Abstract
Hypoxia-inducible factor 1 (HIF-1) is a master regulator of oxygen homeostasis that controls angiogenesis, erythropoiesis, and glycolysis via transcriptional activation of target genes under hypoxic conditions. O2-dependent binding of the von Hippel-Lindau (VHL) tumor suppressor protein targets the HIF-1α subunit for ubiquitination and proteasomal degradation. The activity of the HIF-1α transactivation domains is also O2 regulated by a previously undefined mechanism. Here, we report the identification of factor inhibiting HIF-1 (FIH-1), a protein that binds to HIF-1α and inhibits its transactivation function. In addition, we demonstrate that FIH-1 binds to VHL and that VHL also functions as a transcriptional corepressor that inhibits HIF-1α transactivation function by recruiting histone deacetylases. Involvement of VHL in association with FIH-1 provides a unifying mechanism for the modulation of HIF-1α protein stabilization and transcriptional activation in response to changes in cellular O2 concentration.
Keywords: Corepressor, histone deacetylase, hypoxia, transactivation
Hypoxia-inducible factor 1 (HIF-1) plays an essential role in cellular and systemic oxygen homeostasis (Iyer et al. 1998). HIF-1 activates transcription of genes whose protein products either function to increase O2 availability by promoting erythropoiesis (e.g., erythropoietin) and angiogenesis [e.g., vascular endothelial growth factor (VEGF)] or mediate adaptive intracellular responses to O2 deprivation such as increased glycolytic metabolism (e.g., glucose transporters and glycolytic enzymes) (for review, see Semenza 1999). However, experimental evidence suggests that HIF-1-regulated gene products (e.g., NIP3) may also mediate hypoxia-induced apoptosis under certain conditions (Carmeliet et al. 1998; Bruick 2000). It is not known what determines the battery of genes that are activated by HIF-1 in response to hypoxia in a given cell type and whether this response will lead to adaptation or apoptosis. Because HIF-1 activity is induced by hypoxia in all cell types, the interaction of HIF-1 with other proteins is likely to play a major role in determining its biological activity. Because of the important role played by HIF-1 in cancer and ischemic cardiovascular disease (for review, see Semenza 2000), identification of interacting proteins may also provide novel therapeutic targets.
HIF-1 is a heterodimer consisting of HIF-1α and HIF-1β subunits (Wang et al. 1995; Wang and Semenza 1995). Whereas HIF-1β is constitutively expressed, HIF-1α mRNA expression, protein half-life, and transactivation domain function are all regulated by the cellular O2 concentration in vivo (for review, see Semenza 1999). The C-terminal half of HIF-1α contains several important regulatory domains. First, amino acids 429–608 (Fig. 1A) encompass sequences that mediate O2-dependent ubiquitination of HIF-1α (Huang et al 1998; Sutter et al. 2000) via binding of the von Hippel-Lindau (VHL) tumor suppressor protein, which is the recognition component of an E3 ubiquitin-protein ligase (Cockman et al. 2000; Kamura et al. 2000; Ohh et al. 2000) that targets HIF-1α for proteasomal degradation (Salceda and Caro 1997). Binding of VHL is dependent on hydroxylation of Pro 564 in HIF-1α via an enzymatic process that requires O2 as well as iron and is inhibited by cobalt chloride (Ivan et al. 2001; Jaakkola et al. 2001). VHL also binds to other residues in HIF-1α (Yu et al. 2001), but the functional significance of these interactions remains to be determined. Finally, HIF-1α interacts with p53 (An et al. 1998), which recruits MDM2, another E3 ubiquitin-protein ligase, and this interaction modulates HIF-1α stability under hypoxic conditions (Ravi et al. 2000).
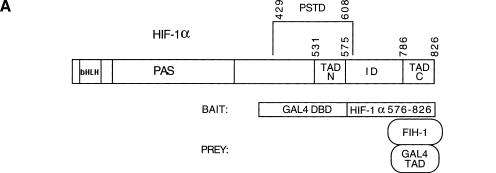
Figure 1.
Identification of FIH-1 by yeast two-hybrid screen. (A) Structure of HIF-1α and the bait and prey proteins. HIF-1α contains basic helix–loop–helix (bHLH) and PAS domains that are required for dimerization and DNA binding, a proline/serine/threonine-rich protein stabilization domain (PSTD), two transactivation domains (TAD-N and TAD-C), and an inhibitory domain (ID) that negatively regulates transactivation domain function under nonhypoxic conditions. For two-hybrid screening, the bait vector encoded a chimeric protein consisting of the DNA-binding domain from the yeast GAL4 transcription factor (GAL4 DBD) fused to residues 576–826 of HIF-1α. The prey vectors encoded the GAL4 transactivation domain (GAL4 TAD) fused to residues encoded by human brain cDNAs, one of which encoded FIH-1. (B) Amino acid sequence of FIH-1 and identification of related protein sequences. TBLASTN searches of GenBank databases were performed using the human FIH-1 sequence (top line). For each GenBank entry only those sequences showing significant similarity to FIH-1 are shown.
In addition to the domain regulating protein stability, HIF-1α contains two transactivation domains (Jiang et al. 1997; Pugh et al. 1997), TAD-N (residues 531–575) and TAD-C (residues 786–826), which bind coactivators including CBP, p300, SRC-1, and TIF-2 (Arany et al. 1996; Ema et al. 1999; Carrero et al. 2000). Residues 576–785 comprise an inhibitory domain the deletion of which increases transactivation domain function especially under nonhypoxic conditions (Jiang et al. 1997). Transactivation domain function is negatively regulated by O2 independently of protein stability. Exposure of cells to hypoxia, cobalt chloride, or the iron chelator desferrioxamine induces both HIF-1α protein stabilization and transactivation (Jiang et al. 1997; Pugh et al. 1997), suggesting the existence of a common molecular mechanism.
We have utilized the yeast two-hybrid assay to screen for proteins that interact with HIF-1α and modulate its biological activity. Here, we report the identification and characterization of FIH-1 (factor inhibiting HIF-1), a negative regulator of HIF-1α transactivation domain function. We demonstrate that FIH interacts with HIF-1α as well as with VHL and that both FIH-1 and VHL inhibit HIF-1α transactivation domain function. The involvement of VHL provides a unifying mechanism for the O2-dependent regulation of HIF-1α protein stability and transcriptional activity.
Results
Identification of FIH-1 in a screen for HIF-1α-interacting proteins
To perform the two-hybrid assay (Fields and Song 1989), his− yeast strain Y190 was cotransformed with a bait vector, encoding the GAL4 DNA-binding domain expressed in-frame with HIF-1α residues 576–826 (encompassing the inhibitory domain and TAD-C) and prey vectors allowing expression of human brain cDNA sequences in-frame with the GAL4 transactivation domain (Fig. 1A). Yeast transformants (2 × 106) were screened for histidine auxotrophy and α-galactosidase (α-gal) expression. The yeast colony demonstrating the strongest α-gal activity was subjected to additional positive and negative screens (as described in Materials and Methods), and both histidine auxotrophy and α-gal activity were found to be dependent on the combined presence of bait and prey vectors.
