Abstract
Set3 is one of two proteins in the yeast Saccharomyces cerevisiae that, like Drosophila Trithorax, contains both SET and PHD domains. We found that Set3 forms a single complex, Set3C, with Snt1, YIL112w, Sif2, Cpr1, and two putative histone deacetylases, Hos2 and NAD-dependent Hst1. Set3C includes NAD-dependent and independent deacetylase activities when assayed in vitro. Homology searches suggest that Set3C is the yeast analog of the mammalian HDAC3/SMRT complex. Set3C represses genes in early/middle of the yeast sporulation program, including the key meiotic regulators ime2 and ndt80. Whereas Hos2 is only found in Set3C, Hst1 is also present in a complex with Sum1, supporting previous characterizations of Hst1 and Sum1 as repressors of middle sporulation genes during vegetative growth. However, Hst1 is not required for meiotic repression by Set3C, thus implying that Set3C (−Hst1) and not Hst1–Sum1, is the meiotic-specific repressor of early/middle sporulation genes.
Keywords: SET domain, repression, sporulation, Hos2, Hst1, deacetylase
Genetic screens in Drosophila have identified several classes of chromatin regulatory proteins including factors involved in position effect variegation (PEV), and members of the Polycomb and trithorax Groups (Pc-G, trx-G) (Pirotta 1998; Wakimoto 1998). Clues as to the underlying mechanisms used by these genetically defined factors have arisen from the identification of several characteristic protein motifs including the SET domain. The SET domain was first identified by its occurrence in Su(var)3–9 (suppressor of position effect variegation 3–9), E(z) (Enhancer of zeste), and Trx (Trithorax) (Jones and Gelbart 1993; Tschiersch et al. 1994; Stassen et al. 1995). Functional data on SET domain proteins relate to chromatin regulation and, in certain cases, epigenetic mechanisms. Most prominently, Clr4, the Schizosaccharomyces pombe homolog of Su(var)3–9, is involved in epigenetic maintenance of centromeres in this yeast (Ekwall et al. 1996). The S. cerevisiae Set1 protein influences silencing at telomeres, and both Set1 and Clr4 influence silencing in mating-type switching in their respective yeasts (Nislow et al. 1997; Ivanova et al. 1998). Drosophila Trx and a mammalian homolog, Mll, play crucial roles in maintenance of homeotic complex gene expression patterns in development (Yu et al. 1995; Ingham 1998).
The SET domains in SU(VAR) 3–9 and Clr4 have been identified recently as histone methyltransferases acting on lysine 9 of histone H3 (Rea et al. 2000). In the same assay, the SET domains of E(z) or Trx homologs did not show histone methyltransferase activity, suggesting that they may act as methyltransferases on other substrates. Other roles for SET domains remain possible. The SET domains of Trx and/or MLL have been shown by two-hybrid and coimmunoprecipitation analyses to participate in several protein–protein interactions including interaction with (1) the SET domain of another trx-G member, Ash1 (Rozovskaia et al. 2000); (2) SBF1, a myotubularin related protein (Cui et al. 1998); and (3) the trx-G-related Swi–Snf complex proteins, INI1 and SNR1 (Rozenblatt-Rozen et al. 1998). Consequently, current data suggests that the SET domain encompasses at least two roles, including histone methyltransferase activity and protein–protein interactions with other factors involved in chromatin regulation.
A total of six genes containing SET domains were found in searches of the S. cerevisiae genome, including the previously studied set1 gene, and now termed set1 through set6 (Fig. 1). Interestingly, two of these six, set3 and set4, also encode a PHD finger. The PHD finger is another domain that was initially identified by its occurrence in Pc-G and trx-G members (Aasland et al. 1995) and is also found in a variety of chromatin regulators and transcriptional cofactors including CBP/p300, ATRX, DNMT3, and the TIF1/KAP-1 family (for review, see Pascual et al. 2000; Capili et al. 2001 for references and the recent determinations of the PHD finger structure). Furthermore, the coincidence of SET domains and PHD fingers is a characteristic displayed by the trx-G proteins, Trx and Ash1 (Stassen et al. 1995; Tripoulas et al. 1996) and their mammalian homologs and analogs, including the NSD/WHSC1 family (Huang et al. 1998; Stec et al. 1998; Angrand et al. 2001). Because there are only 14 genes in S. cerevisiae that encode PHD fingers (http://www.uib.no/aasland/set/), the coincidence of the SET and PHD domains in set3 and set4 was provocative and initiated the work described here on set3.
