Abstract
MS/MS experiments generate multiple, nearly identical spectra of the same peptide in various laboratories, but proteomics researchers typically do not leverage the unidentified spectra produced in other labs to decode spectra generated in their own labs. We propose a spectral archives approach that clusters MS/MS datasets, representing similar spectra by a single consensus spectrum. Spectral archives extend spectral libraries by analyzing both identified and unidentified spectra in the same way and maintaining information about spectra of peptides shared across species and conditions. Thus archives offer both traditional library spectrum similarity-based search capabilities along with novel ways to analyze the data. By developing a clustering tool, MS-Cluster, we generated a spectral archive from ~1.18 billion spectra that greatly exceeds the size of existing spectral repositories. We advocate that publicly available data should be organized into spectral archives, rather than be analyzed as disparate datasets, as is mostly the case today.
Introduction
Recent years have witnessed a dramatic increase in the volume of MS/MS data generated in proteomics laboratories. Analyzing these increasing amounts of data has become a computational challenge, raising the need for improved algorithms.
Mass spectrometry is currently a rather introverted discipline. Researcher A working on, let's say the mouse proteome, has little interest in spectra generated by researcher B who is working on the human proteome. Indeed, since it is not clear how a mouse researcher can benefit from spectra generated by a human researcher, A would rarely bother to analyze spectra generated by B. As a result, there is little motivation to share spectra between different labs. This widespread introversion is particularly troublesome since 75–85% of spectra in a typical MS/MS experiment remain unidentified and thus are essentially discarded. Could a particular unidentified spectrum that showed up in spectral data sets generated by 10 different laboratories working on tumor proteomes be a potential fusion peptide representing a cancer biomarker, even though the spectrum was discarded time and again because none of the labs was able to interpret it? Would it be beneficial to answer the question whether a newly generated spectrum (identified or not) has been seen before by other researchers during the last decade, and under what circumstances? Answering these questions requires a different way of analyzing mass spectrometry data.
The most widely-used method in MS/MS analysis is the identification of peptides using a database search. MS/MS spectra are compared with peptides derived from a database of known protein sequences and the resulting Peptide-Spectrum Matches (PSMs) are evaluated using various scoring methods. As large numbers of confident PSMs are accumulated, it becomes worthwhile to organize them as spectral libraries1,2, which may serve as an additional method for peptide identification. Spectral libraries rely on the reproducible nature of MS/MS data to compare query spectra with the set of previously identified PSMs. While spectral library searches are fast and accurate3,4, they are not able to identify spectra of peptides that are not discovered in standard database searches. However, if identified spectra proved to be useful in the spectral library framework, could we utilize unidentified spectra in a similar way?
We present spectral archives, an approach for detecting and grouping similar spectra, both identified and unidentified, across multiple data sets. Like spectral libraries1,2, spectral archives can be used to perform accurate spectrum similarity-based identifications. However, archives also extend libraries, making it possible to analyze peptides that would typically remain below the radar with traditional database searches, opening new opportunities in areas like proteogenomics or biomarker discovery (see Supplementary Note 1 for further discussion on the relationship between archives and libraries). We believe that spectral archives could change the introverted nature of proteomics by motivating the seemingly unrelated researchers A and B to share their data, since doing so improves the quality of the interpretations of both of their spectral datasets. We argue that publicly available datasets should be organized into spectral archives, rather than be analyzed as disparate datasets, as is the case today. While clustering billions of spectra may look like an intractable problem, here we demonstrate the feasibility of constructing large spectral archives and their use for data analysis.
Spectral archives are generated by clustering spectra. Like libraries, clustering algorithms rely on the notion that MS/MS datasets contain multiple, nearly identical, spectra of the same peptides5–8. Each spectrum in the archive is associated with an identifier describing the experiment ID, file number, scan number, organism from which the sample was taken, the condition (e.g., healthy vs. diseased), experimental settings, etc. All spectra in each cluster are represented by a single consensus spectrum that typically has higher quality than individual spectra. Archives can grow by merging an existing archive with a new spectral dataset. All new spectra that get added to the archive (and all changed consensus spectra) can be searched against a protein database, if one is available, and any confident identifications are assigned to the clusters in the archive. Adding more spectra to the cluster typically increases the signal-to-noise ratio of the cluster's consensus spectrum since the randomly distributed noisy peaks are often canceled out in the consensus spectrum8.
Spectral archives, unlike libraries, include all mass spectra, both identified and unidentified. Though including unidentified clusters in a library-like search does not directly contribute to peptide identifications, it can ultimately have an indirect contribution. One way in which this can happen is by association of unidentified spectra with newly added identified spectra. For example, peptides expressed in low abundance in one lab can generate low-quality spectra. Such a spectrum might get inserted into an archive and remain unidentified until arrival of a new spectrum of the same peptide identified in another lab (e.g., the peptide was highly expressed or the spectrum was acquired on a more accurate instrument). Assume that at this stage the new spectrum, which is confidently identified, gets added to an unidentified cluster in the archive. Then, by virtue of their cluster membership, all spectra in the cluster now gain an identification. As an archive grows and includes many spectra of overlapping peptides, it becomes feasible to make more peptide identifications using advanced methods like spectral networks9.
Construction of community-wide spectral archives, however, requires clustering billions of spectra generated by many laboratories. Existing software tools allow clustering of several million5–7 to tens of millions8 of spectra. Here we describe our MS-Cluster algorithm which is capable of processing billions of spectra (see “Clustering algorithm” in online Methods and Supplementary Note 2 for more details about the algorithm's implementation).