Nucleotide sequence analysis revealed that the prey vector contained a 2.98-kb human cDNA encoding a 349-amino-acid protein that was subsequently designated FIH-1 (GenBank accession no. AF395830). A BLASTN search revealed high sequence similarity to the anonymous full-length cDNA sequence FLJ20615 (GenBank accession no. AK000622) and 87 ESTs (Unigene cluster Hs.14595). Compared with FLJ20615, there were three nucleotide differences in the FIH-1 cDNA that altered the sequence of the encoded protein at amino acid residues 10, 28, and 157. In all three cases, the human genomic DNA sequence in GenBank (see below) was in agreement with the FIH-1 cDNA sequence. EST data indicate that FIH-1 is expressed in multiple cell types derived from adrenal, bladder, brain (adult and fetal), breast, colon, embryo, foreskin, germ cell, heart, kidney, lung, lymph, marrow, muscle, nerve, ovary, parathyroid, prostate, skin, testis, tonsil, and uterus.
TBLASTN searches revealed similarity of FIH-1 to the rat PASS1 protein (GenBank accession no. AF168362.1), proteins encoded by human ESTs FLJ13798 (AK023860.1) and FLJ20656 (AK000663.1), as well as to open reading frames in Caenorhabditis briggsae (AC084551.1) and Drosophila melanogaster (AE003469.2, AE003537.2) genomic DNA, thus defining a family of related proteins that has been conserved through invertebrate and vertebrate evolution (Fig. 1B). ESTs (BF102294.1, BF159006.1, BB264223.1) encoding a putative mouse homolog of FIH-1 were also identified. Analysis of genomic DNA sequences deposited in GenBank revealed that the FIH1 gene is present in contig NT 008635, which is mapped to human chromosome 10q24, and consists of eight exons and seven introns that span 14 kb of DNA.
Interaction of FIH-1 with HIF-1α in vitro
35S-labeled HIF-1α was synthesized in vitro and tested for its interaction with bacterially expressed glutathione-S-transferase (GST) or GST fused to FIH-1. GST pull-down assays revealed that HIF-1α bound to GST–FIH-1 but not to GST alone (Fig. 2A). Next, FIH-1 was synthesized in vitro and tested for its interaction with GST alone or fused to HIF-1α sequences spanning the entire protein. Amino acids 531–826 of HIF-1α interacted with FIH-1, whereas residues 1–329 and 429–608 did not (Fig. 2B). Further analysis of GST fusion proteins containing sequences from the C terminus of HIF-1α revealed that deletions that eliminated all but residues 757–826 did not affect binding to FIH-1 (Fig. 2C). HIF-1α residues 786–826 (TAD-C) did not interact with FIH-1, nor did residues 531–575 (TAD-N), whereas residues 576–784 (inhibitory domain) interacted with FIH-1 but with reduced efficiency compared with residues 531–826 (Fig. 2D). These data indicate that sequences within the inhibitory domain of HIF-1α were required for the interaction with FIH-1 but that optimal binding also required TAD-C.
Figure 2.
Localization of HIF-1α residues interacting with FIH-1. (A) GST and GST–FIH-1 fusion proteins were expressed in E. coli, purified, incubated with 35S-labeled in vitro-translated HIF-1α, captured on glutathione–Sepharose beads, and analyzed by SDS-PAGE and autoradiography. (B–D) GST-fusion proteins containing the indicated HIF-1α residues at their C terminus were incubated with 35S-labeled in vitro-translated FIH-1, captured on glutathione–Sepharose beads, and analyzed as described above. (0) GST only.
FIH-1 inhibits transcriptional activation mediated by HIF-1α in human cells
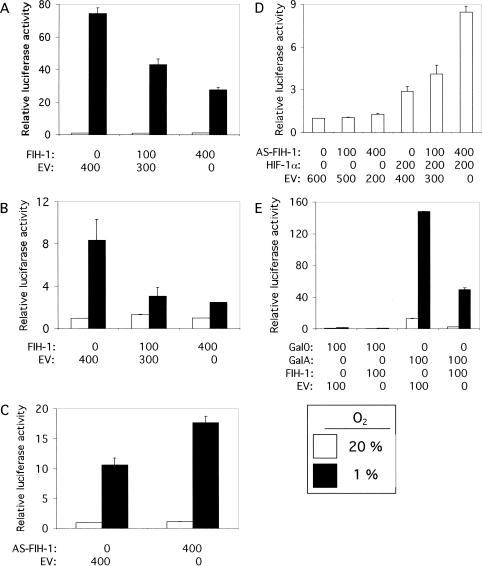
Human embryonic kidney 293 cells were cotransfected with reporter plasmid p2.1, which contains an SV40 promoter-luciferase transcription unit downstream of a 68-bp hypoxia-response element that mediates HIF-1-dependent gene transcription (Semenza et al. 1996). As demonstrated previously, reporter gene expression was markedly increased in cells exposed to 1% O2 relative to cells exposed to 20% O2 (Fig. 3A), reflecting hypoxia-induced HIF-1α stabilization and transactivation. Cotransfection of an expression vector encoding FIH-1 resulted in a dose-dependent inhibition of reporter gene expression at 1% O2. Transcription at 20% O2 was unaffected, as expected because HIF-1α is not detectable in nonhypoxic 293 cells. Similar results were obtained when Hep3B human hepatoblastoma cells were analyzed (Fig. 3B). Transfection of Hep3B cells with an expression vector in which the FIH-1 cDNA was inserted in the antisense orientation resulted in increased reporter gene expression at 1% O2 (Fig. 3C) and similar results were observed in 293 cells (data not shown). Reporter gene expression was unaffected by co-transfection of the antisense FIH-1 expression vector in 293 cells exposed to 20% O2 as expected because HIF-1α is not present (Fig. 3D). However, forced expression of HIF-1α at 20% O2 mediated increased p2.1 expression that was dramatically potentiated by cotransfection of the antisense FIH-1 expression vector. These data suggest that endogenous FIH-1 inhibits the transcriptional activity of HIF-1α in nonhypoxic cells.
Figure 3.
Effect of FIH-1 on HIF-1-mediated reporter gene transcription. Human 293 (A,D) or Hep3B (B,C) cells were cotransfected with pSV-Renilla, a reporter gene containing the SV40 promoter and Renilla luciferase-coding sequences, p2.1, a reporter gene containing a 68-bp hypoxia-response element upstream of the SV40-promoter and firefly luciferase-coding sequences, and the indicated amount of expression vector containing FIH-1 cDNA [inserted in either the sense or antisense (AS) orientation], HIF-1α cDNA, or empty vector (EV). For each expression vector, the amount (in nanograms) of plasmid DNA transfected is indicated. (E) Hep3B cells were co-transfected with pSV-Renilla, reporter pG5ElbLuc, containing five GAL4-binding sites upstream of an adenovirus Elb promoter and firefly luciferase coding sequences, expression vector encoding the GAL4 DNA-binding domain alone (Gal0) or fused to HIF-1α residues 531–826 (GalA), and EV or vector encoding FIH-1. In each panel, cells were exposed to 20% (open bars) or 1% (closed bars) O2 for 16 h and the ratio of firefly:Renilla luciferase activity was determined. The results were normalized to those for cells transfected with EV and exposed to 20% O2 (relative luciferase activity). The mean and standard deviation based on 3–9 independent transfections are shown.
