Abstract
Background
Primary gastric B-cell lymphomas arise from mucosa-associated lymphatic tissue (MALT) in patients with chronic Helicobacter pylori infection. We investigated whether germline variants in the CDH1 gene, coding for E-cadherin, genetically predispose patients to primary gastric B-cell lymphoma.
Design and Methods
Single marker analyses of the CDH1 gene were conducted in patients with primary gastric B-cell lymphoma (n=144), in patients with primary gastric high-grade lymphoma (n=61), and in healthy blood donors (n=361). Twelve single nucleotide polymorphisms were genotyped by TaqMan® technology. Allelic imbalance was tested by pyrosequencing and clone direct sequencing of heterozygote genomic and cDNA. Mutation detection was conducted around the poly-A signal of the CDH1 3′-untranslated region. The influence of the 3′-untranslated region on protein translation was determined by a luciferase reporter assay.
Results
Single marker analyses identified two single nucleotide polymorphisms in strong linkage disequilibrium located in the CDH1 3′-untranslated region. One of them was significantly associated with primary gastric diffuse large B-cell lymphomas after correction for multiple testing and this association was confirmed in an independent sample set. Patients homozygous for the rare T allele (rs1801026) had a 4.9-fold increased risk (95% CI: 1.5–15.9) of developing primary gastric diffuse large B-cell lymphoma. Allelic imbalance and reporter gene assays indicated a putative influence on mRNA stability and/or translational efficacy.
Conclusions
We identified variants in CDH1 as the first potential genetic risk factors for the development of primary gastric diffuse large B-cell lymphomas. One of the potentially causative variants affects allelic CDH1 expression. These findings support the hypothesis that besides somatic alterations of B-cells, germline variants in the CDH1 gene contribute to a predisposition to the development of primary gastric diffuse large B-cell lymphomas.
Keywords: lymphoma, CDH1, H. pylori, allelic imbalance, E-cadherin
Introduction
In 1984, Isaacson and Wright first described four cases of extranodal malignant lymphoma arising from mucosa-associated lymphoid tissue (MALT).1 Helicobacter pylori infection was found in 92% of extranodal marginal zone lymphomas of MALT type (MALT lymphomas). Isaacson and colleagues postulated that H. pylori infection provides the necessary background in which MALT lymphomas might develop.2 Originally, primary gastric diffuse large B-cell lymphomas (high-grade lymphomas) were considered to develop independently of H. pylori infection, but prospective studies provide evidence that a subset of early stage diffuse large B-cell lymphomas may also regress completely after H. pylori eradication.3–5 Fischbach6 and Cavanna et al.7 both reported and reviewed a number of cases of primary gastric diffuse large B-cell lymphomas successfully treated by H. pylori eradication and suggested that patients with primary gastric diffuse large B-cell lymphomas associated with H. pylori should first be managed with an anti-H. pylori treatment. In fact, primary gastric diffuse large B-cell lymphomas are often diagnosed in an advanced tumor stage and lead to death within a few months without treatment.8 In comparison to extranodal marginal zone lymphomas of MALT type (MALT lymphomas, low-grade lymphomas), primary gastric diffuse large B-cell tumors are characterized by more aggressive tumor growth and worse prognosis.9
Besides the virulence of different H. pylori strains, which does not seem to play a decisive role in the development of gastric lymphoma,10 the host genetic background influences the clinical course of H. pylori infection. In preceding studies, we were able to identify variants at TNF and NOD2 gene loci as genetic risk factors for primary extranodal marginal zone lymphoma of MALT type or gastric lymphoma in general,11–12 but a distinct genetic risk factor for primary gastric diffuse large B-cell lymphomas is lacking.
The CDH1 gene, coding for the calcium ion-dependent cell adhesion molecule E-cadherin, plays a pivotal role in epithelial integrity and carcinogenesis. Reduced expression of E-cadherin is regarded as one of the main molecular events triggering cancer invasion and metastasis.13–17 Inactivation is linked to different types of malignant tumors,18–27 especially hereditary diffuse gastric cancer.28–32 Primary gastric extranodal marginal zone lymphomas of MALT type and primary gastric diffuse large B-cell lymphomas express different patterns of adhesion molecules,33 and B-cell tumor growth and clinical aggressiveness are related to the adhesive capacities of the tumor cells.34 E-cadherin is expressed on lymphoma cells of patients with non-Hodgkin’s lymphoma35 and is released into the blood. Serum levels of E-cadherin are of prognostic significance in patients with multiple myeloma, another B-cell-associated hematologic malignancy.36
With the hypothesis that CDH1 could also have in important role in the etiopathogenesis of gastric lymphoma, we conducted a candidate gene association study in patients with primary gastric diffuse large B-cell lymphomas from a European multicenter study.
Design and Methods
Patients from a European multicenter study
We studied patients with gastric lymphoma (median age of 61 years, range 29–75 years; 56% males) participating in an intention-to-treat prospective multicenter study of the German-Austrian-Lymphoma Study Group.9 Caucasian patients with newly diagnosed primary gastric B-cell lymphoma were recruited from March 1993 through March 1996 at 166 centers in Germany and at 13 Austrian centers in this European multicenter study (EMCS). The exclusion criteria were age above 75 years, primary nodal or human immunodeficiency virus-associated lymphoma of any type, and prior or concomitant malignancies including gastric collision tumors. The primary gastric origin was defined histopathologically for low-grade MALT lymphomas and primary gastric diffuse large B-cell lymphomas with evidence of a low-grade component (secondary high-grade lymphoma). Patients with secondary high-grade lymphoma (n=9) were excluded from the analyses due to the ambiguous phenotype. Patients with diffuse large B-cell lymphoma (high-grade lymphoma) without low-grade features were regarded as having primary gastric lymphoma if they met the criteria described by Lewin.37 In all remaining 144 cases, the diagnosis was based on morphological and immunophenotypic analyses. The H. pylori status was assessed by the urease test on samples taken from the gastric antrum and corpus and histological examination of biopsy specimens. The work-up to determine disease stage included the patient’s history and physical examination, blood cell count and serum chemistry, inspection of Waldeyer’s tonsillar ring, chest and small bowel radiography, cervical and abdominal ultrasound, computed tomography of the chest and abdomen, bone-marrow aspirate and biopsy, ileocolonoscopy and upper gastrointestinal endoscopy with biopsies of visible lesions and macroscopically normal mucosa. The stage was defined according to the Ann Arbor staging system38 with modifications described by Musshoff39 and Radaszkiewicz40 (Online Supplementary Table S1).