We also report the construction of a spectral archive from over a billion mass spectra that were acquired at the Pacific Northwest National Laboratory between 2001 and 2009. This data includes samples taken from a diverse set of more than 100 organisms, using various ion-trap based platforms. Beyond demonstrating the feasibility of constructing large archives and their basic utility for run-of-the-mill peptide identification, we present new applications that become possible because a diverse collection of data sets can be analyzed as a whole. For instance, archives have the ability to identify clusters of spectra that come from different organisms. Besides indicating that such spectra are interesting (since they are likely to represent a peptide conserved over multiple species), this fact can be used to reduce the effective protein database size, leading to new confident spectral identifications. We also show that short peptides (shorter than 7 amino acids) can be confidently identified, which is not possible with the Target-Decoy Approach10 (TDA).
Results
Constructing Spectral Archives
We tested the MS-Cluster algorithm on ~1.18 billion spectra acquired at the Pacific Northwest National Laboratories from samples from over 100 organisms (referred to henceforth as the PNNL dataset - see Supplementary Table 1 for more details). MS-Cluster organized the PNNL dataset into ~299 million clusters (Fig. 1a and Supplementary Table 2), a fourfold reduction compared to the number of spectra generated originally. During clustering, the algorithm performed ~3.1×1013 similarity computations which required ~9,200 CPU hours.
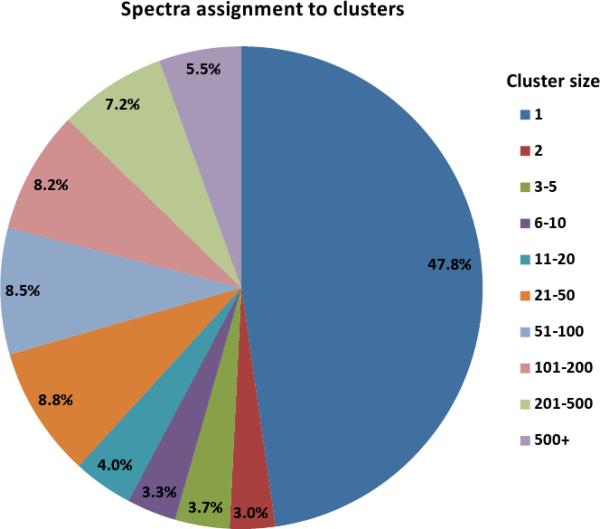
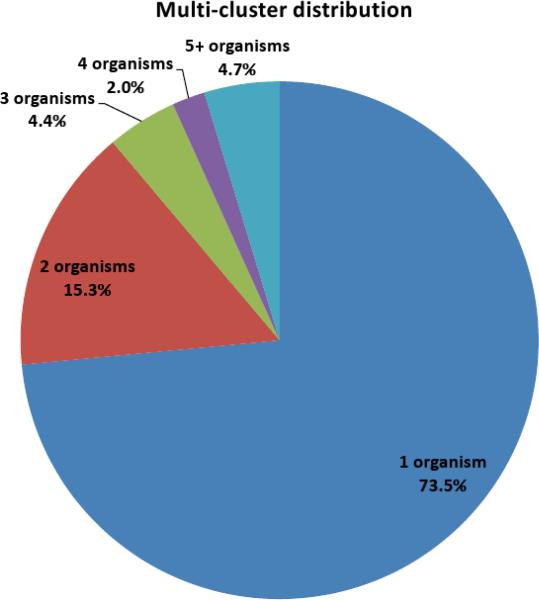
Figure 1.
Clustering of the PNNL dataset. 581 million spectra from the PNNL dataset that passed quality filtration were assigned into 299 million clusters of different sizes. (a) While most spectra form multi-clusters (i.e., clusters containing at least two spectra), most clusters consist of a single spectrum. (b) A breakdown of the clusters according to the number of organisms whose spectra participated in each cluster for each of 21.5 million multi-clusters.
While ~92% of the clusters contain a single spectrum, most of the spectra (52%) belong to clusters of multiple spectra. A quarter of the clusters that contained multiple spectra have spectra from multiple organisms (Fig 1b, Supplementary Table 2). Of these, approximately 58% came from exactly two different organisms. While a large cluster with spectra from many organisms is likely from contaminants (e.g., keratin, trypsin, etc.) or standard proteins added for calibration purposes, clusters with spectra from a small number of organisms likely originate from a peptide common to the different organisms.
Peptide Identifications With Spectral Archives
To evaluate whether spectral archives can increase peptide identifications, we selected a subset of 14.5 million spectra of Shewanella oneidensis, and constructed an archive with them as follows. The dataset was broken into five sets of ~2.9 million spectra. We incrementally added each set of spectra to the archive according to the following procedure: preprocessing the new spectra (including quality filtration), clustering the new data and merging it with the existing archive, searching the new and modified clusters, and updating the archive with the identifications (see online methods for more information). Table 1 summarizes the results of these experiments. At each stage we compared the number of protein and unique peptide identifications made searching the clusters in the archive with the number that could be obtained with a regular database search of the individual spectra. The archive consistently yielded ~5% more unique peptide identifications and ~2% more protein identifications, compared to a regular database search. The False Discovery Rate (FDR) of Cluster-Peptide Matches (CPM) in the archive was updated at each stage to maintain a rate of 2% for the entire archive (simply repeatedly adding batches of CPMs at 2% FDR would ultimately give a higher FDR than 2% for the entire archive8).
Table 1.