To test this hypothesis, Hep3B cells were co-transfected with reporter plasmid pG5ElbLuc, containing five GAL4-binding sites upstream of an adenovirus E1b promoter-luciferase transcription unit, and an expression vector encoding the GAL4 DNA-binding domain alone (Gal0) or in frame with HIF-1α residues 531–826 (GalA). In contrast to HIF-1α, GalA is constitutively expressed (Jiang et al. 1997) because it does not contain the entire domain (residues 429–608) required for O2-dependent ubiquitination and degradation (Huang et al. 1998; Sutter et al. 2000). As shown previously, the HIF-1α sequences in GalA resulted in greatly increased reporter gene transactivation, especially under hypoxic conditions (Fig. 3E). Cotransfection of the FIH-1 expression vector markedly inhibited GalA-mediated transactivation under both nonhypoxic and hypoxic conditions, whereas it had no effect on basal transcription mediated by Gal0.
FIH-1 interacts with VHL in vitro
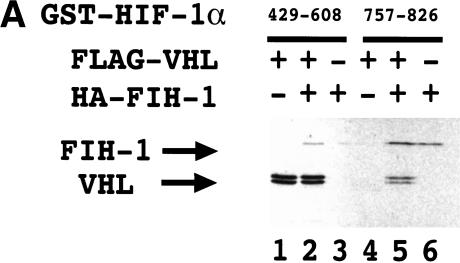
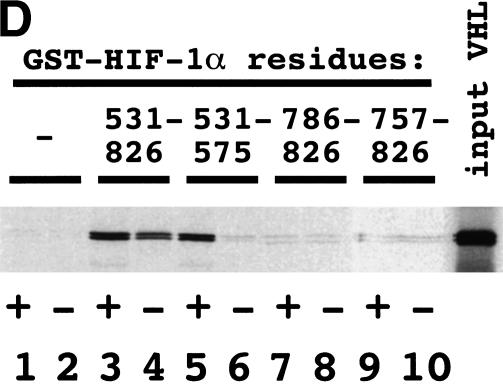
Because FIH-1 and VHL are both negative regulators of HIF-1α, we investigated whether they interact with one another. 35S-labeled VHL bound to a GST–HIF-1α(429–608) fusion protein that had previously been incubated with reticulocyte lysate to induce proline hydroxylation (Fig. 4A) as expected on the basis of the known interaction of VHL with residues 556–574 of HIF-1α (Ivan et al. 2001; Jaakkola et al. 2001). 35S-labeled FIH-1 bound to GST–HIF-1α(429–608) in the presence of VHL (Fig. 4A, lane 2). However, in the absence of VHL, no FIH-1 binding was observed (lane 3), as shown in Figure 2B. Similarly, FIH-1 bound to GST–HIF-1α(757–826), whereas VHL bound in the presence but not in the absence of FIH-1 (lanes 4–6).
Figure 4.
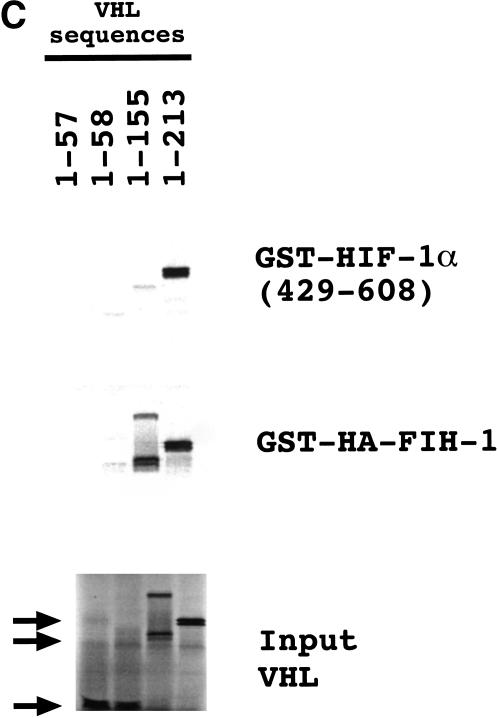
Interaction of FIH-1, HIF-1α, and VHL in vitro. (A) GST-fusion proteins containing HIF-1α residues 429–608 or 757–826 were expressed in E. coli, purified, and incubated with 35S-labeled in vitro-translated FLAG-tagged VHL or HA-tagged FIH-1, captured on glutathione–Sepharose beads, and analyzed by SDS-PAGE and autoradiography. (B) 35S-labeled in vitro-translated FIH-1 residues 1–349 or 126–349 was incubated with unlabeled FLAG–VHL (top) or GST–HIF-1α(531–826) (middle), which were pulled down on beads containing anti-FLAG antibody or glutathione, respectively, and analyzed by SDS-PAGE along with aliquots of the input FIH-1 polypeptides (bottom). (C) 35S-labeled in vitro-translated FLAG–VHL truncated at its C terminus as indicated was incubated with unlabeled lysate-treated GST–HIF-1α(429–608) (top) or GST–HA–FIH-1 (middle), which were captured on glutathione–Sepharose beads and analyzed by SDS-PAGE along with aliquots of the input VHL polypeptides (bottom). (D) GST or the indicated GST–HIF-1α fusion protein was preincubated in reticulocyte lysate (odd-numbered lanes) or buffer (even-numbered lanes), incubated with 35S-labeled FLAG–VHL, captured on glutathione–Sepharose beads, and analyzed as described above.