Patients from the Lymph Node Registry in Kiel
The Lymph Node Registry in Kiel, founded in 1965, serves as a nationwide reference center for hemato-lymphatic diseases in Germany. Using the registry’s database, patients with a diagnosis of primary gastric diffuse large B-cell lymphoma according to the World Health Organization classification were identified retrospectively from 2005–2007. Biopsy samples of normal gastric mucosa, taken at the same time as the pathological biopsy samples, were acquired for 61 patients by contacting the relevant pathologists.
Healthy blood donor study population
The healthy blood donor study population was recruited in the clinical blood donation center of the University Hospital Schleswig-Holstein, Kiel, Germany by the PopGen biobank. Participants were allowed to donate blood if they were serologically negative for human immunodeficiency virus, hepatitis virus and cytomegalovirus and physical examination/history indicated no contraindications to blood donation (e.g. active infections or malignant disorders). Using these criteria, 361 Caucasian age- and sex-matched blood donors from Kiel were genotyped as healthy controls. Parts of the population have been used in other studies without any signs of systematic allelic bias due to the selection criteria.41–44
Data protection and ethics
The study was approved by the Data Protection Commission of Schleswig-Holstein and the ethical committee of the medical faculty of the Christian-Albrechts University of Kiel. Written informed consent was obtained from all individuals prior to enrollment. Personal information associated with medical information and blood samples was rendered anonymous.
Marker selection and genotyping
We prepared genomic DNA from 10 mL samples of fresh or frozen blood using the Blood Gigakit (Invitek, Berlin, Germany) or from paraffin-embedded non-neoplastic tissue using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturers’ recommendations (Lymph Node Registry sample set).
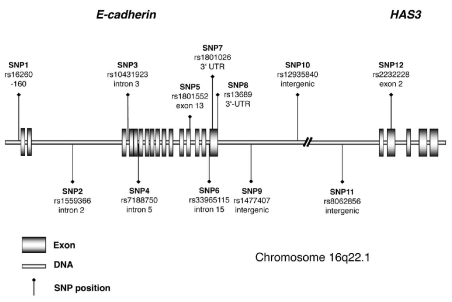
Twelve single nucleotide polymorphisms (SNPs) were selected with the intention of genetically covering the CDH1 locus and the downstream region. No high-density tagging information from the HapMap was available at the time of our studies, hence the 12 SNPs were selected based on other public resources (SNPBrowser from Applied Biosystems, Foster City, CA, USA). We also included the only HapMap-validated coding variant SNP 6 (rs1801552) within the CDH1 gene. The relative localization of the genotyped SNPs on chromosome 16q22.1 is illustrated in Figure 1 (for HapMap CEU LD structure and marker position see Online Supplementary Figure S1). All SNPs were genotyped using functionally tested TaqMan® assays provided by Applied Biosystems. The probe sequences are listed in Online Supplementary Table S2. In brief, genomic DNA was arrayed and dried on 96-well and 384-well microtiter plates. A TaqMan® polymerase chain reaction (PCR) was set up with Genesis pipetting robots (Tecan, Männedorf, Switzerland) and performed as described previously.45 We amplified samples with ABI9700 PCR machines (Applied Biosystems), and measured fluorescence with ABI7700 and ABI7900 fluorometers (Applied Biosystems). PCR cycling conditions were 50°C for 2 min, 95°C for 10 min, 45 cycles at 95°C for 10 s and 60°C for 1 min. All process data were written into and administered by a previously described database-driven laboratory information management system (LIMS).46 Duplicate or related samples were identified and excluded from the analyses, using algorithms implemented in the laboratory information management system. Case and control samples were genotyped on separate bar-coded plates. Visual inspection of the raw intensity data confirmed the absence of batch effects.
Figure 1.
Schematic gene-model of E-cadherin (CDH1) and neighboring HAS3 on chromosome 16q22.1 and relative position of the SNPs analyzed.
Statistical analysis
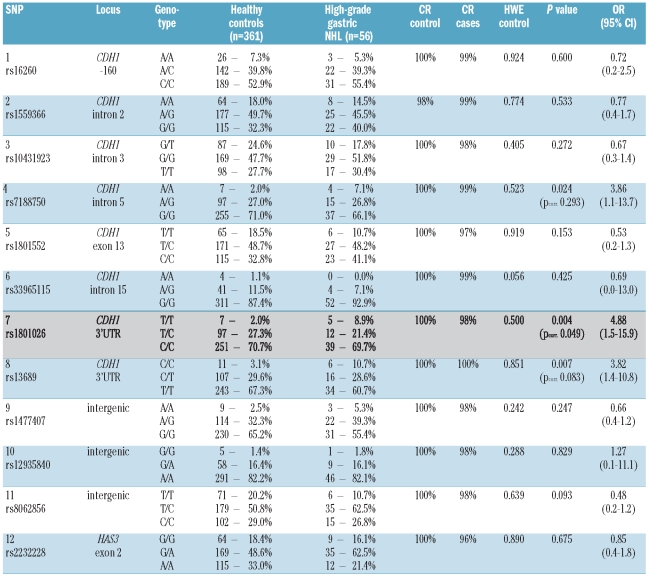
All SNPs had a high call rate (≥ 95% in cases or controls), were not monomorphic, and did not deviate from Hardy-Weinberg equilibrium in the control samples (P>0.05). The detailed call rates and Hardy-Weinberg equilibrium P values are given in Table 1. Two types of association analyses were performed: (i) patients with MALT lymphomas were compared against healthy blood donors; (ii) patients with primary gastric diffuse large B-cell lymphomas were compared against healthy blood donors.
Table 1.