Search results for archive of 14.5 million spectra of Shewanella oneidensis MR-1. For each fraction of the full dataset, the table compares the identifications (number of proteins, peptides and spectra/cluster annotations) made by a regular database search and by searching the clusters in the archive. The searches were done against a database of S. oneidensis protein sequences with false discovery rate of 2%.
| Regular database search | Archive clusters search | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Dataset fraction | Num. spectra searched | Num. protein ids | Num. peptide ids | Num. spectra annotated | Num. clusters searched | Num. protein ids | Num. peptide ids | Num. annotations | |
| Clusters | Spectra | ||||||||
| 1/5 | 2.9M | 2,257 | 28,083 | 0.5M | 0.61M | 2,304 | 29,948 | 0.18M | 0.75M |
| 2/5 | 5.8M | 2,435 | 33,866 | 0.95M | 1.06M | 2,471 | 35,648 | 0.28M | 1.52M |
| 3/5 | 8.7M | 2,518 | 37,093 | 1.42M | 1.49M | 2,566 | 39,355 | 0.37M | 2.34M |
| 4/5 | 11.6M | 2,561 | 39,205 | 1.84M | 1.89M | 2,611 | 41,418 | 0.44M | 3.09M |
| 5/5 | 14.5M | 2,608 | 40,680 | 2.28M | 2.29M | 2,660 | 43,415 | 0.51M | 3.96M |
With the archive we also identified many more spectra (through their cluster membership). At different stages, the archive was able to identify 50–75% more spectra through cluster membership than a regular database search. Our results demonstrate that the fourfold reduction in the number of clusters (as compared to the original number of spectra) does not lead to lost peptide identifications due to clusters mixing spectra of more than one peptide. In practice, MS-Cluster rarely generates such mixed clusters. We ran experiments with several data sets of 1 million spectra; first, we searched their spectra against a database (at 2% FDR), then clustered the data and checked the clusters with identified spectra to see how many clusters had spectra with different peptide annotations. On average, only ~ 1.5% of the clusters were mixed in that way and only ~2% of the peptides identified in the database search were mapped to mixed clusters (data not shown).
Archives Synergize Interpretation Across Labs And Tissues
Spectral archives combine and synergize data originating from different experiments. Unidentified spectra from one dataset can be identified via their membership in clusters with higher quality spectra generated in other experiments involving other tissues or organisms. To demonstrate this synergy we used two spectral datasets: 9.9 million spectra from human plasma (Plasma dataset generated at PNNL) and 7 million spectra from the human HEK293 kidney cell line11 (generated at UCSD). We searched these datasets individually against the human IPI sequence database (v3.68) at 2% peptide level FDR. We then created an archive from both datasets, which enabled us to make additional peptide annotations through cluster membership (e.g., an identified spectrum from the HEK dataset lent its annotation to an unidentified spectrum in the Plasma dataset, and vice versa). To establish a confident peptide identification, we required that spectra exhibit a similarity to the cluster consensus with a p-value of 0.01 or less (see Supplementary Note 3 for more details on p-value computations).
Since samples come from very different tissues (kidney stem cells vs. plasma), there is modest overlap in protein identifications (only 40% of proteins identified in the Plasma dataset are also identified in the HEK dataset, see Table 2). However, by using the archive we were able to add a large number of identifications to both sets (an additional 954 peptide and 114 proteins to the HEK dataset and another 584 peptides and 207 proteins to the Plasma dataset). While it may appear counter-intuitive that using spectra from kidney stem cells boosts the number of protein identifications in a very different plasma sample by 15%, we remark that many “unrecognizable” spectra from plasma exhibit strong similarity to reliably identified spectra from kidney stem cells (and vice versa). In particular, many additional identifications belong to proteins that are differentially expressed between two samples (as evaluated by spectral counts).
Table 2.
Clustering HEK293 and Plasma datasets. The table shows the number of peptide and protein identifications made in each of the individual datasets, and the identifications made with the combined archive: the number of ids that were common to both datasets and the number that were added to each dataset because its spectra were joined into clusters with identified spectra from the other dataset.
| Num. peptides | Num. Proteins | |
|---|---|---|
| HEK 293 | 61,380 | 8,066 |
| Plasma | 18,473 | 1,498 |
| Common to both | 1,003 | 600 |
| Ids added to HEK using archive | 954 | 114 |
| Ids added to Plasma using archive | 584 | 207 |
Identification of Peptides Conserved Across Species
One of the benefits of spectral archives is that the additional information associated with the spectra can be used for more targeted analysis. If a cluster contains spectra from different organisms, we can assume that the corresponding peptide belongs to the intersection of their proteomes, a much smaller search space to consider. Since E-values of peptide identifications are proportional to the database size, a PSM that appears statistically insignificant in a standard MS/MS search often becomes statistically significant in a search against smaller intersection proteome (see12 for the relationship between the database size and FDR).
To test the effectiveness of using intersection proteomes, we analyzed the set of 14.5 million Shewanella oneidensis spectra (Sone) along with 0.95 million spectra from Shewanella frigidimarina (Sfri) and 0.77 million spectra from Shewanella putrefaciens CN-32 (Sput). We also created intersection databases for peptide sequences common to the six-frame translation of the genomes of Sone and Sfri, Sone and Sput and their intersection proteomes (see Supplementary Note 4).
