonas Genetics Center, Duke University. The nitrate-reductase defective strains B215 (mt−) and g1 (mt+) were obtained from Greg Pazour and George Witman (University of Massachusetts Medical Center, Worcester). Cells were cultured with aeration at 23°C on a 13:11 h light/dark cycle in M-medium, and vegetative cells were induced to become gametes by incubation in medium without nitrogen (N-free medium) followed by culturing in continuous light with aeration at room temperature as described previously (Snell 1976).
M-medium (pH 6.8) contained the following components (per liter): 1 mg of H3BO3, 1 mg of ZnSO4 · 7H2O, 0.4 mg of MnSo4 · 4H2O, 0.2 mg of CoCl2 · 6H2O, 0.2 mg of Na2MoO4 · 2H2O, 0.04 mg of CuSO4 (the above trace elements being stored at 4°C as a 1000-fold concentrated stock solution), 0.5 g of sodium citrate dihydrate, 0.01 g of FeCl3 · 6H2O, 0.04 g of CaCl2, 0.3 g of MgSO4 · 7H2O, 0.3 g of NH4NO3, 0.1 g of KH2PO4, and 0.1 g of K2HPO4. It is important to add the constituents in the above order to prevent precipitates from forming. R-medium is the same as M-medium with the addition of 3 g/L of sodium acetate trihydrate and 3 times as much of the phosphate buffers. N-free medium (pH 7.6) is the same as M-medium, but without NH4NO3 and KH2PO4, and only enough K2HPO4 to bring the pH to 7.6.
Plasmid constructs
The plasmid pHSP70A–RBCS2–HSV–GSP1 contains a gsp1 genomic fragment that begins with the ATG coding for the initiation methionine of GSP1, preceded by an initiation methionine and two tandem copies of the Herpes simplex virus (HSV) epitope (QPELAPEDPED) (gsp1 gene, GenBank accession no. AY052652). It is driven by a fused promoter composed of the heat shock protein 70A (HSP70A) promoter and the ribulose bisphosphate carboxylase/oxygenase 2 (RBSC2) promoter from Chlamydomonas (Schroda et al. 2000). This promoter is activated at the light levels used to culture both vegetative cells and gametes. The plasmid pGSP1-9, which encodes a gsp1 genomic fragment in pBluescript SK(−), was generated from a Chlamydomonas genomic library provided by Pete Lefebvre (University of Minnesota, St. Paul). pHSP70A–RBCS2–HSV–GSP1 was constructed by ligating an oligonucleotide encoding two tandem copies of the HSV epitope with EcoRI and XhoI overhangs into the EcoRI–XhoI sites of plasmid pCB745 (M. Schroda and C.F. Beck, pers. comm.). PCR-based methods using pGSP1-9 as a template were used to generate a 183-bp oligonucleotide from a genomic fragment of gsp1. The PCR product contained an internal EcoRV site near the 3′ end and was designed to encode a 3′ KpnI site and a 5′ EcoRI overhang that contained the ATG encoding the initiation methionine of GSP1, which was designed to fit into the open reading frame at the 3′ end of the HSV-encoding portion of the construct. After digestion with EcoRI and KpnI, the PCR product was ligated into the HSP70A–RBSC2–HSV construct. Finally, the EcoRV fragment of pGSP1-9 was ligated into the EcoRV site of the construct to generate pHSP70A–RBSC2–HSV–GSP1. Integrity of the plasmid was confirmed by sequencing from the fused promoter to the 5′ EcoRV site.