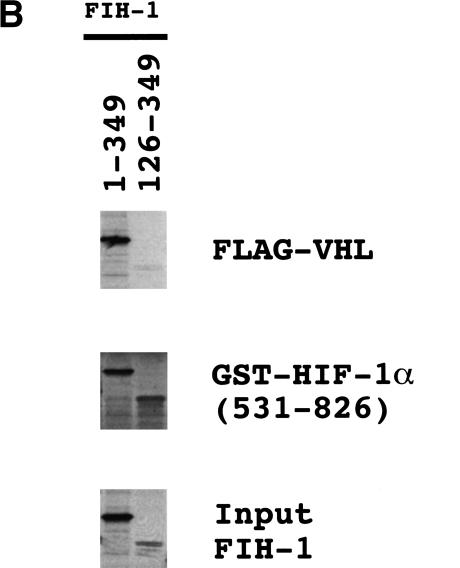
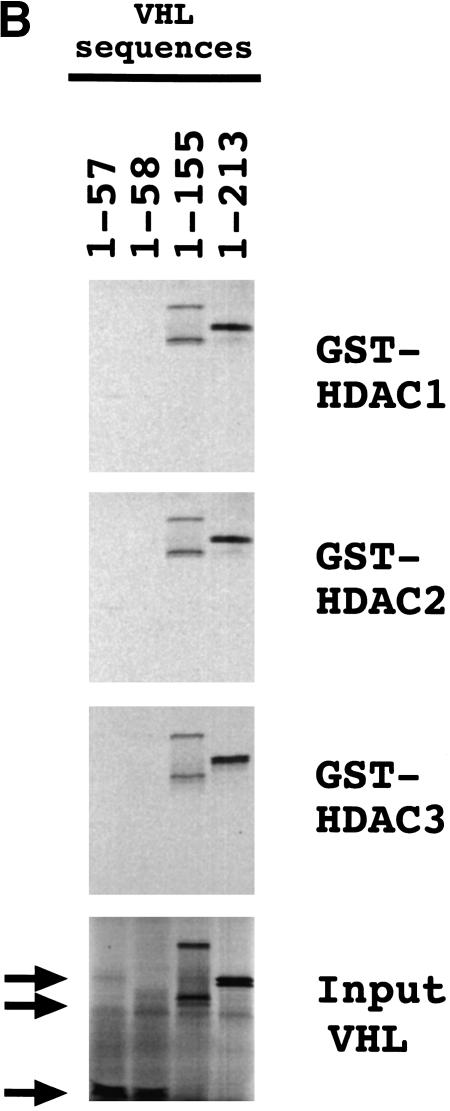
To demonstrate binding directly and to localize the region of FIH-1 that interacted with VHL and with HIF-1α, 35S-labeled FIH-1 that was either full length (1–349) or lacking N-terminal residues (126–349) was synthesized and tested for interaction with FLAG-tagged VHL or GST–HIF-1α(531–826). FIH-1(1–349) bound to FLAG–VHL or GST–HIF-1α(531–826), whereas FIH-1(126–349) bound only to GST–HIF-1α(531–826) (Fig. 4B). These data indicate that VHL and HIF-1α interact with FIH-1 at distinct sites, with the VHL-binding site located N-terminal to the HIF-1α-binding site.
C-terminal truncations of VHL were also tested for interaction with GST–FIH-1 or reticulocyte lysate-treated GST–HIF-1α(429–608). Whereas VHL residues 1–155 were sufficient for binding to FIH-1, residues 1–213 were required for efficient binding to HIF-1α (Fig. 4C). These studies indicate the presence of distinct binding sites that allow the simultaneous interaction of FIH-1, HIF-1α, and VHL.
Finally, we analyzed the interaction of VHL with GST fusion proteins, containing different regions of HIF-1α, that were either untreated or preincubated with reticulocyte lysate. The binding of VHL to GST–HIF-1α(531–575) was strictly dependent on preincubation with reticulocyte lysate (Fig. 4D). VHL did not bind to GST–HIF-1α(786–826) or GST–HIF-1α(757–826) in the presence or absence of lysate. Remarkably, significant binding of VHL to GST–HIF-1α(531–826) was detected in the absence of lysate. These results suggest that VHL may interact with other regions of HIF-1α or with other HIF-1α-binding proteins (e.g., FIH-1) by a mechanism that does not require lysate-dependent hydroxylation.
Interaction of FIH-1 and HIF-1α in human cells
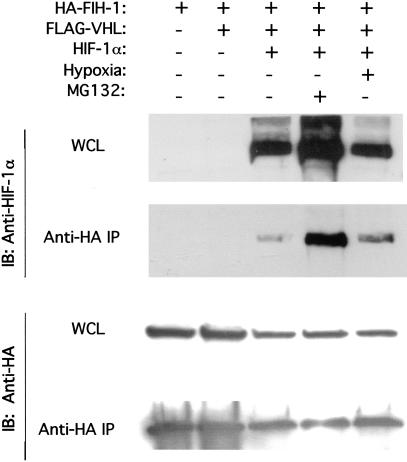
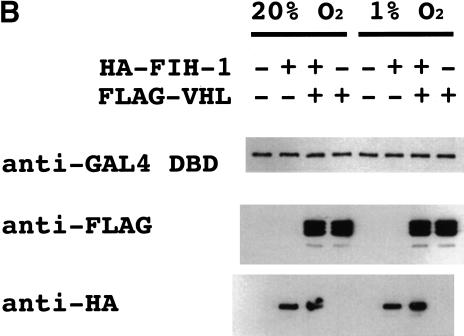
The results of the GST-pulldown and GAL4-transactivation studies suggest that FIH-1 interacts with HIF-1α in human cells. To test this hypothesis directly, 293 cells were cotransfected with expression vectors encoding FIH-1 tagged with hemagglutinin (HA) epitope, FLAG–VHL, and HIF-1α. Aliquots of whole cell lysates were analyzed for expression of the proteins directly or following immunoprecipitation of HA–FIH-1. HIF-1α was present in anti-HA immunoprecipitates from cells overexpressing HA–FIH-1 and HIF-1α (Fig. 5). Exposure of cells to 1% O2 did not alter the interaction of HIF-1α and FIH-1, consistent with the ability of overexpressed FIH-1 to inhibit HIF-1α transactivation domain function at both 20% and 1% O2 (Fig. 3). The proteasome inhibitor MG132 increased HIF-1α expression and the recovery of HIF-1α in anti-HA immunoprecipitates (Fig. 5). These results demonstrate the interaction of FIH-1 and HIF-1α in human cells. However, because the interaction was detected in cells overexpressing these proteins, no conclusion can be drawn as to whether this interaction is regulated by the cellular O2 concentration. HIF-1α protein expression was not regulated, and no interaction of VHL with FIH-1 or HIF-1α could be demonstrated under these conditions (data not shown), suggesting that VHL expression may have been limiting.
Figure 5.
Interaction of FIH-1 and HIF-1α in human cells. Human 293 cells were co-transfected with expression vectors encoding HA–FIH-1, FLAG–VHL, and HIF-1α, as indicated. The transfected cells were untreated, exposed to MG132, or subjected to hypoxia (1% O2) prior to lysis. Aliquots of whole cell lysate (WCL) and anti-HA immunoprecipitate (IP) were analyzed by immunoblot (IB) assay with antibodies that recognize HIF-1α (top) and HA (bottom).