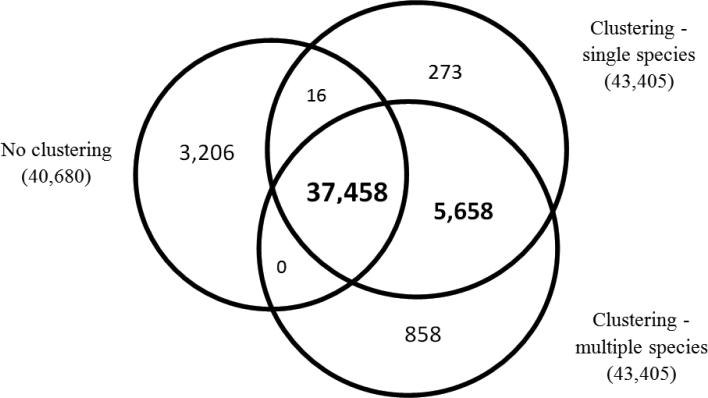
We evaluated three approaches of searching and identifying peptides from the Sone spectral dataset (Fig. 2). The first (“No clustering”) searched all (Sone) spectra against the Sone database. The second (“Clustering - single species”) clustered Sone spectra and searched all clusters against the Sone database. The third (“Clustering - multiple species”) clustered Sone spectra with Sfri and Sput spectra and searched all clusters containing Sone spectra against their appropriate database (clusters with only Sone spectra were searched against the Sone database, clusters with spectra from Sone and Sfri were searched against the intersection of Sone and Sfri, etc.)
Figure 2.
Identification of peptides across different species. The diagram compares the number of peptide identifications made with the Shewanella oneidensis (Sone) data using three methods: No clustering: standard MS/MS search, single-species clustering followed by MS/MS search, and multi-species clustering followed by MS/MS search. Results were processed to maintain a 2% FDR.
The most unique peptide identifications were made with the third method (43,974 vs. 43,405 with clustering and 40,680 with the regular database search). Interestingly, we observed a large difference between the set of peptides identified exclusively with and without clustering (3,206 vs. 5,931). This, again, illustrates the overall positive effect clustering has on the database search scores. Note that the 858 peptides that were identified only by the third method typically had borderline scores, which given the smaller search space of the intersection proteome became sufficient to make a database identification at a fixed peptide-level FDR of 2%.
Short Peptide Identification
Proteomes of every two species share many short peptides. While manual interpretation of short peptides is easier than interpretation of longer peptides, there is currently no software designed for sequencing short (5–6 amino acid) peptides. Moreover, it is not clear how one can evaluate the accuracy of identification of short peptides. In Supplementary Note 5 we discuss some of the problems encountered when sequencing short peptides and demonstrate that spectral archives can be utilized to bootstrap generation of a training set of spectra of short peptides, provide false discovery rates for their identifications, and enable confident identification of nearly 27,000 short peptides (4–6 amino acids), the largest currently available set of spectra of short peptides. Our dataset represents the first large spectral library for identification of short peptides and a training set for developing de novo sequencing approaches aimed at short peptides.
Discussion
The existing MS/MS data analysis methods are usually adequate for basic analysis purposes. Database searches have been extensively optimized for identifying unmodified peptides from databases of protein sequences. Spectral libraries have proven very useful for identifying previously observed peptides in samples from well studied tissues and organisms. However, there are scenarios in which existing methods fall short. We believe that spectral archives can be the cornerstone in the solution of some of these problems, paving the way for new spectral analysis methods.
Standard database searches are not well suited for identifying unexpected protein forms such as proteins with rare PTMs, mutations, alternative splicing variants, etc. Consequently, even when the sample comes from a well-annotated species, a majority of the spectra remain unidentified13. While in principle they might be detected using alternative identification methods such as “blind” searches for PTMs14, mutation-tolerant searches15–17, searches of six-frame translations of genomes18–21, de novo peptide sequencing22–26, spectral networks9, and even searches for fusion peptides27, in practice, it is difficult to perform such computationally-intensive analyses on a large-scale. The question arises, which of these billions of unidentified spectra are interesting candidates for further studying?
One way to single out interesting spectra is to examine large unidentified clusters that are more likely to represent highly expressed peptides (see Supplementary Note 6). Once we rule out the possibility that the spectra belong to a contaminant28, we can apply additional experimental approaches to identify the spectrum (e.g., different fragmentation techniques or multistage mass-spectrometry) or perform a targeted computational analysis (e.g., search for unexpected PTMs).
There are several ways the meta-information associated with the spectra can assist in reducing a large number of unidentified spectra in to a small list of interesting clusters (however, note that the potential applications outlined below are not implemented in the current version of MS-Cluster and require the user to perform additional processing steps as needed). In one scenario, we could take note of clusters that contain unidentified spectra from samples of related organisms; these might originate from an unannotated protein, or include spectra of a conserved peptide with an atypical post-translational modification. In a different scenario, we might have an archive with spectra generated from samples of both healthy and cancerous tissues. An interesting biomarker is one where there is a significant imbalance in the healthy/cancer cluster composition, compared to the ratio observed in the entire archive. Biomarkers could be detected by scanning all the clusters in the archive, whether they are identified or not (not unlike in MS-imaging studies where the identity of a biomarker is often unknown29), and detecting any cluster with a significant composition imbalance. In this way, we can extract a short list of “interesting” clusters that warrant a detailed examination. Note that since cancer often involves abnormal genomic events (mutations, translocations, etc.), these biomarker peptides are unlikely to be present in the protein database, and thus will not be identified in a traditional MS/MS database search.