Genotype counts and frequencies of all markers in patients with primary gastric diffuse large B-cell lymphomas (high-grade gastric lymphoma) from the European multicenter study (EMCS) and healthy blood donors. Alleles are coded according to the plus strand (5′→3′ orientation of CDH1). Differences between n per category and sum of genotype scores per category and SNP are due to missing genotypes (call rate ≥ 95%). P values calculated for carriership of the rare allele. Pcorr. is the P value adjusted for multiple testing using Bonferroni’s correction (n=12). OR: odds ratio; 95% CI: 95% confidence interval. HWE: Hardy-Weinberg equilibrium (Pearson), CR: call rate.
Statistical analyses were performed using the Hardy-Weinberg equilibrium web interface http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl for case-control studies. Pearson’s χ2 tests were used considering a recessive model based on relative genotype frequency distribution in the groups of patients and controls. Due to the potential multiple testing problem given the small sample size we only considered this hypothesis-driven and powerful (one degree of freedom) approach. Odds ratios and 95% confidence intervals were calculated for carriership of the rare allele using the above-described web interface. Linkage disequilibrium values, using the measure r2, were calculated using HAPMAX.47
Mutation detection
For mutation detection around the poly-A signal, primers were designed based on the CDH1 RefSeq NM_004360.3 and the downstream genomic sequence, provided by UCSC Genome Browser (http://genome.ucsc.edu) using ABI Primer Express 2.0 software (Applied Biosystems): forward primer: 5′-CTCAAAGC-CCAGAATCCCC-3′; reverse primer: 5′-ATGAGCCAC-CGCGCC-3′. The reaction contained 5 ng of genomic DNA and 50 μM of primer DNA in 50 mM salt and 1 mM Mg2+. A three-step PCR was performed for 35 cycles: denaturation at 94°C for 20 s, annealing at 55°C for 20 s and extension at 72°C for 30 s. Fluorescent sequencing of amplified fragments (640 bp) was carried out using a BigDye Cycle Sequencing system (Applied Biosystems). Products were analyzed on an ABIPRISM 3700 automated DNA sequencer (Applied Biosystems).
Isolation of complementary DNA
Total RNA was extracted from 7.5 mL peripheral blood and lymphoblastoid cell lines of individuals heterozygous for rs1801026 (SNP 7) using a PAXgene Blood RNA kit (Qiagen, Hilden, Germany). Ten microliters of water containing 1 μg of RNA together with 2.5 μL of RNasin plus RNase inhibitor (40 U/μL, Promega, Mannheim, Germany) and 5 μL of random hexamer primers (50 pmol/μL, Biotez, Berlin, Germany) were incubated for 10 min at 70°C. For the cDNA synthesis, 20 μL M-MLV reverse transcription buffer and 1 μL M-MLV reverse transcription enzyme (Promega), 5 μL dNTP (Amersham Biosciences, Buckinghamshire, UK) and 56.5 μL water were added to the RNA mix and incubated for 2 h at 42°C.
Amplification
PCR was performed using primers for CDH1 (CDHf: 5′-GCTTCAAGAAGCTGGCTGAC-3′; CDHr: 5′-TTTGGACAT-CACCACCATGT-3′) with 10 ng of cDNA or genomic DNA, respectively. Amplification was done in 96-well PCR plates (ABgene, Epsom, UK) containing 25 μL reaction mix/well using ReadyToGo PCR beads (Amersham Biosciences) according to the manufacturer’s recommendation. PCR primers were used as sequencing primers for the direct sequencing of PCR amplicons in Sanger sequencing using the dye terminator chemistry of ABI (BigDye). All primers were from Metabion (Munich, Germany). Individual PCR products were ligated by the topoisomerase reaction into pCR2.1-TOPO (Invitrogen, Karlsruhe, Germany) following the manufacturer’s recommendations. Aliquots of ligated samples were used for transformation of E. coli to obtain clones and as templates for PCR in order to prepare a biotinylated amplicon for pyrosequencing.
Cloning
Cells of E. coli strain TOP10 were transformed with the respective ligation products and plated onto LB-Amp agar. White colonies were picked and individually grown overnight at 37°C in 5 mL of LB broth supplemented with 100 μg/mL ampicillin. Plasmids were isolated and their insertion sequenced with vector primers M13f (5′-GTAAAACGACGGCCAG-3′) and M13r (5′-CAGGAAACAGCTATGAC-3′). Bases at SNP positions were called by manual inspection.
Pyrosequencing
Ligation products were diluted 1:100 and used as templates in a PCR with biotin-labeled primers (btM13r: 5′-CAGGAAACAGCTATGAC-3′; CDHfs: 5′-GTGCTGGGAAAT-GCAGAAATC-3′). PCR products were immobilized on streptavidin-coated paramagnetic beads (Dynabeads® M280, Dynal AS, Oslo, Norway) by mixing 20 μL of the PCR product with 10 μL Dynabeads (10 μg/μL) and 30 μL 2x BW buffer II (10 mM Tris-HCl, 2 M NaCl, 1 mM EDTA, 0.1% Tween 20, pH 7.6). The samples were incubated with shaking at 43°C for 30 min using a thermal mixer. For denaturation and preparation of single-stranded DNA, the samples were transferred into 50 μL NaOH (0.5 M, 1 min) using the multi-magnet PSQ96 Sample Prep Tool (Pyrosequencing AB, Uppsala, Sweden). The samples were washed in 100 μL 1x annealing buffer (20 mM Tris, 5 mM magnesium acetate, pH 7.6) for 1 min. Subsequently, the immobilized templates were transferred to 42 μL 1x annealing buffer plus 3 μL primer CDHfs (5 μM in water) and heated with continuous shaking on a thermal mixer at 80°C for 10 min. After equilibration to room temperature, the sequencing reaction was performed with a PSQ96 SNP reagent kit (Pyrosequencing AB), according to the manufacturer’s recommendation. The nucleotide dispensation order is illustrated in Figure 2.
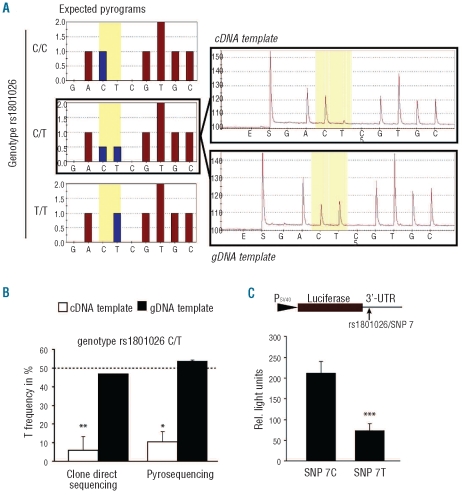
Figure 2.
