Abstract
Epigallocatechin-3-gallate (EGCG), a major component of green tea polyphenols (GTPs), has been reported to down-regulate telomerase activity in breast cancer cells thereby increasing cellular apoptosis and inhibiting cellular proliferation. However, the major concerns with GTPs are their bioavailability and stability under physiological conditions. In the present study, we show that treatments with EGCG and a novel pro-drug of EGCG (pEGCG) dose- and time-dependently inhibited the proliferation of human breast cancer MCF-7 and MDA-MB-231 cells but not normal control MCF10A cells. Further, both EGCG and Pro-EGCG inhibited the transcription of hTERT (human telomerase reverse transcriptase), the catalytic subunit of telomerase, through epigenetic mechanisms in estrogen receptor (ER)-positive MCF-7 and ER-negative MDA-MB-231 cells. The down-regulation of hTERT expression was found to be due to hTERT promoter hypomethylation and histone deacetylations, mediated at least partially through inhibition of DNA methyltransferase and histone acetyltransferase activities, respectively. In addition, we also observed that EGCG and pEGCG can remodel chromatin structures of the hTERT promoter by decreasing the level of acetyl-H3, acetyl-H3K9 and acetyl-H4 to the hTERT promoter. EGCG and pEGCG induced chromatin alterations that facilitated the binding of many hTERT repressors such as MAD1 and E2F-1 to the hTERT regulatory region. Depletion of E2F-1 and MAD1 using siRNA reversed the pEGCG down-regulated hTERT expression and associated cellular apoptosis differently in ER-positive and ER-negative breast cancer cells. Collectively, our data provide new insights into breast cancer prevention through epigenetic modulation of telomerase by using Pro-EGCG, a more stable form of EGCG, as a novel chemopreventive compound.
Keywords: EGCG, pro-drug, telomerase, breast cancer and epigenetics
Introduction
Epigallocatechin-3-gallate (EGCG), a major component of green tea polyphenols (GTPs), has been shown to have anti-cancer, anti-inflammatory and anti-tumor activity in various cancers including breast cancer (1–4). EGCG accounts for more than 50% of the total polyphenols, is the most active component of GTPs, and has been shown to induce apoptosis and cell cycle arrest in many cancer cells without affecting normal cells (1, 4–8). Therefore, it is likely that EGCG imparts its chemopreventive effects through many different mechanisms (8–10). One mechanism includes the inhibition of human telomerase reverse transcriptase (hTERT), the catalytic subunit of telomerase, an important enzyme required for maintenance of telomere length and tumorigenesis (2, 11, 12). Inhibition of telomerase has received wide attention in cancer prevention because of its high expression in cancer cells and very low expression in normal somatic cells (11–13). Furthermore, there has been growing interest in epigenetic regulation by EGCG in chemoprevention due to its DNA methyltransferases (DNMTs) and histone acetyltransferases (HATs) inhibition activities (14–16). The DNMTs inhibition activity of EGCG has been shown to lead to global and local hypomethylation of a number of gene promoters (4, 10, 15).
DNA methylation of the promoter region of a gene has been shown to be an important factor in its ability to bind different transcription factors. hTERT is a promising target for cancer prevention and is regulated by several epigenetic alterations including histone acetylation and methylation at promoter sites (11–13, 17). Furthermore, the hTERT promoter region is paradoxically hypermethylated by specific DNMTs in cancer cells leading to its expression (18). The aberrant methylation pattern in the hTERT 5’-regulatory region prevents the binding of the methylation-sensitive transcriptional factors such as CTCF and E2F-1 to the hTERT promoter (18–20). Further, MAD1 is also a transcription repressor of hTERT that binds to its 5’-CACGTG-3’ sequence (E-box), while c-MYC binds to the same promoter sites to activate hTERT expression (19, 21, 22). Binding of MAD1 recruits mSin3-HDACs complexes, which consequently results in decreased acetylation of histones H3 and H4 at the target gene promoters (22, 23). Therefore, EGCG mediates DNMTs and HATs inhibition which leads to DNA hypomethylation and histone deactylation, respectively, that are central pathways important for hTERT-targeted breast cancer prevention.
We and other investigators have shown that EGCG inhibits telomerase and induces cellular apoptosis in human breast cancer cells (4, 13, 20). However, the effects of EGCG on DNA methylation and histone acetylation in estrogen receptor (ER)-positive [ER (+)] and ER-negative [ER (−)] breast cancers have not yet been well studied. Since EGCG has DNMTs and HATs inhibitory activity, it is important to evaluate the impact of EGCG-induced chromatin modifications and its associated transcriptional factors binding on gene promoters important in cancer prevention such as hTERT. Therefore the present study was undertaken to evaluate EGCG modulation of epigenetic regulation of hTERT expression and its promoter alterations associated with the induction of apoptosis and inhibition of cellular proliferations in both ER (+) and ER (−) human breast cancer cells. In addition, we also used a pro-drug of EGCG (pEGCG, EGCG octaacetate) to enhance the bioavailability and stability of EGCG delivered into the cells (24, 25). Pro-EGCG was synthesized by modifying reactive hydroxyl groups with peracetate groups and found to be converted as well as accumulated into parental EGCG when cultured with human breast cancer cell (24). It was reported that pEGCG was better absorbed into the cells, converted into EGCG, and accumulated in greater quantity than natural EGCG (24). Furthermore, oral administration of pEGCG to CF-1 mice resulted in higher bioavailability compared with equimolar doses of EGCG (26). Our results indicate that pEGCG is more potent than EGCG but shares similar mechanisms as EGCG. Both EGCG and pEGCG inhibit hTERT expression by inducing DNA hypomethylation and promoter deacetylations mediated, at least partially, through inhibition of DNMTs and HATs, respectively. Furthermore, hypomethylation and deacetylation induced by EGCG further recruits hTERT transcriptional repressors such as E2F-1 and MAD1, thereby contributing to inhibition of hTERT expression and induction of cellular apoptosis in human breast cancer cells. Interestingly, the transcriptional binding of E2F-1 and MAD1 to regulate hTERT inhibition and induction of apoptosis occurred differently in ER (+) and ER (−) breast cancer cells. These novel findings are important in developing prevention and therapeutic strategies for ER (+) and ER (−) breast cancers.
Materials and Methods
Materials
EGCG (≥ 95% pure) was purchased from Sigma-Aldrich (St. Louis, Missouri). Purified pro-EGCG (pEGCG, >98% pure) was prepared from EGCG as described previously (25). Structural and molecular differences between EGCG and pEGCG are described in supplementary Fig S1. Both EGCG and pro-EGCG were prepared in DMSO and stored at a stock concentration of 100 mmol/L at −20°C.
Cell culture and cell growth assay
The human breast cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and no authentication was done by the authors. Breast cancer MCF-7 [ER (+)] and MDA-MB-231 [ER (−)] cells as well as normal control MCF10A cells were cultured as monolayer as described previously (27). MCF10A is a non-tumorigenic human breast epithelial cell line and frequently used as a normal human breast control cell type (27–29). After seeding the cells for 24 h, EGCG or pEGCG was added to the culture medium at the indicated concentrations and the maximum concentration of DMSO was 0.1% (v/v) in the medium. Cells treated only with DMSO served as a vehicle control. For cell growth assays, total viable cell numbers were calculated using a hemocytometer and plotted against number of treatment days.
Assay for apoptosis by flowcytometry
Induction of apoptosis in human breast cancer cells caused by EGCG and pEGCG treatments were quantitatively determined by flow cytometry using the Annexin V-conjugated Alexafluor 488 (Alexa488) Apoptosis Vybrant Assay Kit as described previously (30). Following treatment, cells were harvested by brief trypsinization, washed with PBS, and incubated with Alexa488 and propidium iodide for cellular staining in Annexin-binding buffer at room temperature for 10 min in the dark. The stained cells were analyzed by FACS using a FACS-Caliber instrument (BD Biosciences, San Jose, CA) equipped with Cell Quest 3.3 software.