While we advocate that publicly available MS/MS data should be organized in spectral archives to facilitate better sharing and exploration of the data, we acknowledge that there are limitations to the size of archives that can be generated by our method. Though we demonstrated the feasibility of creating archives for billions of spectra, an attempt to cluster a trillion spectra with MS-Cluster is likely to fail. Despite recent algorithmic advances in clustering30, we are not aware of any clustering projects (across all areas of science and engineering) that extend beyond a few billions data points (such as clustering all Internet pages). Two characteristics of MS-Cluster currently present bottlenecks for clustering larger datasets: quadratic running time and the limited discrimination power of the dot-product (as the dataset size increases, one needs a more stringent threshold for combining spectra into clusters). Therefore, when organizing MS/MS data in archives, it might be beneficial to focus on published datasets or datasets from repositories that are more likely to include higher quality spectra and have interesting meta-information.
While we propose spectral archives as a platform for processing data from various laboratories, our experimental data was generated at a single location (PNNL). This was mainly due to the logistics of acquiring such a huge dataset and specific programmatic needs at PNNL. However, we believe the fact that this data was generated over a long period (eight years) using different instrumental platforms and experimental protocols introduced comparable (or even greater) variance into the data compared to a scenario where the data was generated in different laboratories using similar experimental setups.
The MS-Cluster software for constructing spectral archives is freely available for academic and not-profit uses. The source code along with documentation and instructions for creating spectral archives is available for download from http://proteomics.ucsd.edu/Software/MSClustering.html.
We created a web-server that allows users to upload their own spectra and query the PNNL archive in order to retrieve the consensus spectra from the archive that are most similar to their query (along with information on the organisms that contributed the spectra to the retrieved clusters). Its URL is http://proteomics.ucsd.edu/LiveSearch.
Online Methods
Datasets and Database Search
We collected ~approx 1.18 billion MS/MS spectra from over 100 organisms (referred to as the PNNL dataset). This data set was compiled by pooling the ion trap spectra that have been generated at the Richard Smith laboratory at PNNL in 2001–2009. The data was mostly generated on LCQ, LTQ, LTQ-FT and LTQ-Orbitrap instruments. The data contained spectra acquired from samples of ~100 different organisms. The largest datasets came from human, mouse, Shewanella oneidensis. For a complete list of the datasets used in our experiments see Supplementary Table 1. Before clustering, the data was filtered to remove low quality spectra (~50% of the data).
Some of the experiments described below focused on a subset of the PNNL dataset, consisting of ~14.5 million spectra from Shewanella oneidensis MR-1, ~0.95 million spectra from Shewanella frigidimarina, and ~0.77 million spectra from Shewanella putrefaciens CN-32 previously analyzed in31,32 (henceforth referred to as Sone, Sfri, and Sput, respectively). In order to identify peptides in the Shewanella samples, we searched the spectra against their respective genomes along with a set of sequences of common contaminants.
We relied on the InsPecT database search tool33 for peptide identification (release 20081014, using the default search parameters (precursor mass tolerance 2.5 m/z units, fragment ion tolerance 0.5 m/z units). Searches were performed using a shuffled decoy database with 2% FDR.
Filtering Low-Quality Spectra
Not all mass spectra acquired by a mass spectrometer contain a detectable peptide signal that is likely to lead to peptide identification. Some spectra represent non-peptide materials or feature poor fragmentation making them “unidentifiable” by MS/MS database searches. These spectra often lack typical “peak ladders” (consecutive peaks with a mass differences corresponding to the masses of amino acids). Excluding such spectra from the analysis is often beneficial and can lead to increased identification rates and savings in running time34–38.
We developed a quality filtering approach that was originally designed for the PepNovo algorithm23. It is based on training logistic regression models for classification between “good” spectra (that contain a peptide signal) vs. “bad” (noisy spectra). Training spectra are collected from database search results against a database containing both target protein sequences and random decoy sequences; good spectra were selected from the high-scoring confident identifications and for bad spectra we used spectra whose best id was a low-scoring hit to the decoy database. Each spectrum gets converted to a feature vector with various attributes such as: Peak distribution features - number of peaks, number of strong peaks (e.g., ten times grass level), number of weak peaks (grass level), number of peaks in first half of spectrum, etc.; Peak intensity features - proportion of intensity in strong peaks, weak peaks (grass level); Peak ladder features - proportion of peaks that have other peaks that are an amino acid's mass distance, the longest ladder of peaks with amino acid mass differences; Complementary peak features - number of complementary b/y-ion pairs.
Note that grass level is selected so that two thirds of the peaks in the spectrum are above that level. These regression models achieve high classification rates (over 99% accuracy on the training data). In practice it is recommended to start with a low quality threshold to determine that only a small proportion of the good spectra are filtered out. Typically using a threshold of 0.05 filters out approximately 50% of the original set of acquired spectra. At this filtration level only about 2% of the identifiable spectra get filtered out. Note that even though some of the good spectra get thrown out, ultimately the fact that many low-quality spectra get excluded from the analysis often leads to an increase in the number of identification at a given FDR8.
Clustering Algorithm
While the clustering algorithm we used follows the algorithmic approach laid out in8, we had to completely rewrite it and modify its design to be able to process very large datasets while maintaining clustering quality. These changes enabled faster in-memory data processing, achieving 3× speedup compared to the older algorithm. More importantly, the new algorithm has reduced memory requirements and improved memory management, which extend the dataset size that can be processed from tens of millions to several billion spectra.
Our clustering algorithm involves an initial filtration step to remove low quality spectra using a regression model that relies on features that distinguish spectra of non-peptide material or poorly fragmented peptides (described above in “Filtering Low-Quality Spectra”). Typically 40%–55% of the spectra are discarded at this stage.