Inhibition of HIF-1α transactivation domain function by VHL and FIH-1 in human cells
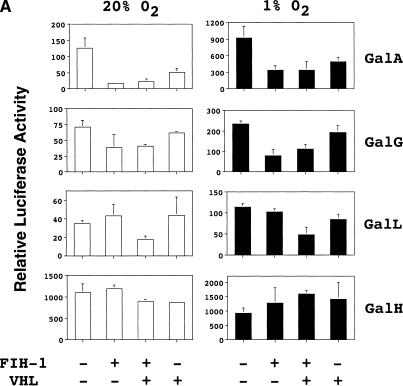
We analyzed the effect of FIH-1, VHL, or both proteins on transactivation mediated by GAL4–HIF-1α fusion proteins that contained binding sites for FIH-1, VHL, both proteins, or neither protein. Hep3B cells were first cotransfected with reporter pG5ElbLuc and expression vector pGalA, which encodes HIF-1α(531–826) and thus includes the binding sites for both VHL and FIH-1. Forced expression of VHL mediated a dramatic inhibition of reporter gene transactivation by GalA in both nonhypoxic and hypoxic cells that was similar to the effect of FIH-1 expression (Fig. 6A). Immunoblot assays demonstrated that GalA protein levels were unaffected by changes in O2 concentration or by overexpression of FIH-1 or VHL (Fig. 6B), demonstrating that these proteins specifically repressed GalA transactivation domain function. Similar assays were performed to analyze the effect of FIH-1 and VHL on transactivation mediated by GalG, which contains HIF-1α(757–826) and includes the binding site for FIH-1 but not for VHL. FIH-1 repressed GalG-mediated transactivation whereas VHL did not (Fig. 6A). Next, we tested GalL, which contains HIF-1α(531–575), and thus includes the binding site for VHL but not for FIH-1. Neither FIH-1 nor VHL alone inhibited GalL-mediated transactivation, whereas the combination of FIH-1 and VHL resulted in significant transcriptional repression. These results suggest that VHL-mediated recruitment of FIH-1 to HIF-1α was required for transcriptional repression of GalL. The effect of VHL alone on GalA may reflect the presence of sufficient endogenous FIH-1 for functional activity when the target protein also contains an FIH-1 binding site, whereas the lower affinity (noncooperative) binding of FIH-1 to VHL alone requires higher FIH-1 levels that are attained by transfection of the expression vector. Finally, we tested pGalH encoding HIF-1α(786–826), which does not include the binding site for either VHL or FIH-1. Notably, transcriptional activation mediated by GalH is not O2 regulated (Jiang et al. 1997; Fig. 6A). Neither FIH-1, VHL, nor the combination of FIH-1 and VHL significantly inhibited GalH-mediated transactivation.
Figure 6.
Functional interaction of FIH-1 and VHL to repress HIF-1α transactivation domain function. (A) Hep3B cells were cotransfected with reporters pSV-Renilla and pG5E1bLuc, expression vector encoding the GAL4 DNA-binding domain alone (Gal0) or a GAL4–HIF-1α fusion protein, and expression vectors encoding no protein, FIH-1, or VHL. The GAL4-fusion proteins (containing the indicated HIF-1α residues) tested were GalA (531–826), GalG (757–826), GalL (531–575), and GalH (786–826). The relative luciferase activity represents the ratio of firefly:Renilla luciferase for each construct normalized to the result for Gal0. (B) Immunoblot analysis of lysates from transfected cells using monoclonal antibodies against the GAL4 DNA-binding domain (DBD), FLAG, and HA to detect expression of GalA (top), FLAG–VHL (middle), and HA–FIH-1 (bottom), respectively.
VHL and FIH-1 interact with histone deacetylases in vitro
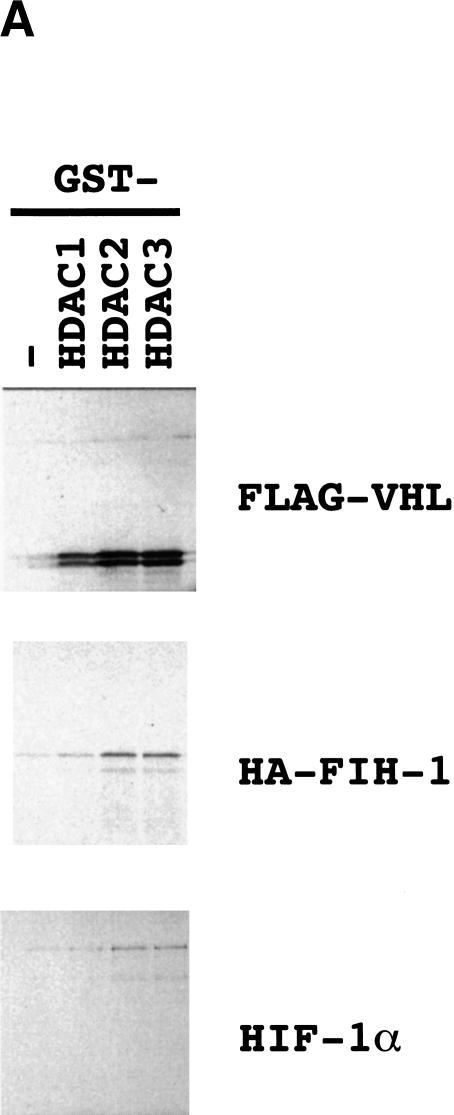
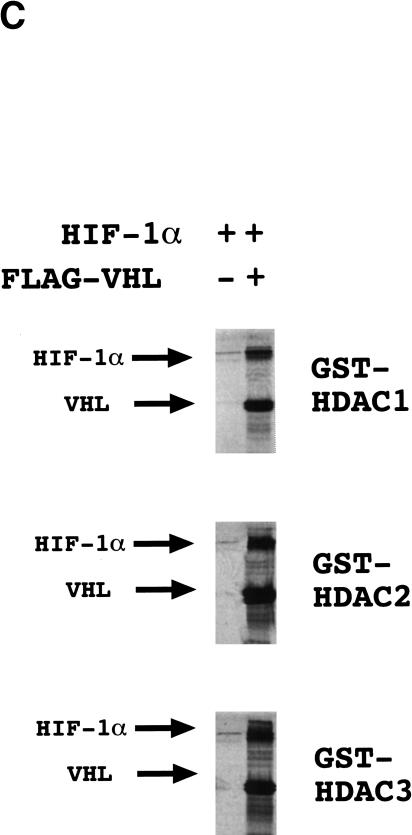
A general property of corepressors is the recruitment of histone deacetylases (HDACs) to DNA-binding transcription factors (for reviews, see Semenza 1998; Cress and Seto 2000). In vitro-translated 35S-labeled VHL interacted with GST–HDAC-1, GST–HDAC-2, and GST–HDAC-3 fusion proteins but not with GST (Fig. 7A). The binding of FIH-1 to HDACs 1–3 was more modest. However, binding of in vitro-translated HDAC-1 or HDAC-3 to GST–HA–FIH-1 but not to GST alone (data not shown) indicates that this interaction is specific. VHL residues 1–213 or 1–155 bound to the GST–HDAC fusion proteins whereas VHL(1–58) and VHL(1–57) did not (Fig. 7B). HIF-1α was pulled down with GST–HDAC fusion proteins very efficiently in the presence of VHL and very inefficiently in its absence (Fig. 7C). Taken together with the cotransfection assays, these studies demonstrate that VHL functions as a corepressor by recruiting HDACs to HIF-1α.