Detection of apoptotic cells by Hoechst staining
Following treatment, cells were harvested and cyto-spinned on microscopic slides using cytospin*4 centrifuge (Thermo scientific, Fisher Scientific Products, Pittsburgh, PA). Slides were washed with PBS and fixed in freshly prepared 0.1% ice-cold paraformaldehyde for 10 min. The cells were then washed with PBS and stained with Hoechst 33342 dye (50 µg/ml) for 1 min in the dark. Hoechst stained slides were randomly pictured under a fluorescence microscope and representative pictures are provided.
Quantification of hTERT expression by RT-PCR and real-time PCR
Total RNA isolation and real-time quantification of hTERT expression were followed as described previously (27). Total RNA (2 µg) was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-rad, Hercules, CA). The hTERT primers are as follows: sense 5’-CGGAAGAGTGTCTGGAGCAA-3’ and anti-sense 5’-GGATGAAGCGGAGTCTGGA-3’. The reaction conditions were 35 cycles at 94°C for 30 sec, 52°C for 30 sec and 72°C for 25 sec. GAPDH was used as an endogenous control. Real-time quantitative PCR was carried out as described earlier with following primers: sense 5’-AGGGGCAAGTCCTACGTCCAGT-3’ and anti-sense 5’-CACCAACAAGAAATCATCCAC C-3’ (27). The calculations for determining the relative level of gene expression were made using the cycle threshold (Ct) method. The mean Ct values from duplicate measurements were used to calculate the expression of the target gene using the formula: fold change in gene expression, 2−ΔΔCt = 2−{ΔCt (treated samples)- ΔCt (untreated control)}, where ΔCt = Ct (hTERT)- Ct (GAPDH).
DNMTs, HDACs and HATs activity assays
Cells were harvested at indicated time points and nuclear extracts were prepared using the nuclear extraction reagent (Pierce, Rockford, IL). The activities of DNMTs (Epigentek, Brooklyn, NY), HDACs (Active Motif, Carlsbad, CA) and HATs (Epigentek) were performed using the colorimetric kit according to the manufacturer’s instruction as described previously (27). The enzymatic activities of DNMTs, HDACs and HATs were detected by a microplate reader at 450 nm.
Bisulfite sequencing analysis
To assess the methylation status of the hTERT promoter, sodium bisulfite methylation sequencing was performed using the EpiTect-Bisulfite modification kit following the manufacture’s protocol (Qiagen, Valencia, CA). Approximately 2 µg of genomic DNA was used for bisulfite modification and then amplified by PCR using Go Taq mix (Promega, Madison, WI). Primers and PCR-conditions were followed as described previously (31). PCR amplified DNA was purified using the QIAquick PCR purification kit (Qiagen) and sequenced using the 3730 DNA Sequencer (Applied Biosystems, Foster City, CA). Percent methylation was calculated using the following formula: Number of methylated CpG×100/ total number of CpG being assessed.
Chromatin immunoprecipitation analysis
Chromatin immunprecipitation (ChIP) analysis was performed using the EZ-ChIP kit according to the manufacturer’s instructions (Upstate Biotechnology, Lake Placid, NY) as described previously (27). The antibodies used in the ChIP assays were ChIP-validated acetyl-histone H3, acetyl-histone H3K9, acetyl-histone H4, dimethyl-histone H3K4, MAD1, c-MYC and E2F-1 (Upstate Biotechnology). No antibody control was also used to check ChIP efficiency. ChIP-purified DNA was quantified by using quantitative-PCR (qPCR) using the Platinum SYBR Green detection system (Invitrogen, Carlsbad, CA) as described earlier (27). The primers for the hTERT promoter were forward-5’-TCCCCTTCACGTCCGGCATT-3’, reverse-5’-AGCGGAGAGAGGTCGAATCG-3’. For MAD1, c-MYC and E2F-1 ChIP-purified DNA was amplified using the following RT-PCR primers: forward, 5’-CTCCGTCCTCCCCTTCAC-3’ and reverse, 5’-CAGCGCTGCCTGAAACTC-3’, with a total of 30 cycles at 94°C for 15 s, 52°C for 30 s, and 72°C for 2 min. After amplification, PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining using Kodak 1D 3.6.1 image software and quantified. Quantitative data were analyzed by optical densitometry using ImageJ Software version 1.36b (http://rsb.info.nih.gov/ij/).
Western blot analysis
For western blot analysis, protein extracts were prepared using the RIPA-lysis buffer (Upstate Biotechnology, Lake Placid, NY) following the manufacturer’s protocol. For immunoblot analysis, 60 µg of protein was resolved on a 10% SDS-PAGE and transferred onto nitrocellulose membrane. After incubation in blocking buffer for 1 h, the membranes were incubated with the primary antibodies specific for E2F-1 and MAD1 (Santa Cruz Biotechnology) and β-actin (Cell Signaling, Danvers, MA). The blot was then washed with TBS and 0.05% (v/v) Tween-20 and incubated with specific secondary antibody conjugated with horseradish peroxidase. Protein bands were then visualized using the ECL detection system following the protocol of the manufacturer. The bands were analyzed by using Kodak 1D 3.6.1 image software.
Small interfering RNA (siRNA) knock-down of E2F-1 and MAD1
Approximately 2 × 105 cells were grown in 100 mm cell culture plates and allowed to incubate overnight. The E2F-1 and MAD1 siRNA (Santa Cruz Biotechnology) were prepared as 10 µM stocks using nuclease-free water. E2F-1 (6 nM) and MAD1 (4 nM) siRNA was delivered to the cells using the Silencer siRNA Transfection kit (Ambion/Applied Biosystems, TX, USA) according to the manufacturer’s instructions. siCONTROL Non-Targeting siRNA (Santa Cruz Biotechnology) was used as a negative control. Cells were harvested and checked for E2F-1 and MAD1 knock-down after 6 and 9 day intervals using western blot analysis. Pro-EGCG (20 µM)-treated and non-treated cells were used to harvest RNA for PCR reactions using total RNA extraction and real time-PCR procedures described in previous sections.
Apoptosis assay in siRNA knockdown cells
Breast cancer cells transfected with E2F-1, MAD1 and control siRNA as well non-transfected cells were treated with 20 µM pEGCG for 9 days. The cells were then lysed with nuclei lysis buffer and assayed for apoptosis using the Cell Death Detection ELISA Kit (Roche, Palo Alto, CA) as described previously (27). Percent apoptosis was calculated using the formula: (100×treatment cell absorbance/control cell absorbance)-100.
Statistical analysis
The statistical significance of differences between the values of treated samples and controls were determined with Kruskal-Wallis with Dunn’s post test using GraphPad Prism version 4.00 for Windows, GraphPad Software, San Diego, California, USA (www.graphpad.com). In each case, P < 0.05 was considered statistically significant.