Nuclear transformation
B215 nit1 vegetative cells (1 × 108 cells) were transformed by a glass-bead method using 1 μg of pHSP70A–RBCS2–HSV–GSP1 DNA that had been linearized by digestion with ScaI and 1 μg of pMN24, a plasmid containing the nitrate–reductase gene that had been linearized by digestion with SalI (Kindle et al. 1989; Tam and Lefebvre 1993). After 7 d of growth on agar plates made with R-medium (Snell 1976) modified to contain 0.003 M KNO3 (potassium nitrate) as the sole nitrogen source, 200 colonies were transferred to wells of 96-well tissue culture dishes containing the selective medium. After several days of growth, samples were transferred to wells of 24-well culture dishes containing 500 μL M-medium; after further growth, vegetatively growing samples from 120 of the clones were assayed for expression of HSV-tagged GSP1 using immunoblotting with an anti-HSV antibody. Four clones expressing HSV-tagged GSP1 along with four adjacent control clones not expressing HSV–GSP1 were selected for further analysis.
Protein detection
GSP1, HSV–GSP1, and ZSP2 proteins were detected by immunoblotting and enhanced chemiluminescence using affinity-purified rabbit anti-GSP1 antibody, murine monoclonal anti-HSV antibody (Novagen), or murine anti-ZSP2 antibody (Suzuki et al. 2000) essentially as described previously for the GSP1 antibody (Kurvari et al. 1998). Incubation with the primary antibodies (1:5000 dilution) was overnight at 4°C and horseradish peroxidase-conjugated, goat anti-rabbit or goat anti-mouse antibodies (1:10,000 dilution) as appropriate were used for the secondary antibodies.
Northern blot hybridization
Total RNA (10 μg of RNA per lane) isolated from 2.5 × 106 cells (Thompson and Mosig 1984) was analyzed by Northern blotting using a NorthernMax kit (Ambion). Randomly primed probes for the hybridizations were prepared with [32P]dATP using a Strip-EZ DNA kit (Ambion). The following fragments were used to prepare probes for hybridization: ezy-1, the 1.6-kb PstI fragment from cDNA 28 (Armbrust et al. 1993); Class V, the 0.45-kb PstI fragment from cDNA 33 (Ferris and Goodenough 1987); zsp2, the 1.1-kb EcoRI–XhoI fragment of cDNA 146 (Class VI) (Ferris and Goodenough 1987); Class VIII (GenBank accession no. AF231333), the 1.65-kb EcoRI fragment of cDNA 27 in pUC118 from P. Ferris (Matters and Goodenough 1992); and zys1A, the 0.94 EcoRI fragment from pZS102-1 (Uchida et al. 1999). The Chlamydomonas beta subunit-like polypeptide (cblp) (Schloss 1990) probe was generated by PCR using genomic DNA as the template. Signal intensities for zys1A were detected and quantified by use of a PhosphorImager (Molecular Dynamics) and standardized by comparison to the signal for cblp from the same blot. Blots were stripped using the Strip-EZ DNA kit.
Acknowledgments
We thank F. Grinnell for helpful comments, U. Goodenough and P. Ferris for invaluable discussions and for providing hybridization probes (including Class VIII cDNA 27 originally generated by G. Matters), M. Misamore for guidance with microscopy, and J. Record and R. Hynecek for assistance in obtaining and characterizing the GSP1 genomic clone. We are indebted to H. Uchida, Y. Nishimura, L. Suzuki, and T. Kuroiwa for providing hybridization probes and the anti-ZSP2 antiserum, and to C. Beck and M. Schroda for providing the ribulose bisphosphate carboxylase/oxygenase 2/heat-shock promoter construct. This work was supported by NIH grant GM-25661 to WJS.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL william.snell@utsouthwestern.edu; FAX (214) 648-8694.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.919501.