Figure 7.
Interaction of VHL and FIH-1 with histone deacetylases. (A) GST and GST–HDAC fusion proteins were incubated with 35S-labeled FLAG–VHL (top), HA–FIH-1 (middle), or HIF-1α (bottom), captured on glutathione–Sepharose beads, and analyzed by SDS-PAGE and autoradiography. (B) GST–HDAC fusion proteins were incubated with 35S-labeled FLAG–VHL truncated at its C terminus as indicated and analyzed as described above. (C) GST–HDAC fusion proteins were incubated with 35S-labeled FLAG–VHL and/or 35S-labeled HIF-1α.
Discussion
In this paper, we report the identification of FIH-1 and demonstrate its interaction with residues 757–826 at the C terminus of HIF-1α, which encompass part of the inhibitory domain and TAD-C. FIH-1 is the founder of a family of proteins with members extending from human, mouse, and rat to worm and fly, which is consistent with the evolutionary conservation of HIF-1α (and VHL) in both vertebrates and invertebrates (Jaakkola et al. 2001). Forced expression of FIH-1 (or VHL) inhibited the transcriptional activity of a reporter gene that was dependent on either the intact HIF-1 heterodimer (p2.1) or isolated HIF-1α transactivation domains (pG5ElbLuc) under hypoxic or nonhypoxic conditions. The effect of expressing antisense FIH-1 RNA (Fig. 3) suggests that FIH-1, like VHL, functions as an O2-dependent negative regulator of HIF-1α. Under conditions of protein overexpression required to detect coimmunoprecipitation of FIH-1 and HIF-1α, regulation of the interaction by cellular O2 concentration could not be demonstrated (Fig. 5). However, VHL becomes limiting under conditions of HIF-1α overexpression (Tanimoto et al. 2000), and this effect probably occurred in the coimmunoprecipitation experiment despite cotransfection of a VHL expression vector because HIF-1α expression was not O2-regulated.
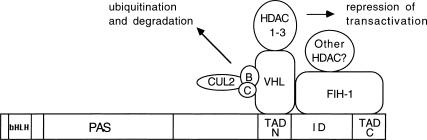
In addition to interacting with HIF-1α, FIH-1 also interacts with VHL, allowing the formation of ternary complexes containing HIF-1α, FIH-1, and VHL (Fig. 8). VHL interacts with HIF-1α and FIH-1 via distinct residues within its β domain and HIF-1α interacts with VHL via TAD-N while interacting with FIH-1 via residues in the inhibitory domain and TAD-C. Because each protein interacts with the other two via independent binding sites, ternary complex formation is likely to involve cooperativity, an hypothesis that is supported by the transactivation studies (Fig. 6A). The failure of VHL to mediate repression of GalL, which contains a binding site for VHL but not for FIH-1, suggests a requirement for functional interaction of VHL and FIH-1 in transcriptional repression of HIF-1α. A critical question that remains to be answered is whether O2-dependent interaction of HIF-1α and VHL is required for recruitment of FIH-1. An alternative but not mutually exclusive hypothesis is that FIH-1 and VHL functionally interact by recruiting distinct HDAC complexes to HIF-1α, as described below.
Figure 8.
Negative regulation of HIF-1α protein stability and transcriptional activity under nonhypoxic conditions mediated by VHL and FIH-1. Elongins B and C and cullin 2 are required for E3 ubiquitin–protein ligase activity, whereas HDACs repress transactivation.
HIF-1-mediated gene transcription is precisely modulated by cellular O2 concentration via regulation of the expression and activity of the HIF-1α subunit (for review, see Semenza 1999). The recent elucidation of the O2-dependent binding of VHL has provided a molecular basis for the regulation of HIF-1α protein stability (Ivan et al. 2001; Jaakkola et al. 2001). Loss of VHL function in renal clear-cell carcinoma (RCC) lines is associated with constitutive expression of HIF-1α protein and of HIF-1 target genes such as GLUT-1 and VEGF (Maxwell et al. 1999). From these results it was not clear whether HIF-1α protein overexpression was sufficient to activate target gene transcription in RCC lines, whether other mutations in these cells eliminated the O2-dependent negative regulation of HIF-1α transactivation, or whether VHL also regulated transactivation. Starting with the identification of FIH-1, we have shown that VHL is an important regulator of HIF-1α TAD function, indicating that VHL controls the two major mechanisms for post-translational regulation of HIF-1α (Fig. 8). These results indicate that loss of VHL function in RCC may be sufficient for constitutive transcription of HIF-1 target genes. Different VHL missense mutations are associated with different disease phenotypes (Clifford et al. 2001; Hoffman et al. 2001). Whereas the effect of specific mutations on HIF-1α and elongin BC binding has been determined, additional genotype–phenotype correlations may be established by analysis of FIH-1 binding to mutant VHL proteins.
The association of VHL with HDAC-1, HDAC-2, and HDAC-3 provides a molecular basis for the repression of HIF-1α transactivation domain function under nonhypoxic conditions. FIH-1 did not interact as strongly with these HDACs, suggesting that it may interact with HDAC-4, HDAC-5, HDAC-6, or HDAC-7. HDAC1–HDAC3 and HDAC4–HDAC7 represent two structurally and functionally distinct groups of proteins (for review, see Cress and Seto 2000). Therefore, the existence of corepressors capable of interacting with each group may provide additional safeguards against inappropriate HIF-1α-mediated transactivation. An alternative but not mutually exclusive possibility is that FIH-1 binds to another corepressor, such as mSin3A, N-CoR, or SMRT, which in turn interacts with HDACs (Kao et al. 2000). Finally, FIH-1 itself may possess HDAC activity. Interestingly, HDAC1 mRNA and protein expression are induced by hypoxia (Kim et al. 2001), suggesting that HDAC1 may represent a HIF-1 target gene and that increased HDAC activity may contribute to the overall decreased rate of transcription in hypoxic cells. The biological role of HDAC1 is complex because of the many transcriptional regulators with which it associates (Cress and Seto 2000) and the net effect of HDAC1 overexpression is to stimulate angiogenesis (Kim et al. 2001). In contrast, VHL and FIH-1 specifically interact with HIF-1α, and overexpression of either of these proteins is predicted to inhibit angiogenesis as a result of decreased HIF-1-mediated VEGF expression.
The results of protein binding and gene transactivation studies suggest that VHL and FIH-1 function cooperatively to repress HIF-1α-mediated transactivation under nonhypoxic conditions. This relationship implies that loss of either VHL or FIH-1 activity may be sufficient to derepress transactivation. This finding is important because of the recent demonstration that phosphatidylinositol-3-kinase (PI3K)/AKT/FRAP signaling stimulated by receptor tyrosine kinases such as HER2 induces HIF-1α protein expression by increasing its rate of synthesis rather than by decreasing its rate of degradation (Laughner et al. 2001). However, activation of the PI3K/AKT/FRAP pathway does not derepress transactivation domain function. In contrast, activation of the MAP kinase pathway has been reported to increase HIF-1 transcriptional activity without affecting HIF-1α protein expression (Richard et al. 1999; Sodhi et al. 2000), and it will be interesting to determine whether this effect is mediated via decreased binding of FIH-1.