Results
pEGCG is more potent than EGCG in inducing apoptosis and inhibiting cellular proliferation of human breast cancer cells
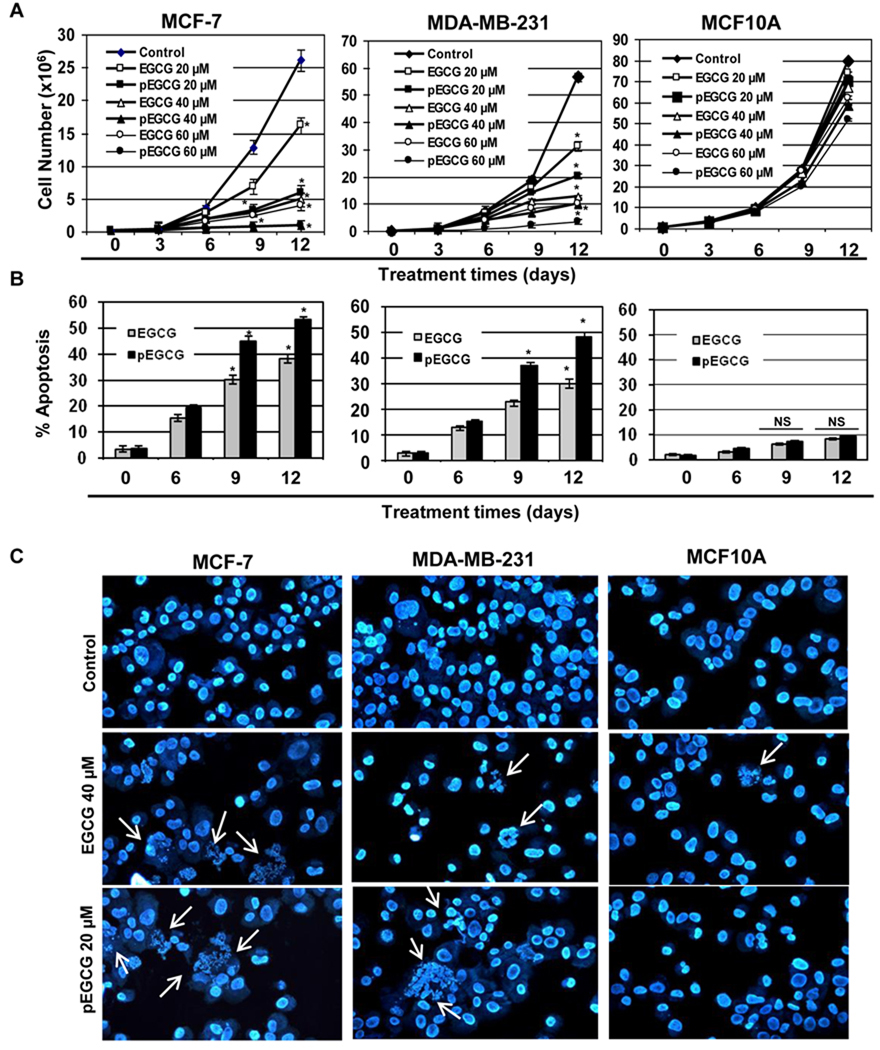
As shown in Fig. 1, human breast cancer MCF-7 (left panel) and MDA-MB-231 (middle panel) cells as well as normal control human breast MCF10A (right panel) cells were treated with the indicated concentrations of EGCG and pEGCG for 3, 6, 9 and 12 days for cell growth assay. We observed a dose- and time-dependent cell growth inhibition with EGCG and pEGCG treatment both in MCF-7 and MDA-MB-231 cells (Fig 1A). Doses of up to 60 µM of EGCG and 40 µM of pEGCG had negligible cell proliferation inhibition activity in control MCF10A cells while these same doses inhibited cellular proliferations for MCF-7 and MDA-MB-231 cells. In addition, significant levels of apoptosis were observed at 9 and 12 days with 20 µM pEGCG treatments for both MCF-7 and MDA-MB-231 cells (Fig 1B). However, EGCG required a higher dose (40 µM) than pEGCG to induce a significant level of apoptosis at the same time intervals in both human breast cancer cells. Both EGCG (40 µM) and pEGCG (20 µM) were found to induce significant apoptosis in both MCF-7 and MDA-MB-231 cells, whereas the equivalent doses were found to have very negligible cellular apoptotic effects on normal MCF10A breast cells (Fig 1B). Further, Hoechast staining analysis clearly demonstrated that treatment with EGCG (40 µM) and pEGCG (20 µM) induced more apoptotic cells in breast cancer cells, whereas negligible apoptotic cells were found in normal MCF10A cells (Fig 1C). Collectively, these results indicate that 40 µM of EGCG and 20 µM of pEGCG, selectively inhibits breast cancer cells. Therefore we chose 40 µM EGCG and 20 µM of pEGCG for further experiments. We used lower doses of EGCG and pEGCG than those previously reported 60 µM IC50 values of EGCG and 50 µM dosage of pEGCG (32, 24, 25), so that we could study the EGCG-induced epigenetic modifications on hTERT regulations without sudden cellular death. In addition, these lower doses should have higher translational potential in cancer chemoprevention as well as drug development and therapy.
Figure 1. EGCG and pEGCG inhibit proliferation of breast cancer cells but have negligible effects on control MCF10A cells.
A) Breast cancer MCF-7 (left panel) and MDA-MB-231 (middle panel) cells as well as control MCF10A cells (right panel) were treated with EGCG and pEGCG (0, 20, 40 and 60 µM) for 3, 6, 9 and 12 days. Growth curve kinetics was obtained by counting the total number of viable cells at the indicated time intervals using trypan blue staining. Results were obtained from three independent experiments, mean ± SD. B) Treatment with EGCG (40 µM) and pEGCG (20 µM) for 6, 9 and 12 days induced cellular apoptosis of human breast cancer MCF-7 (left panel) and MDA-MB-231 (middle panel) cells in a time-dependent manner. Control MCF10A cells (right panel) did not show a significant level of apoptosis at the same dosages and times of treatment. Percent apoptosis was assayed by using the Annexin V-Alexa Fluor 488 Apoptosis Vybrant Assay Kit. The experiment was repeated two times and each point indicates the mean ± SD. Statistical significance, *P < 0.05, NS-not significant. C) Treatment with EGCG (40 µM) and pEGCG (20 µM) for 9 days induced the cellular apoptosis in MCF-7 (left panel) and MDA-MB-231 (middle panel) cells but not in control MCF10A (right panel) cells. Apoptotic cells were detected by immunofluorescence staining under a fluorescence microscope, and are shown as multinucleated cells (white arrows-indicated). Representative photographs are shown from three repeated experiments.
EGCG and pEGCG inhibits hTERT expression in breast cancer cells
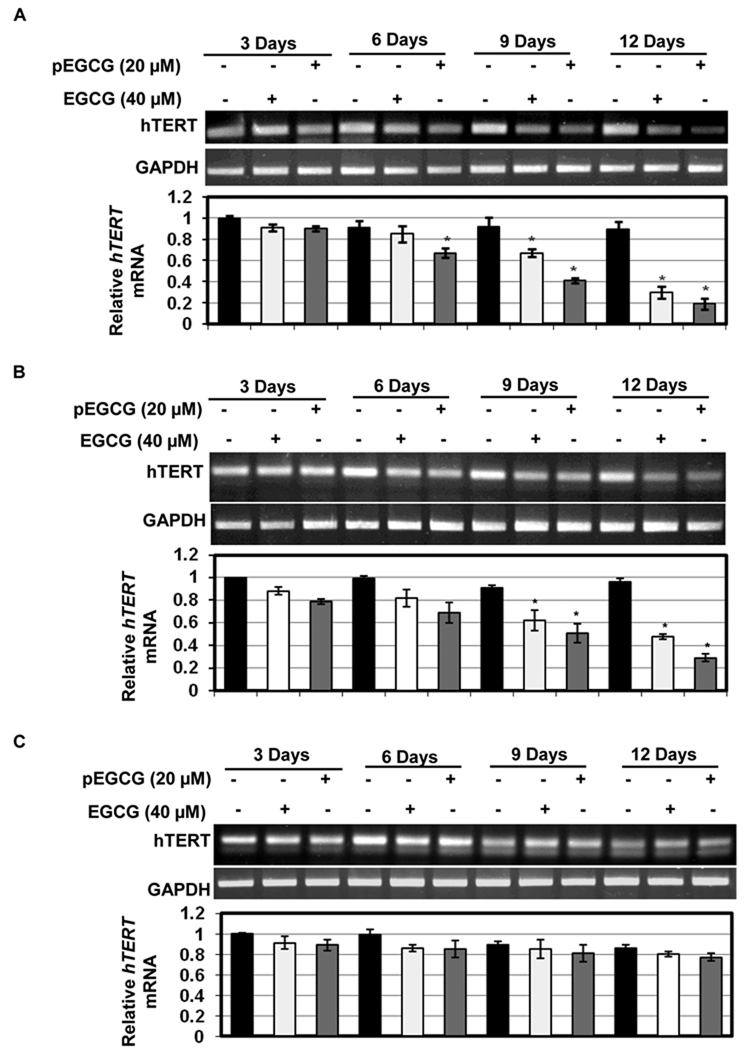
More than 90% of the cancer cells express higher levels of hTERT, the key catalytic subunit of telomerase, which serves as an important target for cancer chemoprevention (11–13, 33). Therefore we investigated the effect of EGCG and pEGCG on hTERT expression in MCF-7 (Fig 2A) and MDA-MB-231 (Fig 2B) human breast cancer cells as well as normal breast MCF10A cells (Fig 2C) by using conventional RT-PCR and real-time PCR. As shown in Fig. 2, treatment with EGCG (40 µM) and pEGCG (20 µM) time-dependently inhibited hTERT expression in both types of human breast cancer cells, while very negligible hTERT inhibitory activity was found in normal MCF10A cells. This is consistent with previous findings that inhibition of hTERT by chemopreventive compounds in cancer cells but not in normal cells is one of the important contributing factors in cancer chemoprevention (20, 27, 33). Further, 20 µM of pEGCG inhibited more hTERT expression than 40 µM of EGCG at similar time points although both compounds inhibited significant levels of hTERT at 9 and 12 days of treatments. These results indicate that pEGCG is more potent than EGCG and both compounds act on hTERT leading to its down-regulation specifically in breast cancer cells, which may play a critical role in inhibition of cancer cell proliferation and survival.