SNP 7 (rs1801026) is associated with allelic imbalance of CDH1 mRNA expression and decreased allelic reporter gene activity. (A) Expected and observed pyrograms for SNP 7 for genomic DNA (gDNA) and complementary DNA (cDNA). In contrast to the expected pyrograms found in the gDNA template, the cDNA template revealed a pronounced allelic imbalance in rs1801026 heterozygous cells. (B) Allelic imbalance was confirmed by clone direct sequencing (see Design and Methods). The dashed line indicates the expected T frequency in heterozygous individuals (50%). (C) HeLa cells were transfected with luciferase reporter gene constructs containing the 3′-UTR of CDH1 driven by a constitutive SV40-promoter. Luciferase activities were determined by a dual luciferase assay (n=3). *P<0.05; **P<0.01; ***P<0.005.
All primers were from Metabion. All amplifications were done in 96-well PCR plates (ABgene) containing 25 μL reaction mix/well using ReadyToGo PCR beads (Amersham Biosciences) according to the manufacturer’s recommendations with the following conditions: 1 cycle of 94°C for 3 min, followed by 45 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 45 s and a final elongation step (72°C for 10 min).
Luciferase reporter gene assay
To analyze the effect of the SNP rs1801026 (SNP 7) on mRNA stability, the 3′-untranslated region (3′-UTR) of CDH1 was amplified and cloned directly to the 3′ side of the luciferase gene of the vector pGL3-control (Promega). SNP variants were introduced by site-directed mutagenesis following the instructions in the manufacturer’s manual (QuickChange site-directed mutagenesis kit; Stratagene, La Jolla, CA, USA). After sequence verification, plasmids were transfected into HeLa S3 cells by using FuGene6 (Roche). Twenty-four hours after transfection, cells were lysed and luciferase activity was measured by a dual luciferase assay (Promega) in a GeniosPRO plate reader (Tecan) and results normalized against pRL-TK (Clontech, Saint-Germain-en-Laye, France).
Results
Association analyses
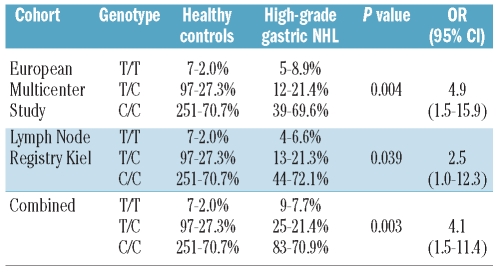
In total, 12 SNPs were genotyped across the CDH1 gene locus (8 SNPs) and the downstream region (Figure 1). Single marker analyses identified SNP 7 (rs1801026), located in the 3′-UTR, as the SNP significantly associated with primary gastric diffuse large B-cell lymphomas. Patients homozygous for the rare T allele of SNP 7 in the 3′-UTR had a 4.9-fold (95% CI: 1.5–15.9) increased risk of developing high-grade lymphoma (Pcorr.=0.049) (Table 1). This association signal was confirmed in the independent cohort of primary gastric diffuse large B-cell lymphomas from the Lymph Node Registry in Kiel (P=0.039, OR 3.5, 95% CI: 1.0–12.3) (Table 2) and findings were robust when corrected for multiple testing. SNP 8 (rs13689), a second marker in the 3′-UTR, was also associated with high-grade lymphomas (P=0.007, OR 3.8, CI 95%: 1.4–10.8), but the association did not remain statistically significant after correction for multiple testing. It should be pointed out that we used an overly conservative test to correct for multiple testing which shows, given the still significant findings, the robustness of our association findings. Bonferroni’s correction, the method employed, assumes that all observations are independent from another. This is certainly not the case as shown by the correlation between the different SNPs. In order to exclude any neighboring candidate gene in linkage disequilibrium, three intergenic SNPs located more telomeric of the 3′-UTR of CDH1 and one SNP in the neighboring hyaluronan synthase 3 (HAS3) gene (Figure 1) were genotyped, but did not show any association with lymphoma (Table 1). No significant associations were identified at any SNP between (low-grade) MALT lymphomas and healthy blood donors (Online Supplementary Table S3).
Table 2.
Genotype counts and frequencies of SNP 7 (rs1801026) in patients with primary gastric diffuse large B-cell lymphomas (high-grade gastric lymphoma) from the European multicenter study (EMCS) and the Lymph Node Registry and healthy controls. Alleles are coded according to the plus strand (5′→3′ orientation of CDH1). Single marker analysis identified rs1801026 (SNP 7) in the 3′-UTR associated with high-grade gastric lymphoma. Homozygosity for the rare allele was tested in cases and controls assuming a recessive model of inheritance. P values calculated for carriership of the rare allele. OR: odds ratio; 95% CI: 95% confidence interval.
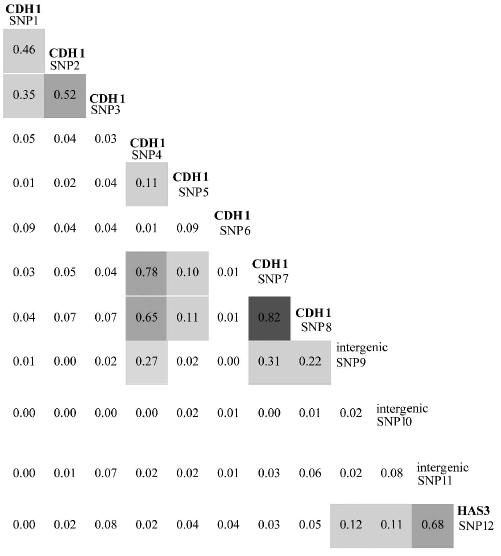
Pairwise linkage disequilibrium between SNP 7 and SNP 8 in the 3′-UTR was strong (r2=0.82). SNP 4 was also in moderate linkage disequilibrium with SNP 7 and 8 (r2=0.78 and 0.65, respectively), which explains its weak association with gastric diffuse large B-cell lymphomas in the single marker analysis. Linkage disequilibrium between other CDH1 SNPs and the 3′-neighbouring intergenic markers or the HAS3 SNP (rs2232228) was less than 0.5 (Table 3). The association signal is, therefore, most likely confined to the 3′-UTR of the CDH1 gene.