References
- Armbrust EV, Ferris PJ, Goodenough UW. A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell. 1993;74:801–811. doi: 10.1016/0092-8674(93)90460-8. [DOI] [PubMed] [Google Scholar]
- Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc Natl Acad Sci. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Craig S, Dennis E, Peacock W. Ovule and embryo development, apomixis and fertilization. Curr Opin Plant Biol. 1998;1:26–31. doi: 10.1016/s1369-5266(98)80123-4. [DOI] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW. Transcription of novel genes, including a gene linked to the mating-type locus, induced by Chlamydomonas fertilization. Mol Cell Biol. 1987;7:2360–2366. doi: 10.1128/mcb.7.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics. 1997;146:859–869. doi: 10.1093/genetics/146.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Woessner JP, Goodenough UW. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol Biol Cell. 1996;7:1235–1248. doi: 10.1091/mbc.7.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann I, Colwin AL, Colwin LH. Fine-structural aspects of fertilization in Chlamydomonas reinhardi. J Cell Sci. 1968;3:115–128. doi: 10.1242/jcs.3.1.115. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. Homeo boxes in the study of development. Science. 1987;236:1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Goodenough UW. Cyclic AMP enhances the sexual agglutinability of Chlamydomonas flagella. J Cell Biol. 1989;109:247–252. doi: 10.1083/jcb.109.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Ferris PJ. Genetic regulation of development in Chlamydomonas. In: Loomis W, editor. Genetic regulation of development. New York, NY: Alan Liss; 1987. pp. 171–189. [Google Scholar]
- Goodenough UW, Armbrust E, Campbell AM, Ferris P. Molecular genetics of sexuality in Chlamydomonas. Annu Rev Plant Physiol. 1995;46:21–44. [Google Scholar]
- Harada JJ. Signaling in plant embryogenesis. Curr Opin Plant Biol. 1999;2:23–27. doi: 10.1016/s1369-5266(99)80005-3. [DOI] [PubMed] [Google Scholar]
- Hunnicutt GR, Kosfiszer MG, Snell WJ. Cell body and flagellar agglutinins in Chlamydomonas reinhardtii: The cell body plasma membrane is a reservoir for agglutinins whose migration to the flagella is regulated by a functional barrier. J Cell Biol. 1990;111:1605–1616. doi: 10.1083/jcb.111.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Wilmut I. Developmental biology. Don't clone humans! Science. 2001;291:2552. doi: 10.1126/science.1060463. [DOI] [PubMed] [Google Scholar]
- Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Kamper J, Reichmann M, Romeis T, Bolker M, Kahmann R. Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109:2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB, et al. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama H, Takano H, Suzuki L, Uchida H, Kawano S, Kuroiwa H, Kuroiwa T. Characterization of Chlamydomonas reinhardtii zygote-specific cDNAs that encode novel proteins containing ankyrin repeats and WW domains. Plant Physiol. 1999;119:873–884. doi: 10.1104/pp.119.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurvari V, Grishin NV, Snell WJ. A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J Cell Biol. 1998;143:1971–1980. doi: 10.1083/jcb.143.7.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE. Mechanisms and control of embryonic genome activation in mammalian embryos. Int Rev Cytol. 1999;193:71–124. doi: 10.1016/s0074-7696(08)61779-9. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Matters GL, Goodenough UW. A gene/pseudogene tandem duplication encodes a cysteine-rich protein expressed during zygote development in Chlamydomonas reinhardtii. Mol Gen Genet. 1992;232:81–88. doi: 10.1007/BF00299140. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Misumi O, Matsunaga S, Higashiyama T, Yokota A, Kuroiwa T. The active digestion of uniparental chloroplast DNA in a single zygote of Chlamydomonas reinhardtii is revealed by using the optical tweezer. Proc Natl Acad Sci. 1999;96:12577–12582. doi: 10.1073/pnas.96.22.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Snell WJ. Regulated targeting of a protein kinase into an intact flagellum. An aurora/Ipl1p-like protein kinase translocates from the cell body into the flagella during gamete activation in Chlamydomonas. J Biol Chem. 2000a;275:24106–24114. doi: 10.1074/jbc.M002686200. [DOI] [PubMed] [Google Scholar]