Figure 2. EGCG and pEGCG inhibits hTERT in human breast cancer cells.
Both EGCG (40 µM) and pEGCG (20 µM) inhibit hTERT mRNA expression in MCF-7 (panel A) and MDA-MB-231 (panel B) human breast cancer cells, but negligible hTERT inhibition effects were found in control MCF10A (panel C) cells. The indicated breast cell types were treated with EGCG or pEGCG for 3, 6, 9 and 12 days. After treatment periods, relative mRNA levels of hTERT in each sample were assessed using conventional gel-based PCR and quantified using real-time PCR. Data are in triplicates from three independent experiments and were normalized to GAPDH. The values were plotted against respective controls as relative fold of induction ± SD. Significance against respective nontreated control, *P<0.05.
EGCG and pEGCG induced hTERT hypomethylation and altered epigenetic-modulating enzyme activities
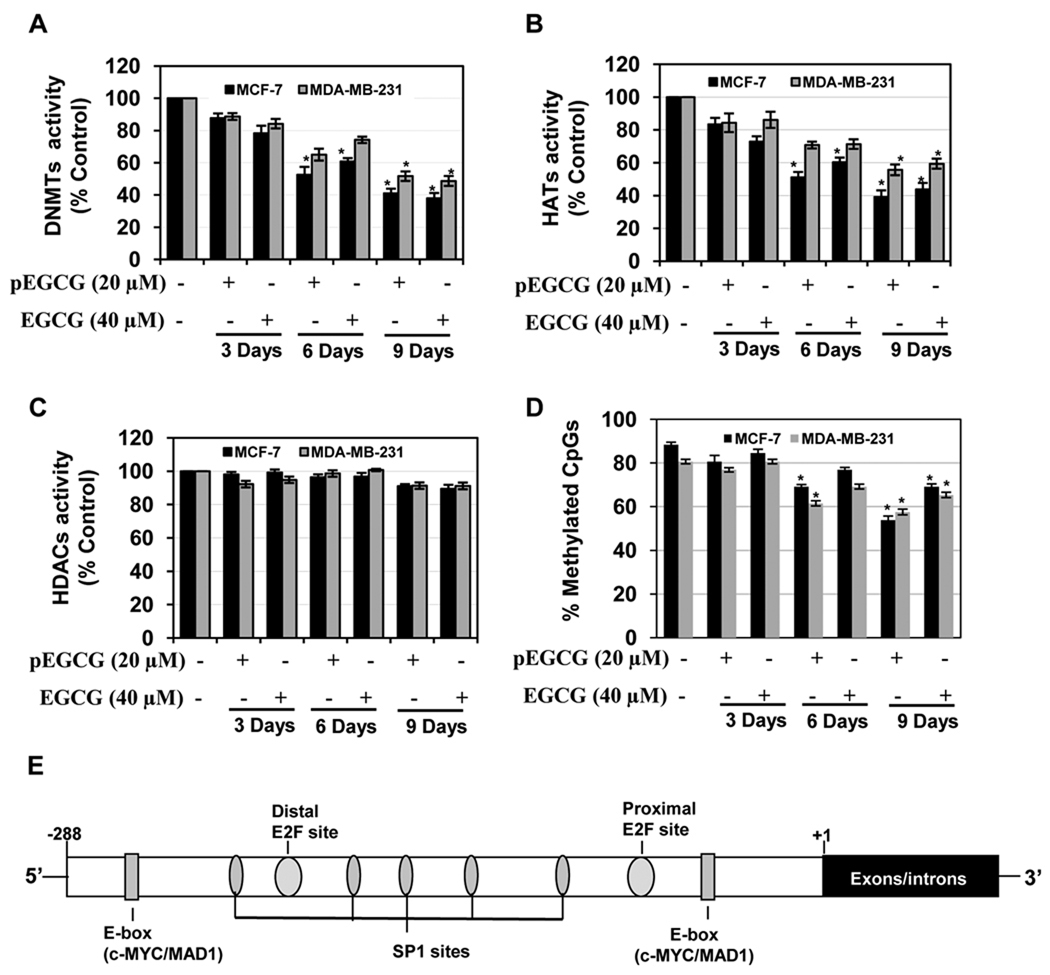
Since hTERT is one of the most epigenetically regulated genes (11–13, 17), we assessed epigenetic-modulating enzymatic activity of the DNMTs (Fig 3A), HATs (Fig 3B) and HDACs (Fig 3C) in MCF-7 and MDA-MB-231 breast cancer cells, using EGCG or pEGCG treatments. Interestingly, both EGCG and pEGCG at the indicated concentrations significantly inhibited DNMTs and HATs activities at 6 and 9 days of treatment in human breast cancer cells. However, we did not find any significant alteration with HDACs activity in these breast cancer cells with EGCG- and pEGCG-treatments. EGCG inhibition of DNMTs activity might be due to the direct binding of EGCG to the active site of the DNMTs as reported previously (15). Furthermore, EGCG was also reported to have inhibitory activity of the HATs in HeLa nuclear extracts (16). Since the hTERT promoter is hypermethylated in most cancer cells for its transcriptional activation, we assessed the methylation status of the hTERT promoter region from −288 to −31 covering 26 CpG dinucleotides and various overlapping transcription factor binding sites (Fig 3E). We used bisulfite-sequencing to detect the hTERT methylation patterns of EGCG- and pEGCG-treated human breast cancer cells. As shown in Fig 3D, control untreated MCF-7 and MDA-MB-231 breast cancer cells maintain a high level of methylation at promoter sites at 87.6±3.24 % and 79.4±2.29 %, respectively, whereas treatment with EGCG and pEGCG considerably reduced promoter methylation in a time-dependent manner. The EGCG- and pEGCG-mediated inhibition of DNMTs expression could be an important contributing factor in facilitating demethylation of hTERT promoter, which leads to transcriptional repression of hTERT expression (11–13).
Figure 3. EGCG and pEGCG altered epigenetic-modulating enzymes activity and hTERT promoter methylation in breast cancer cells.
A) EGCG (40 µM) and pEGCG (20 µM) inhibits DNMTs (panel A) and HATs (panel B) activity but no significant effects on HDACs (panel C) activity were found in human breast cancer cells. MCF-7 and MDA-MB-231 cells were treated with the indicated concentration of EGCG and pEGCG for 3, 6 and 9 days. DNMTs, HATs and HDACs activities were assayed and compared with mean percent control obtained from respective time intervals. Control values did not change considerably over the treatment times. Values are representative of three independent experiments and are represented as percent control ± SD; statistical significance, *P<0.05. D) EGCG (40 µM) and pEGCG (20 µM) induced hTERT promoter DNA hypomethylation in human breast cancer cells as assayed by bisulfite sequencing. Breast cancer MCF-7 and MDA-MB-231 cells were treated with the indicated concentration of EGCG and pEGCG for 3, 6 and 9 days. Percent methylation was obtained by dividing the number of methylated CpGs by the total number of CpGs (26) in the indicated hTERT promoter region assessed. Values are representative of three independent experiments and are represented as percent control ± SD; statistical significance, *P<0.05. E) The hTERT transcription factors such as E2F-1, Sp1 and c-MYC/MAD1 (E-box-binding sites) are shown in the hTERT promoter.