A common finding in brain tumors is loss-of-function mutations in the gene encoding PTEN, a phosphatase that negatively regulates PI3K signaling (for review, see Di Cristofano and Pandolfi 2000). Genetic manipulation of PTEN expression has been shown to modulate HIF-1α expression in glioma and prostate cancer cell lines (Zhong et al. 2000; Zundel et al. 2000). Another common genetic alteration in high-grade gliomas is deletion of chromosome 10q23–q26 (Bigner and Vogelstein 1990), which encompasses the FIH1 locus at 10q24. Thus, FIH-1 loss-of-function may contribute to increased HIF-1-mediated transactivation of downstream target genes such as VEGF in gliomas and other human cancers (Semenza 2000; Zagzag et al. 2000).
The complex functional relationship between HIF-1α, FIH-1, and VHL documented in this study and the finding that binding of VHL to the intact HIF-1α regulatory domain (residues 531–826) does not require preincubation of HIF-1α with cellular lysate (as a source of putative prolyl hydroxylase activity; Fig. 4D), suggest that negative regulation of HIF-1α by VHL may be controlled by factors in addition to the O2-dependent hydroxylation of Pro 564 (Ivan et al. 2001; Jaakkola et al. 2001), a conclusion that is supported by other recent studies (Yu et al. 2001). Most importantly, the results reported here establish a unifying mechanism for the modulation of HIF-1α protein stabilization and transcriptional activation in response to changes in cellular O2 concentration that is mediated by VHL in association with FIH-1.
Materials and methods
Yeast two-hybrid system and bait vector construction
A cDNA encoding FIH-1 was isolated by use of the yeast two-hybrid assay (MATCHMAKER Two-hybrid System 2, Clontech). The bait vector pGAL4–HIF-1α(576–826) was constructed by PCR amplification of HIF-1α cDNA sequences (using forward and reverse primers containing NdeI and BamHI restriction sites, respectively), restriction endonuclease digestion, and ligation into the vector pAS2–1. The prey vectors (human brain MATCHMAKER cDNA library cloned into plasmid pACT II; Clontech) encoded fusion proteins consisting of the GAL4 transactivation domain followed by amino acids encoded by human brain cDNA sequences.
Library screening
The physical interaction of the bait and prey proteins within yeast cells functionally reconstitutes an active GAL4 transcription factor, resulting in the expression of genes that contain upstream GAL4-binding sites and mediate histidine auxotrophy (his+) and α-gal expression. To screen for such yeast cells, Saccharomyces cerevisiae strain Y190 was transformed by use of the LiAc/PEG method. YPD medium (850 mL) was inoculated with 150 mL of overnight yeast culture and grown to OD600 = 0.5. The cells were pelleted by centrifugation (5 min at 1,000g), resuspended in 8 mL of TE/LiAc solution, and exposed to 300 μg of pGAL4–HIF-1α(576–826), 600 μg of pACT II/human brain cDNA, and 20 mg of herring testes DNA (Clontech). The cells were agitated at 30°C for 30 min, mixed with 7 mL of DMSO, heat-shocked for 15 min at 30°C, pelleted, resuspended in 10 mL of TE, and plated (200 μL per 15-cm dish) onto media lacking tryptophan, leucine and histidine and supplemented with 15 mM 3-amino-1,2,4-triazole (Sigma). A 2 mg/mL solution of X-α-gal in dimethyl formamide (Clontech) had been previously spread onto the surface of each plate. The his+ and α-gal-expressing colonies were selected over a period of 12 d.
Purification of his+/α-gal-expressing clones and identification of false positives
his+ and α-gal-expressing colonies were subjected to three rounds of colony purification. Then, an individual colony was selected from the final master plate and grown in liquid medium lacking leucine to select for the presence of the prey vector. The resulting cell suspension was then spread onto medium lacking leucine and supplemented with 10 μg/mL cycloheximide to cure clones of the bait vector for identification of clones containing prey vector encoding a protein that could autonomously activate the α-gal reporter gene (i.e., a false positive). Individual colonies were then picked from these cycloheximide plates and grown in liquid culture (lacking leucine), and the prey vector was isolated by the glass bead method (Hoffman and Winston 1987) for transformation of Escherichia coli DH5α cells. The transformed cells were inoculated onto agar plates supplemented with ampicillin, a single colony was used to inoculate LB medium, and plasmid DNA was isolated from the resulting bacterial culture. Finally, the two-hybrid interaction was confirmed by retransformation of yeast strain Y190 with the bait vector and purified prey plasmid to demonstrate that the resulting transformants were again his+ and expressed α-gal.
In vitro interaction (GST pull-down) assays
The pGEX-5X-1-HIF-1α expression plasmids were made by inserting the EcoRI–SalI fragment from mammalian expression plasmid pGalA, pGalG, pGalH, or pGalL (Jiang et al. 1997) into EcoRI- and XhoI-digested pGEX-5X-1 (Amersham Pharmacia Biotech). The expression plasmid pGEX-5X-1-FIH-1 was constructed by insertion of a BglII–EcoRI fragment from pACT II-FIH-1 into BamHI- and EcoRI-digested pGEX-5X-1. pCR3.1-HA–FIH-1 was constructed by insertion of PCR-amplified HA–FIH-1 DNA sequence into pCR3.1. To construct pCR3.1-HA–FIH-1(126–349), a PCR product was generated from template pCR3.1-HA–FIH-1 (primers: 5′-CGGACCATGGCTTAC CCATACGATGTTCCAGATTACGCTTACAGTGCCAGCACC CACAA-3′ and 5′-TACTAGCGCTTGGAGTCTCCTGTCC TCATC-3′) and ligated into pCR3.1. The pGST–HDAC1, pGST–HDAC2, and pGST–HDAC3 expression plasmids (Yang et al. 1997) were kindly provided by W.-M. Yang and E. Seto (University of South Florida).
To prepare GST fusion proteins, E. coli strain BL21-Gold(DE3)pLysS (Stratagene) was transformed with a pGEX expression vector and treated for 4 h with 0.5 mM isopropyl-D-thiogalactoside. Pelleted cells were lysed by sonication in PBS containing 1% Triton X-100 and Complete protease inhibitor cocktail (Roche). After centrifugation, supernatants were applied to glutathione–Sepharose 4B (Amersham Pharmacia Biotech). GST fusion proteins were eluted with 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0) and stored at −80°C. The concentration and purity of eluates were determined by the Bradford method and by SDS-PAGE.