- ————— Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr Opin Microbiol. 2000b;3:596–602. doi: 10.1016/s1369-5274(00)00146-6. [DOI] [PubMed] [Google Scholar]
- Sager R, Ramanis Z. The mechanisms of maternal inheritance in Chlamydomonas: Biochemical and genetic studies. Theor Appl Genet. 1973;43:101–108. doi: 10.1007/BF00306558. [DOI] [PubMed] [Google Scholar]
- Schloss JA. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol Gen Genet. 1990;221:443–452. doi: 10.1007/BF00259410. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blocker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Snell WJ. Mating in Chlamydomonas: A system for the study of specific cell adhesion. I. Ultrastructural and electrophoretic analyses of flagellar surface components involved in adhesion. J Cell Biol. 1976;68:48–69. doi: 10.1083/jcb.68.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spit A, Hyland RH, Mellor EJC, Casselton LA. A role for heterodimerization in nuclear localization of a homeodomain protein. Proc Natl Acad Sci. 1998;95:6228–6233. doi: 10.1073/pnas.95.11.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki L, Woessner JP, Uchida H, Kuroiwa H, Yuasa Y, Waffenschmidt S, Goodenough UW, Kuroiwa T. Zygote-specific protein with hydroxyproline-rich glycoprotein domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii (Chlorophyta) J Phycol. 2000;36:571–583. doi: 10.1046/j.1529-8817.2000.99112.x. [DOI] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesarik J, Mendoza C. In vitro fertilization by intracytoplasmic sperm injection. Bioessays. 1999;21:791–801. doi: 10.1002/(SICI)1521-1878(199909)21:9<791::AID-BIES11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Thompson RJ, Mosig G. Light and genetic determinants in the control of specific chloroplast transcripts in Chlamydomonas reinhardtii. Plant Physiol. 1984;76:1–6. doi: 10.1104/pp.76.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H, Kawano S, Sato N, Kuroiwa T. Isolation and characterization of novel genes which are expressed during the very early stage of zygote formation in Chlamydomonas reinhardtii. Curr Genet. 1993;24:296–300. doi: 10.1007/BF00336779. [DOI] [PubMed] [Google Scholar]
- Uchida H, Suzuki L, Anai T, Doi K, Takano H, Yamashita H, Oka T, Kawano S, Tomizawa KI, Kawazu T, et al. A pair of invertedly repeated genes in Chlamydomonas reinhardtii encodes a zygote-specific protein whose expression is UV-sensitive. Curr Genet. 1999;36:232–240. doi: 10.1007/s002940050495. [DOI] [PubMed] [Google Scholar]
- Vershon AK, Pierce M. Transcriptional regulation of meiosis in yeast. Curr Opin Cell Biol. 2000;12:334–339. doi: 10.1016/s0955-0674(00)00104-6. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U. Delayed activation of the paternal genome during seed development. Nature. 2000;404:91–94. doi: 10.1038/35003595. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Yanagimachi R. Mouse cloning with nucleus donor cells of different age and type. Mol Reprod Dev. 2001;58:376–383. doi: 10.1002/1098-2795(20010401)58:4<376::AID-MRD4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Wassarman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat Cell Biol. 2001;3:E59–E64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- Wegener D, Beck CF. Identification of novel genes specifically expressed in Chlamydomonas reinhardtii zygotes. Plant Mol Biol. 1991;16:937–946. doi: 10.1007/BF00016066. [DOI] [PubMed] [Google Scholar]
- Wilson NF, Foglesong MJ, Snell WJ. The Chlamydomonas mating type plus fertilization tubule, a prototypic cell fusion organelle: Isolation, characterization, and in vitro adhesion to mating type minus gametes. J Cell Biol. 1997;137:1537–1553. doi: 10.1083/jcb.137.7.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NF, O'Connell JS, Lu M, Snell WJ. Flagellar adhesion between mt(+) and mt(−) Chlamydomonas gametes regulates phosphorylation of the mt(+)-specific homeodomain protein GSP1. J Biol Chem. 1999;274:34383–34388. doi: 10.1074/jbc.274.48.34383. [DOI] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW. Molecular characterization of a zygote wall protein: An extensin-like molecule in Chlamydomonas reinhardtii. Plant Cell. 1989;1:901–911. doi: 10.1105/tpc.1.9.901. [DOI] [PMC free article] [PubMed] [Google Scholar]