EGCG and pEGCG induced chromatin modifications and binding of transcriptional repressors of the hTERT promoter
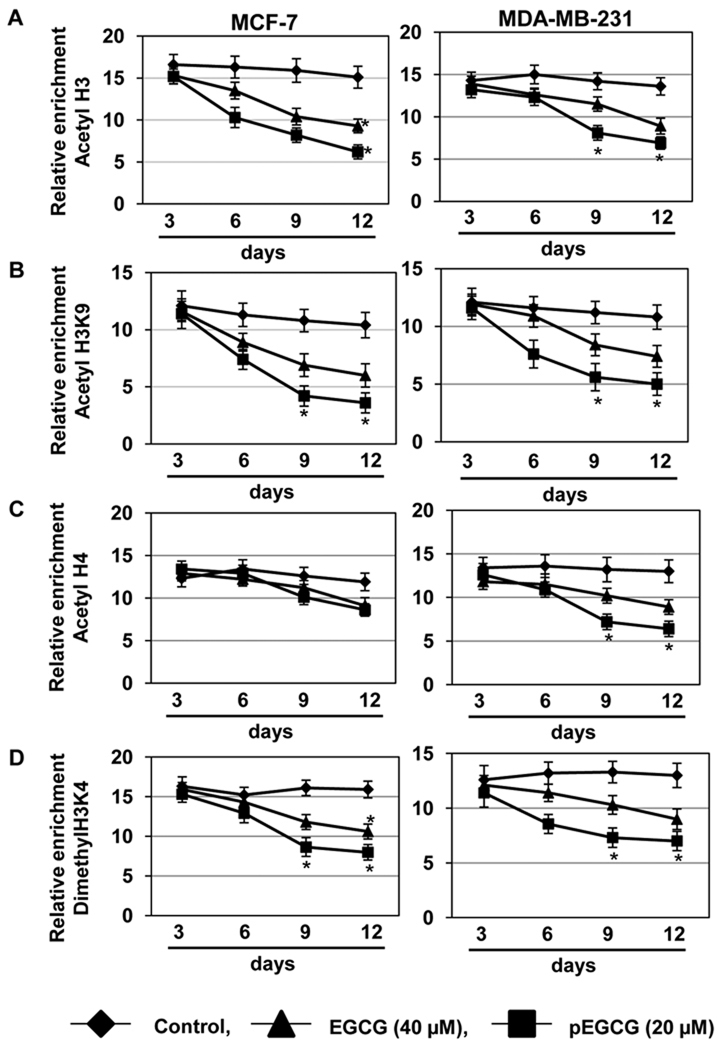
Previous studies have shown that hTERT expression is often modulated by epigenetic processes such as DNA methylations and histone acetylations (11–13, 17). We observed that EGCG and pEGCG time-dependently inhibited HATs activity without altering HDACs activity in human breast cancer cells (Fig 3). Decreased HATs activity is often associated with histone hypoacetylation at the hTERT promoter, which is associated with transcriptional repression of hTERT expression (12, 22). Therefore, we sought to determine changes in histone modifications of the hTERT regulatory region by EGCG- and pEGCG-treatment in MCF-7 and MDA-MB-231 cells. EGCG and pEGCG treatments resulted in a time-dependent decrease in the acetylation of transcriptionally active chromatin markers; acetylated histone H3 (ac-H3), H3 at lysine 9 (ac-H3K9) and ac-H4 in both MCF-7 and MDA-MB-231 cells (Fig 4A–C). We also found a decrease in the methylation status of active histone markers such as dimethyl-H3 lysine 4 (di-me-H3K4) in MCF-7 and MDA-MB-231 cells with EGCG and pEGCG treatments (Fig 4D). These changes of histone acetylation and deacetylation allow transcriptional factors binding into the hTERT regulatory region by maintaining a repressive environment (16–18, 22). Active and inactive chromatin modulations can control the antagonistic binding of MAD1 and c-MYC to the two E-boxes of the hTERT promoter, which are a major repressors and activators, respectively, of hTERT (11, 12, 19). Indeed, we found that the MAD1 repressor of hTERT is increased in its binding in response to EGCG and pEGCG whereas the c-MYC activator is decreased in its binding to the hTERT promoter in MCF-7 and MDA-MB-231 (Fig 5). Further, EGCG- and pEGCG-induced promoter hypomethylation led to the binding of the methylation-sensitive hTERT repressor, E2F-1, to the hTERT promoter. Collectively, these results suggest that the EGCG- and pEGCG-induced chromatin modifications affect binding of key transcriptional repressors to the hTERT promoter and inhibition of DNMTs mediated CpG hypomethylation at the hTERT regulatory region contributed to hTERT down-regulation in both MCF-7 and MDA-MB-231 breast cancer cells.
Figure 4. EGCG and pEGCG induced histone modification changes of the hTERT promoter in breast cancer cells.
A) Breast cancer MCF-7 (left panel) and MDA-MB-231 (right panel) cells were treated with EGCG (40 µM) and pEGCG (20 µM) for 3, 6, 9 and 12 days, and analyzed by ChIP-qPCR assays using chromatin markers including acetyl-H3 (panel A) acetyl-H3K9 (panel B), acetyl-H4 (panel C) and dimethyl-H3K4 (panel D) in the promoter region of hTERT. No antibody controls were also assessed to verify the ChIP efficiency. qPCR primers and conditions were used as described in Materials and Methods. The×axis represents the different treatment time in days, and the y axis represents the relative enrichment of individual binding factors [the percentage of immunoprecipitates compared with the corresponding input samples (defined as 100)]. The experiment was repeated three times with triplicates in real-time PCR and each point indicates the mean ± SD; statistical significance, *P<0.05.
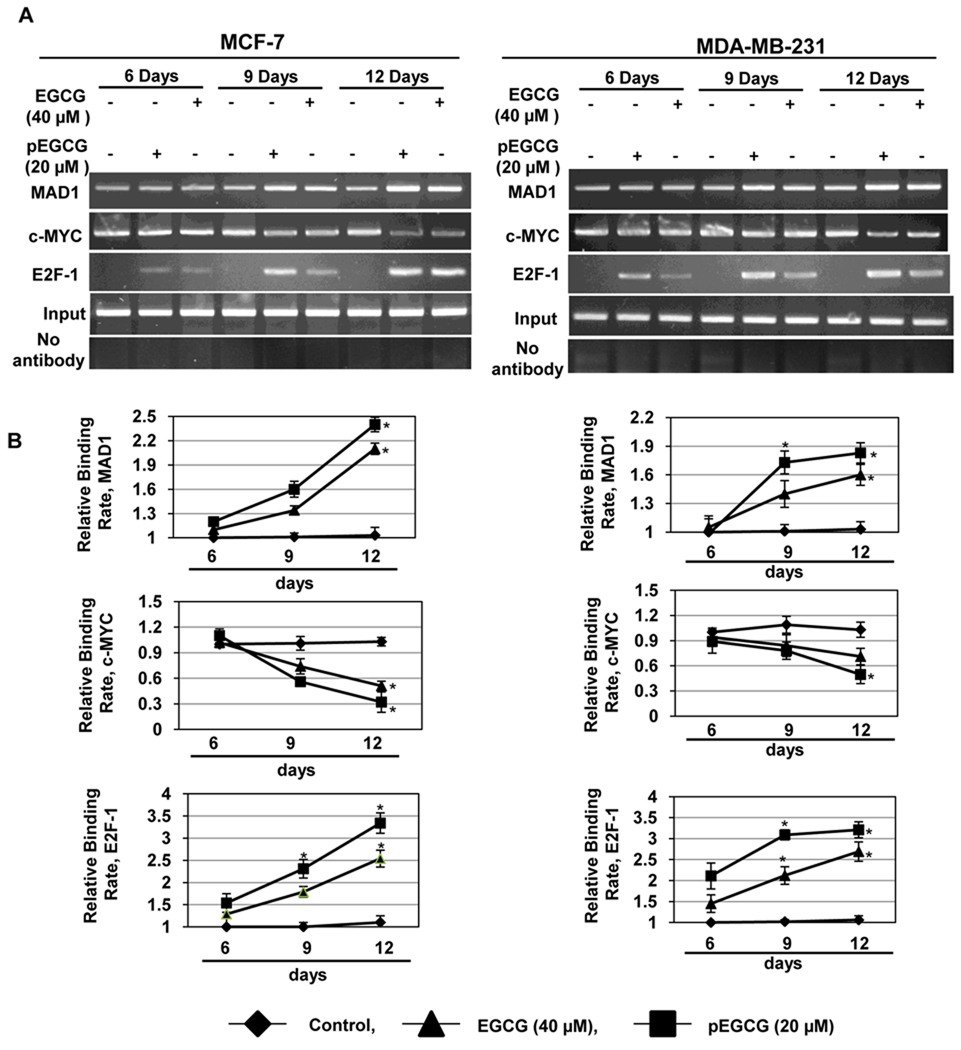
Figure 5. EGCG and pEGCG altered binding of transcriptional factors to the hTERT promoter in breast cancer cells.