Table 3.
Pairwise linkage disequilibrium coefficient r2 of all markers calculated using HAPMAX. Darker gray shading indicates a higher linkage disequilibrium coefficient.
Mutation detection
We conducted mutation detection around the poly-A signal to identify a possible functional variant that could be in linkage disequilibrium with SNP 7 and 8. An amplicon of 640 bp was screened by genomic re-sequencing in 46 unrelated individuals (15 patients with low-grade MALT lymphoma, 15 patients with high-grade lymphoma and 16 H. pylori-infected patients) without detection of any new variants. We, therefore, performed further functional investigations of the strongest associated 3′-UTR SNP 7 (rs1801026).
mRNA instability/translational efficacy
To determine whether the lymphoma-associated SNP 7 (rs1801026) could potentially lead to reduced CDH1 mRNA stability, we investigated the balance of the C and T alleles in cDNA versus genomic DNA. Genomic DNA and total RNA extracted from a panel of lymphoblastoid cell lines and peripheral blood were isolated and mRNA was reverse-transcribed to cDNA using oligo(dT). We identified five heterozygous cDNA samples for rs1801026 (C/T), suitable for allelic expression analysis. Using two different methods, clone sequencing and pyrosequencing, we detected a significant allelic imbalance with an allelic mRNA ratio (T/C), which was consistent between the two methods 0.06±0.07 (by clone sequencing) and 0.1±0.05 (by pyrosequencing) (Figure 2). Control experiments with genomic DNA showed values close to the expected ratio of 0.5 (Figure 2). The putatively lower mRNA stability of the T allele containing 3′-UTR could be confirmed by a luciferase reporter gene assay. The 3′-UTR of CDH1 was cloned directly behind the open reading frame of a luciferase gene with a constitutive promoter (SV40). Comparison of the relative expressions/luciferase activities of the constructs containing T and C alleles showed that the normalized luciferase activity of the T allele-containing construct was 2.5 to 3-fold lower than that of the C allele (Figure 2), likely corresponding to reduced mRNA stability or translational efficacy and subsequent diminished protein levels compared to the C allele.
Discussion
H. pylori is the first formally recognized bacterial carcinogen and is one of the most successful human pathogens, as over half of the world’s population is colonized with this Gram-negative bacterium. H. pylori infection is a key factor in the etiology of various gastrointestinal diseases, ranging from chronic active gastritis without clinical symptoms to peptic ulceration, gastric adenocarcinoma, and gastric MALT lymphoma.48 Besides a chronic inflammatory response, down-regulation of the cell adhesion molecule E-cadherin can be found in gastric biopsy samples from H. pylori-infected stomachs.49 More precisely, H. pylori has been found to target adherens junction regulatory proteins resulting in increased rates of migration in human gastric epithelial cells. At the same time, E-cadherin is translocated from the plasma membrane to intracellular vesicles.50 CDH1 methylation was found in 85% of gastric cancer specimens and was associated with depth of tumor invasion and regional nodal metastasis. Since normal mucosa did not show CDH1 methylation, gene-silencing by pro-motor methylation is an early event in gastric carcinogenesis, and is initiated by H. pylori infection.51
In general, partial or total loss of CDH1 is associated with changes or loss of differentiation characteristics, acquisition of cell invasiveness, increased tumor grade, metastasis and poor prognosis52–56 and is linked specifically to hereditary diffuse gastric cancer.28–32 Given its suppressive effect on tumor cell invasion and metastasis, CDH1 is often termed a “metastasis suppressor” gene.55
The influence of CDH1 on the etiopathogenesis of lymphoproliferative disorders is less clear. It is tempting to speculate that loss of epithelial integrity per se - even in the absence of H. pylori - may contribute to malignant transformation of lymphocytic cells. However, the CDH1 gene was found to be expressed in a subset of human bone marrow mononuclear cells and plays a role in hematopoiesis.57 Hypermethylation of CDH1 is associated with leukemia.55 E-cadherin is expressed on lymphoma cells of patients with non-Hodgkin’s lymphoma35 and is released into the blood. Serum levels of (soluble) E-cadherin are of prognostic significance in patients with multiple myeloma, another B-cell-associated hematologic malignancy.36 Recently, it was shown that E-cadherin-expressing monocyte-derived inflammatory dendritic cells promote intestinal inflammation and are, therefore, putative therapeutic targets for the treatment of inflammatory bowel diseases.58
The CDH1 gene (OMIM 192090) maps to chromosome 16q22.1 and comprises 16 exons spanning approximately 100 kb of genomic DNA. Exon 16 spans 2269 bp and codes almost entirely for an unusually long 3′-UTR. In this study, we genotyped 12 SNP in order to cover the broad linkage disequilibrium structure of the region of CDH1 (Online Supplementary Figure S1) and identified two polymorphisms in high linkage disequilibrium in the 3′-UTR of the CDH1 gene as the first detected genetic risk factors for primary gastric diffuse large B-cell lymphomas using DNA isolated from blood and tissue samples. The genotyping results (SNP 7, Table 2) of the EMCS and the Lymph Node Registry sample sets showed very similar allele frequencies and, therefore, strongly argue for no differences at our genetic locus of interest between blood- or tissue-derived DNA. As with most other association findings, we cannot, of course, exclude other rare private variants also contributing to the disease risk at this locus. Meanwhile, several additional rare coding variants have been added to the dbSNP database, which were identified in the 1000 Genomes Project.59 Our functional in vitro experiments suggest that one of the identified variants (SNP 7) may affect mRNA stability. A simultaneous effect on mRNA levels with CDH1 promotor hypermethylation in vivo cannot be excluded. Lower CDH1 expression levels could be detected by immuohistochemistry in biopsies from patients with MALT lymphoma (C/C versus T/T; data not shown). Although the evidence for mRNA instability is suggestive, it cannot be completely excluded that the SNP also affects translational efficacy or contributes to differential nuclear RNA processing or export.