Following that, we use a bottom-up heuristic hierarchical clustering approach to join similar spectra that are likely to have originated from the same peptide. Each cluster of spectra is represented by a single consensus spectrum that contains the peaks shared by different spectra in the cluster (see below “Constructing Consensus Spectra” for more details on the creation of the consensus spectrum). Clustering reduces the number of spectra that need to be analyzed (yielding up to a tenfold reduction8) and results in “cleaner” consensus spectra with increased signal-to-noise ratio. Clustering often yields 5–10% more peptide identifications, at a given false discovery rate, compared to the number of peptides identified without using it8. See Supplementary Note 2 for more details about the running time of MS-Cluster.
We note that our current implementation focuses on ion-trap data. Using our algorithm with other types of instruments that display significantly different fragmentation patterns can require retraining of models for optimal results (namely spectrum quality models and empirical distributions of spectral similarities).
Spectral Similarity
In order to cluster mass spectra we need to determine the similarity between them. We use the normalized dot-product, which has previously been found to work well1,4,5,6,39–42. To calculate the normalized dot-product of two mass spectra S and S', we first reduce each spectrum to a vector. Since the computation of the spectral similarity is a major part of the clustering algorithm, restricting the dimensionality of these vectors can reduce the running time. To construct such vectors we first select the k highest intensity peaks from S and S' (we assume that S and S' have similar precursor masses). Joining these two sets of masses yields a set of masses M={m1,…,mt}, where k ≤t≤2k. M may contain less than 2k masses because the spectra may share some of their peaks which will produce duplicate masses and each pair of duplicate masses is only represented once (masses within 0.5 m/z units are considered duplicates). Finally, we reduce the spectrum S to a vector s=s1,…,stby assigning to each si the intensity found at mass mi in S if mi was one of the top k peaks in S, otherwise 0 is given to that position. Similarly, we fill s' using the intensities of the peaks in S'. In our experiments we found that for these similarity computations it is optimal to set k=(Precursor m/z)/50. Increasing k to larger values did not improve the performance of the similarity measure and in fact with larger values of k, we started to note a certain decrease in performance.
Once spectra S and S' are converted to vectors, their normalized dot-product is given by
The normalized dot-product takes values between 0 (when spectra do not share any selected peaks) and 1. Dot-products were initially used for measuring similarity between mass spectra of chemical compounds, whose mass spectra typically contain a small number of peaks1. Directly applying this measure to spectra of peptides can yield suboptimal results since a small number of strong peaks in the spectrum can dominate the outcome of the spectral similarity computation. Scaling peak intensities has been shown to improve the quality of the similarity computations1. One method that has been suggested is to scale a peak's intensity according to the square root of the intensity1,42,43. The scaling method we found most suitable for spectral clustering was to first normalize the peak intensities to bring the total spectrum's intensity to 1000 and then fill the dot-product vectors with 1+ln(si), where ln(si) is the natural logarithm of the selected peak i's normalized intensity.
Constructing Consensus Spectra
A common approach for creating a representative spectrum for a cluster is to use a consensus spectrum3–6,8,9,42, which is generated by aggregating the spectra in the cluster. Our method for creating a consensus spectrum involves several steps:
Spectra merging - Given the cluster's spectra, we create a single merged peak list for all the spectra, and sort the list according to the peaks' masses. The list is then scanned and when a pair of consecutive peaks in the list have a mass difference below a specified tolerance, the peaks are consolidated to a single peak with a mass that equals the weighted average of the joined peaks' masses and an intensity that equals the sum of the joined peaks' intensities. The two peaks are replaced by the new consolidated peak and the scanning process continues from the new peaks (this way during a single scanning multiple peaks can be consolidated into one). To increase the accuracy of the peak joining, the process is repeated several times with an increasing tolerance threshold (the final threshold we used was 0.33 m/z units). This is done to avoid erroneous peak merging due to isotopic peaks.
Intensity Normalization - To increase the signal-to-noise ratio in the spectrum, we take advantage of the fact that peaks corresponding to genuine fragments are likely to appear in many of the cluster's spectra. Thus for each peak i in the consensus spectrum, we take note of ni the number peaks from the original spectra that were merged to create peak i (each spectrum can contribute at most 1 to the count ni) and also to N, the total number of spectra in the cluster. We then compute the probability of pi observing ni peaks from a total of N spectra at random according to the binomial probability model (using a probability of observing a random peak of 0.1 which is a typical probability for low-resolution spectra). Each peak's intensity is then multiplied by p. This action has the effect of greatly reducing the intensity of spurious peaks that only appear in a small number of spectra.
Peak Filtering - After normalization, the peaks are scanned using a sliding window of width 200 m/z units, keeping the top ten peaks in each such window.
Along with the mass and intensity of each peak, the consensus spectrum also stores the number of spectra in which each peak appeared (ni) and the cluster size (N). Therefore, while maintaining spectral archives we do not need to store the peak lists of the individual spectra in each cluster, but we are still able to create consensus spectra by merging spectra and normalizing intensities as described above.
Creating and Updating Spectral Archives
Spectral archives are created by the MS-Cluster algorithm. An additional command line argument can be supplied to the program to instruct it to create an archive. When creating an archive, the program outputs the clusters in a binary file format in increments of 1 m/z unit (i.e., spectra with the same nominal precursor m/z value are stored in a single file). Storing the data this way makes it easier to process and search the archive. In addition, the program also outputs text based files for managing the archive. Thus, the time required to generate an archive is practically the same as the time required to cluster the dataset.