A) Breast cancer MCF-7 (left panel) and MDA-MB-231 (right panel) cells were treated with EGCG (40 µM) and pEGCG (20 µM) for 6, 9 and 12 days, and ChIP-assayed using transcriptional factors such as c-MYC, MAD1 and E2F-1 in the promoter region of hTERT. No antibody controls were also assessed to verify the ChIP efficiency. PCR primers and conditions were used as described in Materials and Methods. Photographs are representative of an experiment that was repeated in triplicates. B) ChIP data were calculated from the corresponding DNA fragments amplified by PCR using Kodak 1D 3.6.1 image software; columns, mean; bars, SD; statistical significance, *P<0.05. The relative binding ratio was calculated as the ratio between the net intensity of each bound sample divided by the input and the untreated control sample divided by the input (bound/input)/(control/input).
E2F-1 and MAD1 knockdown differentially regulates pEGCG-inhibited hTERT expression in ER (+) and ER (−) human breast cancer cells
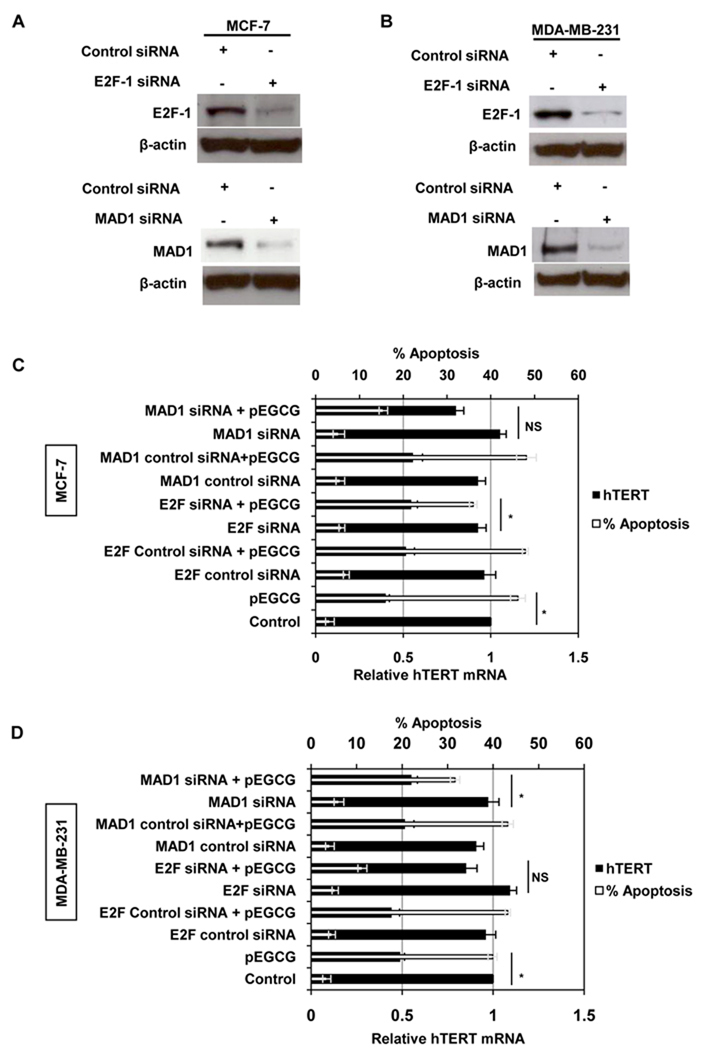
We found that EGCG and pEGCG induced transcriptional repression of hTERT expression, at least partially, by altering the binding of repressor proteins such as MAD1 and E2F-1 to the hTERT promoter (Fig 5). Therefore, we transiently transfected E2F-1 and MAD1 siRNA into the MCF-7 and MDA-MB-231 cells. Transfection of E2F-1 and MAD1 siRNA for 9 days considerably knocked down their expressions in both MCF-7 (Fig 6A, left panel) and MDA-MB-231 (Fig 6B, right panel) cells without inducing any significant level of cellular toxicity (data not shown). Treatment with pEGCG (20 µM) of ER (+) MCF-7 (Fig 6C) and ER (−) MDA-MB-231 (Fig 6D) breast cancer cells significantly inhibited hTERT expression and induced cellular apoptosis as observed in Fig 2A and Fig 2B, respectively. Surprisingly, pEGCG inhibition of hTERT expression and its associated enhanced cellular apoptosis were significantly restored in E2F-1 knockdown-MCF-7 cells but not in MDA-MB-231 cells. In contrast, pEGCG inhibition of hTERT expression was significantly restored by MAD1 knockdown-MDA-MB-231 cells but not in MCF-7 cells. Collectively, our data revealed for the first time that pEGCG inhibits hTERT expression and induced cellular apoptosis at least partially through the binding of E2F-1 and MAD1 to the hTERT promoter for ER (+) and ER (−) human breast cancer cells, respectively. Studies have indicated that ERα directly regulates E2F-1 expression and knockdown of E2F-1 blocks estrogen regulation in E2F-1 target genes (34, 35). Therefore, the observed hTERT restoration by E2F-1 knockdown in MCF-7 cells but not in MDA-MB-231 cells is likely, at least in part, due to the ERα expression. However, in ER (−) breast cancer cells, pEGCG-induced binding of MAD1 plays an important role in hTERT regulation apparently due to lack of ERα expression (Fig 6D). Collectively, our results indicate that knockdown of hTERT repressors can reverse the inhibitory effect of pEGCG on hTERT expression and, perhaps most importantly, reverses the antiapoptotic effects of pEGCG. Therefore, these new findings suggest that the impedance of repressor binding to the hTERT promoter due to their epigenetic effects leads to cancer cell apoptotic properties by pEGCG.
Figure 6. E2F-1 and MAD1 knockdown differentially regulate pEGCG-inhibited hTERT expression in ER (+) and ER (−) breast cancer cells.
Breast cancer ER (+) MCF-7 (Panel A) and ER (−) MDA-MB-231 (Panel B) cells were subjected to treatments with 6 and 4 nM of E2F-1 and MAD1 siRNA, respectively, or control siRNA fragments. Effects of siRNA interference with E2F-1 and MAD1 gene expression was assayed by western blot analysis after 9 days using specific antibodies (panel A). Data shown are representative of the three separate experiments. E2F-1 and MAD1 siRNA-transfected cells were treated with 20 µM pEGCG for 9 days and analyzed for hTERT mRNA expression by RT-PCR as well as apoptosis by ELISA in MCF-7 (Panel C) and MDA-MB-231 (Panel D) breast cancer cells. Data are in triplicates from two independent experiments and were normalized to GAPDH for calculating relative hTERT mRNA. Statistical significance *P<0.05; NS-not significant.