[35S]methionine-labeled proteins were generated in reticulocyte lysates using the TNT T7 coupled transcription/translation system (Promega). Ten microliters of in vitro-translated 35S-labeled protein was mixed with 4 μg of GST or GST fusion protein in a final volume of 200 μL of binding buffer [Dulbecco's PBS at pH 7.4 (Cellgro), 0.1% Tween-20] and incubated for 2 h at 4°C with rotation followed by the addition of 10 μL of glutathione–Sepharose 4B beads (Pharmacia) and a further incubation at 4°C for 1 h. The beads were pelleted, washed three times in binding buffer, pelleted, resuspended in Laemmli sample buffer, and analyzed by SDS-PAGE.
[35S]methionine-labeled proteins were produced in reticulocyte lysates programmed with plasmids encoding HIF-1α, HA–FIH-1, or FLAG–VHL sequences. C-terminal deletion mutants of VHL were made as follows: pcDNA3.1-FLAG–VHL (Feldman et al. 1999; kindly provided by J. Frydman, Stanford University, CA) was digested with NotI, SacII, or AccI. Where indicated, bacterially produced GST fusion proteins were preincubated with 10 μL of rabbit reticulocyte lysate for 30 min at 30°C. Five microliters of the indicated lysate (10 μL in the case of HIF-1α) and/or 5 μg of the indicated recombinant proteins was mixed in 150 μL of NETN buffer (150 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl at pH 8.0, 0.5% NP-40). After 90 min at 4°C, 20 μL of glutathione–Sepharose 4B or 20 μL of anti-FLAG M2 monoclonal antibody–agarose affinity gel (Sigma) was added. After 30 min of mixing on a rotator, beads were washed three times with NETN buffer. Proteins were eluted in Laemmli sample buffer and analyzed by SDS-PAGE followed by autoradiography.
Transient expression assay
HEK293 and Hep3B cells were seeded onto 24-well plates at 4 × 104 cells per well. The following day, the cells were transfected with plasmid DNAs using Fugene-6 (Roche) as per the manufacturer's protocol. After 8 h, the medium in each well was replaced, and the cells were exposed to 95% air and 5% CO2 or to 1% O2, 5% CO2, and balance N2 for 16 h. Cells were lysed, and the luciferase activities were determined by use of the Dual-Luciferase Reporter Assay System (Promega) as per the manufacturer's protocol.
Immunoprecipitation assay
Cells were harvested in 200 μL of lysis buffer (Dulbecco's PBS at pH 7.4, 0.1% Tween-20, 1 mM sodium orthovanadate, and Complete protease inhibitor) and drawn through a 20G needle four times. The lysate was incubated on ice for 1 h followed by centrifugation at 14,000 rpm for 15 min. The cleared lysates were brought to a volume of 1 mL with lysis buffer followed by a 2-h incubation with 20 μL of anti-HA affinity matrix beads (Roche) at 4°C on a rotator. The beads were then washed three times with lysis buffer. Protein was eluted by the addition of Laemmli sample buffer and analyzed by SDS-PAGE and autoradiography.
Acknowledgments
We thank Kelly Chiles and Shannon Berg-Dixon for technical assistance. We are grateful to the laboratory of Edward Benz for generously providing yeast two-hybrid reagents and protocols. We thank Judith Frydberg, Edward Seto, and Wen-Ming Yang for generous gifts of expression plasmids. This work was supported by grant R01-DK39869 from the National Institutes of Health and by a grant from Boehrïnger Ingelheim Austria to G.L.S. K.H. was supported in part by the Yamanouchi Foundation for Research on Metabolic Disorders and by a fellowship from the Uehara Memorial Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gsemenza@jhmi.edu; FAX (410) 955-0484.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.924501.
References
- An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1α. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigner SH, Vogelstein B. Cytogenetics and molecular genetics of malignant gliomas and medulloblastoma. Brain Pathol. 1990;1:12–18. doi: 10.1111/j.1750-3639.1990.tb00633.x. [DOI] [PubMed] [Google Scholar]
- Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci. 2000;97:9082–9087. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert J-M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation, and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Carrero P, Okamato K, Coumailleau P, O'Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol Cell Biol. 2000;20:402–415. doi: 10.1128/mcb.20.1.402-415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ, Maher ER. Contrasting effects on HIF-1α regulation by disease-causing pVHL mutations correlate with patterns of tumorigenesis in von Hippel-Lindau disease. Hum Mol Genet. 2001;10:1029–1038. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- Cockman ME, Masson N, Mole DR, Jaakkola P, Chang GW, Clifford SC, Maher ER, Pugh CW, Ratcliffe PJ, Maxwell PH. Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J BiolChem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: Their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman DE, Thuylasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TriC. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hoffman MA, Ohh M, Yang H, Klco JM, Ivan M, Kaelin WG., Jr von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10:1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes & Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jiang B-H, Zheng JZ, Leung SW, Roe R, Semenza GL. Transactivation and inhibitory domains of hypoxia-inducible factor 1α: Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF-1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H-Y, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Kwon HJ, Lee YM, Baek JH, Jang J-E, Lee S-W, Moon E-J, Kim H-S, Lee S-K, Chung HY, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nature Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. Her2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: Novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang G W, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG., Jr Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- Pugh CW, O'Rourke JF, Nagao M, Gleadle JM, Ratcliffe PJ. Activation of hypoxia-inducible factor-1: Definition of regulatory domains within the α subunit. J Biol Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1α. Genes & Dev. 2000;14:34–44. [PMC free article] [PubMed] [Google Scholar]
- Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- Salceda S, Caro J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions: Its stabilization by hypoxia depends upon redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Transcription factors and human disease. New York, NY: Oxford University Press; 1998. [Google Scholar]
- ————— Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- ————— HIF-1: One highly involved factor. Genes & Dev. 2000;14:1983–1991. [PubMed] [Google Scholar]
- Semenza GL, Jiang B-H, Leung SW, Passantino R, Concordet J-P, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Sodhi A, Montaner S, Patel V, Zohar M, Bais C, Mesri EA, Gutkind JS. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1α. Cancer Res. 2000;60:4873–4880. [PubMed] [Google Scholar]
- Sutter CH, Laughner E, Semenza GL. HIF-1α protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc Natl Acad Sci. 2000;97:4748–4753. doi: 10.1073/pnas.080072497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor 1α by the von Hippel Lindau tumor suppressor protein. EMBO J. 2000;19:4298–4309. doi: 10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang B-H, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-M, Yao Y-L, Sun J-M, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Yu F, White SB, Zhao Q, Lee FS. Dynamic, site-specific interaction of hypoxia-inducible factor 1α with the von Hippel-Lindau tumor suppressor protein. Cancer Res. 2001;61:4136–4142. [PubMed] [Google Scholar]
- Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1α in human brain tumors: Association with angiogenesis, invasion, and progression. Cancer. 2000;88:2606–2618. [PubMed] [Google Scholar]
- Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: Implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk AR, Ryan HE, Johnson RS, Jefferson AB, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes & Dev. 2000;14:391–396. [PMC free article] [PubMed] [Google Scholar]