Many clinically relevant mRNAs are regulated by differential RNA stability,60 and the aberrant control of RNA stability has been implicated in the development of cancer and chronic inflammatory responses.61 AU-rich elements of the 3′-UTR have the ability to promote rapid deadenylation of mRNA transcripts. AU-rich elements are grouped into three classes according to their sequence features and decay characteristics.62 Whereas group I and II are characterized by the pentamer AUUUA and its reiteration, group III contains AU-rich regions lacking the hallmark AUUUA. Given the large sequence variation of AU-rich elements, a plethora of AU-rich element-binding proteins have been identified. Binding of these factors can have positive or negative effects on stability, translation and localization of the mRNA.61,63 The 3′-UTR of the CDH1 transcript is characterized by an abundance of AU-rich elements and one could speculate that the SNP identified may interfere with AU-rich element-binding proteins (Online Supplementary Table S4).
Annotation of SNP 7 (rs1801026) and SNP 8 (rs13689) in the genomic sequence of CDH1 shows that SNP 7 is located 141 bp upstream of the poly-A signal and SNP 8 918 bp downstream (Online Supplementary Table S4). Genetic variations in the vicinity of the poly-A signal may directly interfere with RNA-polymerase binding sites and thus impair the polyadenylation of the 3′ end of the mRNA.
Keirsebilck et al. investigated the influence of the 3′-UTR of CDH1 on gene expression in vivo.64 The CDH1−/− mouse mesenchymal tumor cell line MO4 was transfected with several plasmids expressing mouse CDH1 cDNA. These plasmids differed from each other by the extent of CDH1-specific 3′-UTR sequences. Transfectants were isolated which expressed functional E-cadherin in a homogeneous way. In syngenic mice, such MO4-Ecad transfectants invariably produced malignant fibrosarcoma-like tumors, which were completely E-cadherin-negative at the protein level. Northern blotting revealed that CDH1 mRNA expression was down-regulated in some but not all MO4-Ecad tumors. Down-regulation was caused by mRNA instability triggered by particular 3′-UTR sequences.64
As we demonstrated a significant association of SNP 7 homozygosity with gastric lymphoma (under a recessive model), it is conceivable that, despite the allelic imbalance of CDH1 expression in the heterozygous state, the mRNA levels of subjects carrying the wild-type allele are sufficient to compensate for the functional defect.
In conclusion, our findings support the hypothesis that besides somatic alterations of B cells, germline variation in the CDH1 gene contributes to a predisposition to primary gastric diffuse large B-cell lymphoma.
Acknowledgments
the authors would like to thank Tanja Kaacksteen, Melanie Schlapkohl, Yvonne Görlich, Ilona Urbach, Dorina Ölsner, Tanja Wesse and Rainer Vogler for their expert technical assistance. We also thank two anonymous reviewers for helpful comments.
Footnotes
Funding: the work was supported by the NGFN plus ‘Network on Environmental Disorders’, the Cluster of excellence ‘Inflammation at Interfaces’ and the DFG SFB877, as well as an internal grant from the University of S.H.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Isaacson P, Wright DH. Extranodal malignant lymphoma arising from mucosa-associated lymphoid tissue. Cancer. 1984;53 (11):2515–24. doi: 10.1002/1097-0142(19840601)53:11<2515::aid-cncr2820531125>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 2.Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338(8776):1175–6. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]
- 3.Chen LT, Lin JT, Tai JJ, Chen GH, Yeh HZ, Yang SS, et al. Long-term results of anti-Helicobacter pylori therapy in early-stage gastric high-grade transformed MALT lymphoma. J Natl Cancer Inst. 2005;97(18):1345–53. doi: 10.1093/jnci/dji277. [DOI] [PubMed] [Google Scholar]
- 4.Chen LT, Lin JT, Shyu RY, Jan CM, Chen CL, Chiang IP, et al. Prospective study of Helicobacter pylori eradication therapy in stage I(E) high-grade mucosa-associated lymphoid tissue lymphoma of the stomach. J Clin Oncol. 2001;19(22):4245–51. doi: 10.1200/JCO.2001.19.22.4245. [DOI] [PubMed] [Google Scholar]
- 5.Morgner A, Miehlke S, Fischbach W, Schmitt W, Muller-Hermelink H, Greiner A, et al. Complete remission of primary high-grade B-cell gastric lymphoma after cure of Helicobacter pylori infection. J Clin Oncol. 2001;19(7):2041–8. doi: 10.1200/JCO.2001.19.7.2041. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach W. Gastric mucosa-associated lymphoid tissue lymphoma: a challenge for endoscopy. Gastrointest Endosc. 2008;68 (4):632–4. doi: 10.1016/j.gie.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Cavanna L, Pagani R, Seghini P, Zangrandi A, Paties C. High grade B-cell gastric lymphoma with complete pathologic remission after eradication of Helicobacter pylori infection: report of a case and review of the literature. World J Surg Oncol. 2008;6:35. doi: 10.1186/1477-7819-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andriani A, Zullo A, Di Raimondo F, Patti C, Tedeschi L, Recine U, et al. Clinical and endoscopic presentation of primary gastric lymphoma: a multicentre study. Aliment Pharmacol Ther. 2006;23(6):721–6. doi: 10.1111/j.1365-2036.2006.02826.x. [DOI] [PubMed] [Google Scholar]
- 9.Fischbach W, Dragosics B, Kolve-Goebeler ME, Ohmann C, Greiner A, Yang Q, et al. Primary gastric B-cell lymphoma: results of a prospective multicenter study. The German-Austrian Gastrointestinal Lymphoma Study Group. Gastroenterology. 2000;119(5):1191–202. doi: 10.1053/gast.2000.19579. [DOI] [PubMed] [Google Scholar]