Discussion
In accordance with previous findings, EGCG dose- and time-dependently inhibited the growth of both ER (+) and ER (−) human breast cancer cells (1, 4, 20, 24). pEGCG was found to be more potent than EGCG in inhibiting cellular proliferations and inducing cellular apoptosis in both ER (+) and ER (−) human breast cancer cells. Studies have clearly shown that pEGCG has increased stability compared to EGCG and is converted into parental EGCG in cultures with ~2.4 fold greater recovery in cells than EGCG after 72 h treatment (24, 26). Further pEGCG treatment increased the bioavailability of EGCG in breast, esophageal and colon cancer cells (24, 26), which further substantiates our present findings.
We have shown that both EGCG and pEGCG time-dependently inhibited hTERT expression in both ER (+) and ER (−) human breast cancer cells but not in normal cells. This is in accordance with previous findings that EGCG inhibited telomerase through epigenetic modifications and induced cellular apoptosis in lung and ER (+) breast cancer cells (2, 4, 20). However, studies have been largely obscure on hTERT regulations in ER (−) human breast cancer cells. Interestingly, we found that EGCG and pEGCG inhibited hTERT expression similarly in both ER (+) and ER (−) breast cancer cells, although hTERT is a one of the targets for ligand-activated ER, and the presence of ER is a contributing factor, at least partially, for hTERT activation (36, 37). Therefore, it is likely that hTERT regulation in ER (+) and ER (−) breast cancer cells might act through different mechanisms. Further, epigenetic regulation of hTERT is actively involved in cellular proliferation and apoptosis in various cancer cells. In most of the cancer cells the hTERT regulatory region is hypermethylated, which is associated with increased hTERT expression, whereas demethylation of this region inhibits hTERT transcription (18, 38). This phenomenon is opposite to the general model of gene activation, in which the presence of methylated cytosines in a promoter typically inhibits gene transcription (10, 39, 40).
Previously, we have shown that genistein and EGCG result in down-regulation of the DNMTs which is directly associated with repression of hTERTexpression through hTERT promoter demethylation in breast cancer cells (20, 31). The EGCG-mediated DNMTs inhibition might be due to the possible direct interaction of EGCG with the DNMTs active site (15). Numerous studies have also reported that DNA methylation plays important roles in hTERTtranscriptional regulation (10–12, 31). Together, our results suggest that EGCG-induced down-regulation of DNMTs expression is not only involved in the demethylation processes of the hTERT control region in the process of anti-carcinogenesis, but also enhances binding of methylation-sensitive transcription factors such as E2F-1 to the hTERT regulatory region (23). Further, our ChIP-analysis confirmed that EGCG- and pEGCG-induced demethylation at the CpG dinucleotides of the hTERT promoter resulted in an increased binding of E2F-1 to the hTERT proximal promoter which leads to repression of hTERT transcription.
In general, chromatin acetylation and deacetylation are catalyzed by HATs and HDACs, respectively, which play an important role in transcriptional regulations of hTERT expression (10, 16). EGCG was reported to have HATs inhibitory activity in HeLa nuclear extracts (16). Similarly, we found that EGCG-treatment significantly inhibited HATs activities in human breast cancer cells; however, we did not find any significant alterations in HDACs activities. By contrast, sulforaphane (SFN), an isothiocyanate present in the cruciferous vegetables, also has hTERT inhibitory activity in human breast cancer cells, which specifically inhibits HDACs but not HATs activities (27). Unlike with the use of SFN, we found an EGCG-induced time-dependent decrease of transcriptional active chromatin markers such as ac-H3, ac-H3K9 and ac-H4 in human breast cancer cells. Chromatin remodeling resulting from reversible acetylation of histones has been suggested to be a critical component of transcriptional regulation of hTERT expression (22). Histone acetylation and decetylation-modulated chromatin structure can be accessed with a number of transcription factors, including c-MYC and MAD1, which often regulates gene expression by recruiting HATs and HDACs, respectively (22). Our results also suggest that EGCG-induced MAD1 binding might recruit RBP2, a histone demethylase, to the hTERT promoter and reduced hTERT mRNA expression is accompanied by H3K4-demethylation (19). In addition, hTERT expression in normal and malignant human cells was found to have an inverse correlation with MAD1 expression (20, 41, 42). The MAD1-induced repression of hTERT transcription is mediated by the N-terminal SID of Mad1 that recruits HDACs to chromatin (41). Furthermore, there is a switch from MYC/MAX to MAD1/MAX binding and a decrease in histone acetylation at the hTERT promoter during HL60 differentiation (22). Taken together, it is apparent that DNMTs-induced promoter demethylation and chromatin remodeling alter binding of hTERT transcriptional repressors to the hTERT promoter is closely linked to the control of hTERT expression by EGCG and pEGCG in human breast cancer cells.
Our functional studies with E2F-1 and MAD1 siRNA revealed for the first time that pEGCG–inhibited hTERT expression and associated apoptosis is differently regulated in ER (+) MCF-7 and ER (−) MDA-MB-231 breast cancer cells. Previous studies have shown the EGCG can induce cellular apoptosis and inhibits cellular proliferations in both ER (+) and ER (−) human breast cancer cells by similar mechanisms (1, 4). In addition, the present study also confirmed the fact that EGCG and pEGCG induced cellular apoptosis in both ER (+) and ER (−) human breast cancer cells but regulates hTERT through different transcriptional repressors. Our siRNA study revealed that E2F-1 is an important transcriptional repressor required for ER (+) cancer cells. However, in ER (−) breast cancer cells MAD1 appears to play an important role in hTERT repression and associated apoptosis. This might be partially due to the fact that estrogen stimulates ER-c-Myc protein interactions and lack of ER induces MAD1, c-MYC antagonist, for the target gene suppression (43).
In the present study, we not only used a pro-drug of EGCG (pEGCG) to enhance the bioavailability and stability of EGCG but also explored the possible epigenetic mechanisms involved in hTERT repression. It is important to point out that hTERT gene control is unique and the proposed mode of action is not the only way EGCG inhibits cancer cell growth. The optimal concentrations of EGCG and pEGCG used in this study are lower than the many other studies have used previously (1, 4, 15, 20). Furthermore, the concentrations we used selectively inhibited cellular proliferation and induced apoptosis in human breast cancer cells but not in control MCF10A cells as shown in Fig 1. Studies have shown that EGCG and pEGCG up to 50 mg/kg/day were well tolerated in experimental animals without noticeable toxicity (24, 44). Another important finding is that for the first time we showed that pEGCG treatment-induced chromatin changes resulted in a differentially-regulated transcriptional repressor binding to the hTERT promoter in ER (+) and ER (−) human breast cancer cells. These findings have important implications for the application of EGCG in cancer prevention and drug development for ER (−) human breast tumors. However, further studies with spontaneous multistage breast tumor-producing mouse models such as C3(1)/SV40 and Her2/neu are necessary to study the in vivo effect of EGCG and pEGCG during different stages of breast cancer progression. These in vivo mouse models will not only produce breast tumors which closely resemble the development, progression and morphology of human breast tumors (45, 46) but also allow studying long-term bioavailability of EGCG, which is best suited for cancer chemoprevention models.
Supplementary Material
Acknowledgements
We thank Dr. Yuanyuan Li for her support and critical reading of the manuscript.
Financial Support: This work was supported by grant R01 CA129415 from the National Cancer Institute, National Institutes of Health and the National Science and Engineering Research Council of Canada (THC).