Spectral archives are intended to be continuously updated as additional spectra become available. Adding a new batch of spectra to the archive is done by first clustering the new data and creating an archive for it, and then merging the main archive with the newly generated one. Since these steps require the processing of all the data in the archive, it is more efficient to add spectra in large batches.
Besides storing peak information for the clusters, the archive can be used to store peptide identifications (for clusters that were searched against a database). This is done by adding text-based files that map clusters (using their unique cluster id) to peptide sequences and protein names. This identification information is added to the archive search results when query spectra are compared with the archive. If additional meta information is used, it can also be kept as text files along with the archive. Since peptide identifications and meta-information involve using outside resources and might have an arbitrary structure, the management and updating of this information is not performed by the clustering algorithm and is the responsibility of the user.
Constructing a Spectral archive of the PNNL dataset
Clustering was performed with default parameters: precursor m/z window of 2 m/z units, peak tolerance of ε=0.34 m/z units, and a probability for mixed clusters p=0.05 that gave a good tradeoff between cluster sizes and the number of peptide identifications that were obtained. A precursor m/z window of 2 m/z units forces that algorithm to consider every pair of spectra with precursor m/z values within 2 m/z units. This wide 2 m/z unit window was used to demonstrate the robustness of the algorithm even with low resolution precursor mass measurements. High-resolution precursor mass measurements would greatly reduce the number of spectra pairs that need to be compared, making the clustering faster and more accurate, and would also allow us to use lower similarity threshold for joining spectra. Another way to reduce the number of compared spectra is to consider the spectra's normalized elution times44 and only cluster together spectra with elution times that fall within a specified window.
Due to the large computational time involved in creating the archive for over one billion spectra, we relied on a standard Linux-based computational grid to perform this task. We split the 1.18 billion spectra in the PNNL dataset into 35 batches according to the precursor m/z of the spectra. Each batch was run on a single computing node of a large Linux-based computational grid at the Center for Computational Mass Spectrometry at UCSD (each node with at most 6 GB of RAM). The whole clustering job required approximately 430 CPU days to complete.
Supplementary Material
Acknowledgements
The authors would like to thank Ian Kaufman for his assistance in running the experiments on the computational grid. This work was supported by National Institutes of Health Grant 1-P41-RR024851 from the National Center for Research Resources. This work used measurements based upon capabilities developed by the Department of Energy, Office of Biological and Environmental Research, and National Center for Research Resources (grant RR18522) at conducted the Environmental Molecular Sciences Laboratory, a DOE/BER national scientific user facility located at Pacific Northwest National Laboratory in Richland, Washington.
References
- [1].Stein SE, Scott DR. Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Mass. Spectrom. 1994;5:859–866. doi: 10.1016/1044-0305(94)87009-8. [DOI] [PubMed] [Google Scholar]
- [2].Yates JR, III, Morgan SF, Gatlin CL, Griffin PR, Eng JK. Method to compare collision-induced dissociation spectra of peptides: Potential for library searching and subtractive analysis. Anal. Chem. 1998;70:3557–3565. doi: 10.1021/ac980122y. [DOI] [PubMed] [Google Scholar]
- [3].Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J. of Proteome Research. 2006;5:1843–1849. doi: 10.1021/pr0602085. [DOI] [PubMed] [Google Scholar]
- [4].Lam H, et al. Development and validation of a spectral library searching method for peptide identification from ms/ms. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- [5].Beer I, Barnea E, Ziv T, Admon A. Improving large-scale proteomics by clustering of mass spectrometry data. Proteomics. 2004;4:950–60. doi: 10.1002/pmic.200300652. [DOI] [PubMed] [Google Scholar]
- [6].Tabb DL, Thompson MR, Khalsa-Moyers G, VerBerkmoes NC, McDonald WH. MS2Grouper: Group assessment and synthetic replacement of duplicate proteomic tandem mass spectra. J. Am. Soc. Mass Spec. 2005;16:1250–1261. doi: 10.1016/j.jasms.2005.04.010. [DOI] [PubMed] [Google Scholar]
- [7].Flikka K, et al. Implementation and application of a versatile clustering tool for tandem mass spectrometry data. Proteomics. 2007;7:3245–3258. doi: 10.1002/pmic.200700160. [DOI] [PubMed] [Google Scholar]
- [8].Frank AM, et al. Clustering millions of tandem mass spectra. J. Proteome Res. 2008;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bandeira N, Tsur D, Frank A, Pevzner P. Protein identification by spectral networks analysis. PNAS. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [11].Tanner S, et al. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gupta N, Pevzner PA. False discovery rates of protein identifications: A strike against the two peptide rule. J. Proteome Research. 2009;8:4173–4181. doi: 10.1021/pr9004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Searle BC, Turner M, Nesvizhskii AI. Improving sensitivity by probabilistically combining results from multiple ms/ms search methodologies. J. Proteome Res. 2008;7:245–253. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- [14].Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications via blind search of mass-spectra. Nature Biotech. 2005;23:1562–2567. doi: 10.1038/nbt1168. [DOI] [PubMed] [Google Scholar]