- 10.Lehours P, Menard A, Dupouy S, Bergey B, Richy F, Zerbib F, et al. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect Immun. 2004;72 (2):880–8. doi: 10.1128/IAI.72.2.880-888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmig S, Fischbach W, Goebeler-Kolve ME, Folsch UR, Hampe J, Schreiber S. A functional promotor polymorphism of TNF-alpha is associated with primary gastric B-cell lymphoma. Am J Gastroenterol. 2005;100(12):2644–9. doi: 10.1111/j.1572-0241.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 12.Rosenstiel P, Huse K, Till A, Hampe J, Hellmig S, Sina C, et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling path-ways. Proc Natl Acad Sci USA. 2006;103(9):3280–5. doi: 10.1073/pnas.0505423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Celebiler Cavusoglu A, Kilic Y, Saydam S, Canda T, Baskan Z, Sevinc AI, et al. Predicting invasive phenotype with CDH1, CDH13, CD44, and TIMP3 gene expression in primary breast cancer. Cancer Sci. 2009;100(12):2341–5. doi: 10.1111/j.1349-7006.2009.01333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo C, Liu QG, Yang W, Zhang ZL, Yao YM. Relation among p130Cas, E-cadherin and beta-catenin expression, clinicopathologic significance and prognosis in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2008;7(5):490–6. [PubMed] [Google Scholar]
- 15.Mareel M, Bracke M, Van Roy F. Cancer metastasis: negative regulation by an invasion-suppressor complex. Cancer Detect Prev. 1995;19(5):451–64. [PubMed] [Google Scholar]
- 16.Pena C, Garcia JM, Silva J, Garcia V, Rodriguez R, Alonso I, et al. E-cadherin and vitamin D receptor regulation by SNAIL and ZEB1 in colon cancer: clinicopathological correlations. Hum Mol Genet. 2005;14 (22):3361–70. doi: 10.1093/hmg/ddi366. [DOI] [PubMed] [Google Scholar]
- 17.Zhai B, Yan HX, Liu SQ, Chen L, Wu MC, Wang HY. Reduced expression of E-cadherin/catenin complex in hepatocellular carcinomas. World J Gastroenterol. 2008;14 (37):5665–73. doi: 10.3748/wjg.14.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinheiro H, Bordeira-Carrico R, Seixas S, Carvalho J, Senz J, Oliveira P, et al. Allele specific CDH1 down-regulation and hereditary diffuse gastric cancer. Hum Mol Genet. 2010;19(5):943–52. doi: 10.1093/hmg/ddp537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayrbaeurl B, Keller G, Schauer W, Burgstaller S, Czompo M, Hoebling W, et al. Germline mutation of the E-cadherin gene in three sibling cases with advanced gastric cancer: clinical consequences for the other family members. Eur J Gastroenterol Hepatol. 2010;22(3):306–10. doi: 10.1097/MEG.0b013e32832bab9a. [DOI] [PubMed] [Google Scholar]
- 20.Hebbard PC, Macmillan A, Huntsman D, Kaurah P, Carneiro F, Wen X, et al. Prophylactic total gastrectomy (PTG) for hereditary diffuse gastric cancer (HDGC): the Newfoundland experience with 23 patients. Ann Surg Oncol. 2009;16(7):1890–5. doi: 10.1245/s10434-009-0471-z. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira C, Senz J, Kaurah P, Pinheiro H, Sanges R, Haegert A, et al. Germline CDH1 deletions in hereditary diffuse gastric cancer families. Hum Mol Genet. 2009;18(9):1545–55. doi: 10.1093/hmg/ddp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caron O, Schielke A, Svrcek M, Flejou JF, Garzon J, Olschwang S, et al. Usefulness of prophylactic gastrectomy in a novel large hereditary diffuse gastric cancer (HDGC) family. Am J Gastroenterol. 2008;103(8):2160–1. doi: 10.1111/j.1572-0241.2008.01982_17.x. [DOI] [PubMed] [Google Scholar]
- 23.Richards FM, McKee SA, Rajpar MH, Cole TR, Evans DG, Jankowski JA, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum Mol Genet. 1999;8(4):607–10. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 24.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392(6674):402–5. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 25.Rocha AS, Soares P, Fonseca E, Cameselle-Teijeiro J, Oliveira MC, Sobrinho-Simoes M. E-cadherin loss rather than beta-catenin alterations is a common feature of poorly differentiated thyroid carcinomas. Histopathology. 2003;42(6):580–7. doi: 10.1046/j.1365-2559.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Ma X, Zhu QG, Li LC, Chen Z, Ye ZQ. Association between a C/A single nucleotide polymorphism of the E-cadherin gene promoter and transitional cell carcinoma of the bladder. J Urol. 2003;170(4 Pt 1):1379–82. doi: 10.1097/01.ju.0000084297.43710.e9. [DOI] [PubMed] [Google Scholar]
- 27.Tsukino H, Kuroda Y, Nakao H, Imai H, Inatomi H, Kohshi K, et al. E-cadherin gene polymorphism and risk of urothelial cancer. Cancer Lett. 2003;195(1):53–8. doi: 10.1016/s0304-3835(03)00130-7. [DOI] [PubMed] [Google Scholar]
- 28.Guilford P, Humar B, Blair V. Hereditary diffuse gastric cancer: translation of CDH1 germline mutations into clinical practice. Gastric Cancer. 2010;13(1):1–10. doi: 10.1007/s10120-009-0531-x. [DOI] [PubMed] [Google Scholar]
- 29.Guilford P, Blair V, More H, Humar B. A short guide to hereditary diffuse gastric cancer. Hered Cancer Clin Pract. 2007;5 (4):183–94. doi: 10.1186/1897-4287-5-4-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks-Wilson AR, Kaurah P, Suriano G, Leach S, Senz J, Grehan N, et al. Germline E-cadherin mutations in hereditary diffuse gastric cancer: assessment of 42 new families and review of genetic screening criteria. J Med Genet. 2004;41(7):508–17. doi: 10.1136/jmg.2004.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedrazzani C, Corso G, Marrelli D, Roviello F. E-cadherin and hereditary diffuse gastric cancer. Surgery. 2007;142(5):645–57. doi: 10.1016/j.surg.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Kaurah P, MacMillan A, Boyd N, Senz J, De Luca A, Chun N, et al. Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA. 2007;297(21):2360–72. doi: 10.1001/jama.297.21.2360. [DOI] [PubMed] [Google Scholar]
- 33.Sato Y, Ito M, Morise K, Saito Y, Kusugami K. Expression of adhesion molecules in primary B-cell gastric lymphoma and lymphoid follicles. Virchows Arch. 1996;429(6):377–82. doi: 10.1007/BF00198443. [DOI] [PubMed] [Google Scholar]