References
- 1.Roy AM, Baliga MS, Katiyar SK. Epigallocatechin-3-gallate induces apoptosis in estrogen receptor-negative human breast carcinoma cells via modulation in protein expression of p53 and Bax and caspase-3 activation. Mol Cancer Ther. 2005;4:81–90. [PubMed] [Google Scholar]
- 2.Sadava D, Whitlock E, Kane SE. The green tea polyphenol, epigallocatechin-3-gallate inhibits telomerase and induces apoptosis in drug-resistant lung cancer cells. Biochem Biophys Res Commun. 2007;360:233–237. doi: 10.1016/j.bbrc.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Tran PL, Kim SA, Choi HS, Yoon JH, Ahn SG. Epigallocatechin-3-gallate suppresses the expression of HSP70 and HSP90 and exhibits anti-tumor activity in vitro and in vivo. BMC Cancer. 2010;10:276. doi: 10.1186/1471-2407-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittal A, Pate MS, Wylie RC, Tollefsbol TO, Katiyar SK. EGCG down-regulates telomerase in human breast carcinoma MCF-7 cells, leading to suppression of cell viability and induction of apoptosis. Int J Oncol. 2004;24:703–710. [PubMed] [Google Scholar]
- 5.Lin J, Liang Y. Cancer chemoprevention by tea polyphenols. Proc Natl Sci Counc Repub China B. 2000;24:1–13. [PubMed] [Google Scholar]
- 6.Ahmad N, Feyes D, Nieminen A, Agarwal R, Mukhtar H. Green tea constituent epigallocatechin-3-gallate and induction of apoptosis and cell cycle arrest in human carcinoma cells. J Natl Cancer Inst. 1997;89:1881–1886. doi: 10.1093/jnci/89.24.1881. [DOI] [PubMed] [Google Scholar]
- 7.Gu B, Ding Q, Xia G, Fang Z. EGCG inhibits growth and induces apoptosis in renal cell carcinoma through TFPI-2 overexpression. Oncol Rep. 2009;21:635–640. [PubMed] [Google Scholar]
- 8.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (−)-epigallocatechin-3-gallate. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 9.Fassina G, Venè R, Morini M, Minghelli S, Benelli R, Noonan D, et al. Mechanisms of inhibition of tumor angiogenesis and vascular tumor growth by epigallocatechin-3-gallate. Clin Cancer Res. 2004;10:4865–4873. doi: 10.1158/1078-0432.CCR-03-0672. [DOI] [PubMed] [Google Scholar]
- 10.Meeran SM, Ahmed A, Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenet. 2010;1:101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham AP, Love WK, Zhang RW, Andrews LG, Tollefsbol TO. Telomerase inhibition in cancer therapeutics: molecular-based approaches. Curr Med Chem. 2006;13:2875–2888. doi: 10.2174/092986706778521887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Lai S, Andrews L, Tollefsbol T. Genetic and epigenetic modulation of telomerase activity in development and disease. Gene. 2004;340:1–10. doi: 10.1016/j.gene.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Naasani I, Seimiya H, Tsuruo T. Telomerase inhibition, telomere shortening, and senescence of cancer cells by tea catechins. Biochem Biophys Res Commun. 1998;249:391–396. doi: 10.1006/bbrc.1998.9075. [DOI] [PubMed] [Google Scholar]
- 14.Fang M, Chen D, Yang C. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 15.Fang M, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 16.Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, Kang HB, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69:583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 17.Kyo S, Takakura M, Fujiwara T, Inoue M. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renaud S, Loukinov D, Abdullaev Z, Guilleret I, Bosman F, Lobanenkov V, et al. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007;35:1245–1256. doi: 10.1093/nar/gkl1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge Z, Li W, Wang N, Liu C, Zhu Q, Björkholm M, et al. Chromatin remodeling: recruitment of histone demethylase RBP2 by Mad1 for transcriptional repression of a Myc target gene, telomerase reverse transcriptase. FASEB J. 2010;24:579–586. doi: 10.1096/fj.09-140087. [DOI] [PubMed] [Google Scholar]
- 20.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103:509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Günes C, Lichtsteiner S, Vasserot AP, Englert C. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 2000;60:2116–2121. [PubMed] [Google Scholar]
- 22.Xu D, Popov N, Hou M, Wang Q, Björkholm M, Gruber A, et al. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci U S A. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowe DL, Nguyen DC, Tsang KJ, Kyo S. E2F-1 represses transcription of the human telomerase reverse transcriptase gene. Nucleic Acids Res. 2001;29:2789–2794. doi: 10.1093/nar/29.13.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landis-Piwowar KR, Huo C, Chen D, Milacic V, Shi G, Chan TH, et al. A novel prodrug of the green tea polyphenol (−)-epigallocatechin-3-gallate as a potential anticancer agent. Cancer Res. 2007;67:4303–4310. doi: 10.1158/0008-5472.CAN-06-4699. [DOI] [PubMed] [Google Scholar]
- 25.Lam WH, Kazi A, Kuhn DJ, Chow LM, Chan AS, Dou QP, et al. A potential prodrug for a green tea polyphenol proteasome inhibitor: evaluation of the peracetate ester of (−)-epigallocatechin gallate [(−)-EGCG]. Bioorg Med Chem. 2004;12:5587–5593. doi: 10.1016/j.bmc.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Lambert JD, Sang S, Hong J, Kwon SJ, Lee MJ, Ho CT, et al. Peracetylation as a means of enhancing in vitro bioactivity and bioavailability of epigallocatechin-3-gallate. Drug Metab Dispos. 2006;34:2111–2116. doi: 10.1124/dmd.106.011460. [DOI] [PubMed] [Google Scholar]
- 27.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciftci K, Su J, Trovitch P. Growth factors and chemotherapeutic modulation of breast cancer cells. J Pharm Pharmacol. 2003;55:1135–1141. doi: 10.1211/002235703322277177. [DOI] [PubMed] [Google Scholar]
- 29.Golubovskaya V, Virnig C, Cance W. TAE226-induced apoptosis in breast cancer cells with overexpressed Src or EGFR. Mol Carcinog. 2008;47:222–234. doi: 10.1002/mc.20380. [DOI] [PubMed] [Google Scholar]
- 30.Meeran SM, Katiyar S, Katiyar SK. Berberine-induced apoptosis in human prostate cancer cells is initiated by reactive oxygen species generation. Toxicol Appl Pharmacol. 2008;229:33–43. doi: 10.1016/j.taap.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Liu L, Andrews L, Tollefsbol T. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009;125:286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Henning SM, Heber D. Limitations of MTT and MTS-based assays for measurement of antiproliferative activity of green tea polyphenols. PLoS One. 2010;5:e10202. doi: 10.1371/journal.pone.0010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naasani I, Oh-Hashi F, Oh-Hara T, Feng W, Johnston J, Chan K, et al. Blocking telomerase by dietary polyphenols is a major mechanism for limiting the growth of human cancer cells in vitro and in vivo. Cancer Res. 2003;63:824–830. [PubMed] [Google Scholar]
- 34.Louie MC, McClellan A, Siewit C, Kawabata L. Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol Cancer Res. 2010;8:343–352. doi: 10.1158/1541-7786.MCR-09-0395. [DOI] [PubMed] [Google Scholar]
- 35.Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21:2112–2123. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- 36.Nanni S, Narducci M, Della Pietra L, Moretti F, Grasselli A, De Carli P, et al. Signaling through estrogen receptors modulates telomerase activity in human prostate cancer. J Clin Invest. 2002;110:219–227. doi: 10.1172/JCI15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misiti S, Nanni S, Fontemaggi G, Cong YS, Wen J, Hirte HW, et al. Induction of hTERT expression and telomerase activity by estrogens in human ovary epithelium cells. Mol Cell Biol. 2000;20:3764–3771. doi: 10.1128/mcb.20.11.3764-3771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zinn R, Pruitt K, Eguchi S, Baylin S, Herman J. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 39.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 40.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 41.Cong YS, Bacchetti S. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J Biol Chem. 2000;275:35665–35668. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]
- 42.Oh S, Song YH, Yim J, Kim TK. Identification of Mad as a repressor of the human telomerase (hTERT) gene. Oncogene. 2000;19:1485–1490. doi: 10.1038/sj.onc.1203439. [DOI] [PubMed] [Google Scholar]
- 43.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Sun DK, Chen D, Cui QC, Gu YY, Jiang Te, et al. Antitumor activity of novel fluoro-substituted (−)-epigallocatechin-3-gallate analogs. Cancer Lett. 2010;292:48–53. doi: 10.1016/j.canlet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19:1020–1027. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 46.Rossi C, Di Lena A, La Sorda R, Lattanzio R, Antolini L, Patassini C, et al. Intestinal tumour chemoprevention with the antioxidant lipoic acid stimulates the growth of breast cancer. Eur J Cancer. 2008;44:2696–2704. doi: 10.1016/j.ejca.2008.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.