- [15].Shevchenko A, et al. Charting the proteomes of organisms with unsequenced genomes by MALDI Quadrupole Time-of Flight Mass Spectrometry and BLAST homology searching. Anal. Chem. 2001;73:1917–1926. doi: 10.1021/ac0013709. [DOI] [PubMed] [Google Scholar]
- [16].Han Y, Ma B, Zhang K. SPIDER: software for protein identification from sequence tags with de novo sequencing error. J Bioinform. Comput. Biol. 2005;3:697–716. doi: 10.1142/s0219720005001247. [DOI] [PubMed] [Google Scholar]
- [17].Waridel P, et al. Sequence similarity-driven proteomics in organisms with unknown genomes by lc-ms/ms and automated de novo sequencing. Proteomics. 2007;7:2318–29. doi: 10.1002/pmic.200700003. [DOI] [PubMed] [Google Scholar]
- [18].Choudhary JS, Blackstock WP, Creasy DM, Cottrell JS. Matching peptide mass spectra to EST and genomic DNA databases. Trends Biotechnol. 2001;19:S17–S22. doi: 10.1016/S0167-7799(01)01795-4. [DOI] [PubMed] [Google Scholar]
- [19].Jaffe JD, Berg HC, Church GM. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- [20].Desiere F, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Siepel A, et al. Targeted discovery of novel human exons by comparative genomics. Genome Res. 2007;17:1763–73. doi: 10.1101/gr.7128207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ma B, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- [23].Frank A, Pevzner P. Pepnovo: De novo peptide sequencing via probabilistic network modeling. Anal. Chem. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- [24].Savitski MM, Nielsen ML, Kjeldsen F, Zubarev RA. Proteomics-grade de novo sequencing approach. J. Proteome Res. 2005;4:2348–2354. doi: 10.1021/pr050288x. [DOI] [PubMed] [Google Scholar]
- [25].Shen Y, et al. De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Anal. Chem. 2008;80:7742–7754. doi: 10.1021/ac801123p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim S, Gupta N, Bandeira N, Pevzner PA. Spectral dictionaries: Integrating de novo peptide sequencing with database search of tandem mass spectra. Mol. Cell. Proteomics. 2009;8:53–69. doi: 10.1074/mcp.M800103-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ng J, Pevzner PA. Algorithm for identification of fusion proteins via mass spectrometry. J. Proteome Res. 2008;7:89–95. doi: 10.1021/pr070214g. [DOI] [PubMed] [Google Scholar]
- [28].Junqueira M, et al. Separating the wheat from the chaff: Unbiased filtering of background tandem mass spectra improves protein identification. J. Proteome Res. 2008;7:3382–3395. doi: 10.1021/pr800140v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu B, et al. Identification of early intestinal neoplasia protein biomarkers using laser capture microdissection and MALDI MS. Mol. Cell. Proteomics. 2009;8:936–45. doi: 10.1074/mcp.M800345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Andoni A, Indyk P. Near-optimal hashing algorithms for approximate nearest neighbor in high dimensions. Commun. ACM. 2008;51(Special issue: Breakthrough Research):117–122. [Google Scholar]
- [31].Masselon C, et al. Targeted comparative proteomics by liquid chromatography-tandem fourier ion cyclotron resonance mass spectrometry. Anal. Chem. 2005;77:400–406. doi: 10.1021/ac049043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gupta N, et al. Whole proteome analysis of post-translational modifications: applications of massspectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tanner S, et al. Inspect: Fast and accurate identification of post-translationally modified peptides from tandem mass spectra. Anal. Chem. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- [34].Bern M, Goldberg D, McDonald WH, Yates JR., III Automatic Quality Assessment of Peptide Tandem Mass Spectra. Bioinformatics. 2004;20:i49–i54. doi: 10.1093/bioinformatics/bth947. [DOI] [PubMed] [Google Scholar]
- [35].Flikka K, Martens L, Vandekerckhove J, Gevaert K, Eidhammer I. Improving the reliability and throughput of mass spectrometry-based proteomics by spectrum quality filtering. Proteomics. 2006;6:2086–2094. doi: 10.1002/pmic.200500309. [DOI] [PubMed] [Google Scholar]
- [36].Nesvizhskii AI, et al. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol Cell Proteomics. 2006;5:652–670. doi: 10.1074/mcp.M500319-MCP200. [DOI] [PubMed] [Google Scholar]
- [37].Wong J, Sullivan M, Cartwright H, Cagney G. msmseval: tandem mass spectral quality assignment for high-throughput proteomics. BMC Bioinformatics. 2007;8:51. doi: 10.1186/1471-2105-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Salmi J, et al. Quality classification of tandem mass spectrometry data. Bioinformatics. 2007;22:400–406. doi: 10.1093/bioinformatics/bti829. [DOI] [PubMed] [Google Scholar]
- [39].Wan XK, Vidavsky I, Gross ML. Comparing similar spectra: from similarity index to spectral contrast angle. J. Am. Soc. Mass. Spectrom. 2002;13:85–88. doi: 10.1016/S1044-0305(01)00327-0. [DOI] [PubMed] [Google Scholar]
- [40].Tabb DL, MacCoss MJ, Wu CC, Anderson SD, Yates JR., III Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal. Chem. 2003;75:2470–2477. doi: 10.1021/ac026424o. [DOI] [PubMed] [Google Scholar]
- [41].Ramakrishnan SR, et al. A fast coarse filtering method for peptide identification by mass spectrometry. Bioinformatics. 2006;22:1524–1531. doi: 10.1093/bioinformatics/btl118. [DOI] [PubMed] [Google Scholar]
- [42].Liu J, et al. Methods for peptide identification by spectral comparison. Proteome Sci. 2007;5:3. doi: 10.1186/1477-5956-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Frewen FB, Merrihew GE, Wu CC, Stafford Noble W, MacCoss MJ. Analysis of peptide ms/ms spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- [44].Jaitly N, et al. Robust algorithm for alignment of liquid chromatography-mass spectrometry analyses in an accurate mass and time tag data analysis pipeline. Anal. Chem. 2006;78:7397–7409. doi: 10.1021/ac052197p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.