- 34.Jacob MC, Agrawal S, Chaperot L, Giroux C, Gressin R, Le Marc’Hadour F, et al. Quantification of cellular adhesion molecules on malignant B cells from non-Hodgkin’s lymphoma. Leukemia. 1999;13 (9):1428–33. doi: 10.1038/sj.leu.2401517. [DOI] [PubMed] [Google Scholar]
- 35.Takubo T, Kanashima H, Terada Y, Shibata H, Aoyama Y, Nakamae H, et al. E-cadherin expression in lymph nodes of three patients with non-Hodgkin’s lymphoma. Haematologia (Budap) 2002;32 (1):67–72. doi: 10.1163/156855902760262781. [DOI] [PubMed] [Google Scholar]
- 36.Syrigos KN, Harrington KJ, Karayiannakis AJ, Baibas N, Katirtzoglou N, Roussou P. Circulating soluble E-cadherin levels are of prognostic significance in patients with multiple myeloma. Anticancer Res. 2004;24 (3b):2027–31. [PubMed] [Google Scholar]
- 37.Lewin KJ, Ranchod M, Dorfman RF. Lymphomas of the gastrointestinal tract: a study of 117 cases presenting with gastrointestinal disease. Cancer. 1978;42(2):693–707. doi: 10.1002/1097-0142(197808)42:2<693::aid-cncr2820420241>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 38.Carbone PP, Kaplan HS, Musshoff K, Smithers DW, Tubiana M, Radaszkiewicz T, et al. Report of the Committee on Hodgkin’s Disease Staging Classification. Cancer Res. 1971;31(11):1860–1. [PubMed] [Google Scholar]
- 39.Musshoff K. Clinical staging classification of non-Hodgkin’s lymphomas (author’s transl) Strahlentherapie. 1977;153(4):218–21. [PubMed] [Google Scholar]
- 40.Radaszkiewicz T, Dragosics B, Bauer P. Gastrointestinal malignant lymphomas of the mucosa-associated lymphoid tissue: factors relevant to prognosis. Gastroenterology. 1992;102(5):1628–38. doi: 10.1016/0016-5085(92)91723-h. [DOI] [PubMed] [Google Scholar]
- 41.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40(6):713–5. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 42.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40(11):1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 43.Franke A, Balschun T, Sina C, Ellinghaus D, Hasler R, Mayr G, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat Genet. 2010;42(4):292–4. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 44.Karlsen TH, Franke A, Melum E, Kaser A, Hov JR, Balschun T, et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 2010;138(3):1102–11. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 45.Hampe J, Wollstein A, Lu T, Frevel HJ, Will M, Manaster C, et al. An integrated system for high throughput TaqMan based SNP genotyping. Bioinformatics. 2001;17(7):654–5. doi: 10.1093/bioinformatics/17.7.654. [DOI] [PubMed] [Google Scholar]
- 46.Teuber M, Koch WA, Manaster C, Wächter S, Hampe J, Schreiber S. Improving quality control and workflow management in high-throughput single-nucleotide polymorphism genotyping environments. 2005;10(1):43–7. [Google Scholar]
- 47.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 48.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terres AM, Pajares JM, O’Toole D, Ahern S, Kelleher D. H pylori infection is associated with downregulation of E-cadherin, a molecule involved in epithelial cell adhesion and proliferation control. J Clin Pathol. 1998;51(5):410–2. doi: 10.1136/jcp.51.5.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conlin VS, Curtis SB, Zhao Y, Moore ED, Smith VC, Meloche RM, et al. Helicobacter pylori infection targets adherens junction regulatory proteins and results in increased rates of migration in human gastric epithelial cells. Infect Immun. 2004;72(9):5181–92. doi: 10.1128/IAI.72.9.5181-5192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, et al. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52(4):502–6. doi: 10.1136/gut.52.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caldeira JR, Prando EC, Quevedo FC, Neto FA, Rainho CA, Rogatto SR. CDH1 promoter hypermethylation and E-cadherin protein expression in infiltrating breast cancer. BMC Cancer. 2006;6:48. doi: 10.1186/1471-2407-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lapyckyj L, Castillo LF, Matos ML, Gabrielli NM, Luthy IA, Vazquez-Levin MH. Expression analysis of epithelial cadherin and related proteins in IBH-6 and IBH-4 human breast cancer cell lines. J Cell Physiol. 2010;222(3):596–605. doi: 10.1002/jcp.21974. [DOI] [PubMed] [Google Scholar]
- 54.Margineanu E, Cotrutz CE, Cotrutz C. Correlation between E-cadherin abnormal expressions in different types of cancer and the process of metastasis. Rev Med Chir Soc Med Nat Iasi. 2008;112(2):432–6. [PubMed] [Google Scholar]
- 55.Melki JR, Vincent PC, Brown RD, Clark SJ. Hypermethylation of E-cadherin in leukemia. Blood. 2000;95(10):3208–13. [PubMed] [Google Scholar]
- 56.Paul R, Ewing CM, Jarrard DF, Isaacs WB. The cadherin cell-cell adhesion pathway in prostate cancer progression. Br J Urol. 1997;79 (Suppl 1):37–43. doi: 10.1111/j.1464-410x.1997.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 57.Armeanu S, Buhring HJ, Reuss-Borst M, Muller CA, Klein G. E-cadherin is functionally involved in the maturation of the erythroid lineage. J Cell Biol. 1995;131(1):243–9. doi: 10.1083/jcb.131.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siddiqui KR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32(4):557–67. doi: 10.1016/j.immuni.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467 (7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20 (11):465–70. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 61.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2(4):237–46. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 62.Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17(8):4611–21. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, et al. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14(5):571–83. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Keirsebilck A, Van Hoorde L, Gao Y, De Bruyne G, Bruyneel E, Vermassen P, et al. Mechanisms of downregulation of transfected E-cadherin cDNA during formation of invasive tumors in syngeneic mice. Invasion Metastasis. 1998;18(1):44–56. doi: 10.1159/000024498. [DOI] [PubMed] [Google Scholar]