Figure 1.
SET domain proteins in S. cerevisiae. (A) Cartoons of the six yeast SET domain proteins identified. Each sequence was retrieved from SGD (http://genome-www.stanford.edu/Saccharomyces/) by the systematic yeast protein names given. The length of each protein and the position in each protein of the PHD fingers and SET domains are also indicated. Set5 and Set6 have large insertions (173 and 192 amino acids, respectively) indicated as light green boxes in the SET icon. E-values obtained when using the Set1 SET domain as a probe in a PSI-BLAST search (four iterations; Altschul et al. 1997) against the yeast proteome, are indicated. (B) A multiple alignment of the six SET domains in yeast. The alignment was generated with CLUSTAL_X (Thompson et al. 1997) and subsequently manually refined. The alignment is color coded to highlight conserved features using the color scheme implemented in CLUSTAL_X. The two positions in Set5 and Set6 with large insertions are indicated. Below the alignment is shown a consensus featuring positions conserved in at least five of the six sequences. The six positions in the alignment that correspond to the motif common to SET domains and methyltransferases (Rea et al. 2000) are indicated with red dots. The alignment includes the C-terminal SET domain-associated cysteine cluster, postSET (position 203–217 in the alignment).
After analyzing set3 in several conventional ways without significant insight, we applied proteomic techniques to Set3 (Rigaut et al. 1999; Shevchenko et al. 1999). The identified Set3 complex (Set3C) includes two potential histone deacetylases, Hos2 and Hst1. Hos2 is a class I histone deacetylase like Rpd3 and the mammalian HDACs 1–3 (Rundlett et al. 1996). Hst1 is a member of the recently identified Sir2 class of NAD-dependent deacetylases (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000) and has been linked previously to repression of sporulation genes (Xie et al. 1999; Lindgren et al. 2000). Here we identify a role for Set3C in repression of the sporulation gene program.
Results
Phenotypic analyses of Set3 disruption in haploid cells
Disruptions in haploid cells of set3, set4, or both together, resulted in normal viability with no obvious phenotype (data not shown). The previous finding that set1 influences silencing at telomere and mating type loci (Nislow et al. 1997) prompted similar tests of set3 disruptions on these processes. A moderate reduction in silencing of a telomeric ura3 (10-fold reduced growth on 5-fluoro-orotic acid plates), and no effect on silencing of a lacZ reporter gene integrated at HML, was found in Δset3 strains (data not shown). We also screened Δset3 strains for sensitivity to the alkylating agents MMS or EMS, UV irradiation, bleomycin, hydroxyurea, and heat, and found no viability differences with wild-type yeast (data not shown). Therefore, it is unlikely that Set3 has a direct role in silencing at telomeres and mating-type loci, DNA repair, or stress responses. To approach set3 function from a different angle, we examined the protein interaction partners of Set3.
Identification of Set3 protein–protein interactions
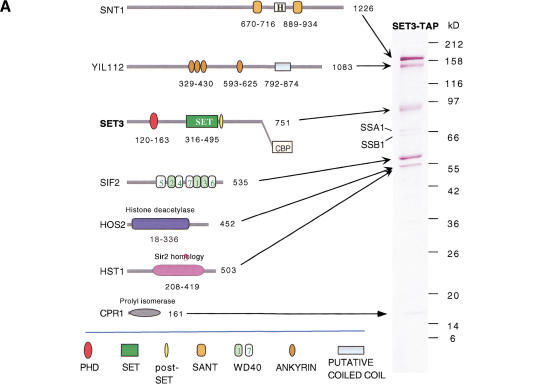
The TAP method (Rigaut et al. 1999) was used to determine protein–protein interactions with Set3. This method relies on fusion of the TAP tag, a double-specificity tag, to the protein of interest by knock-in to the endogenous coding sequence, followed by two rounds of affinity chromatography. All purified proteins were identified by mass spectrometry (MS; Shevchenko et al. 1996a,b). Purification of C-terminally tagged Set3 (Set3–TAP) resulted in specific copurification of seven proteins, now termed Set3C, which were not observed in purifications of wild-type or unrelated TAP-tagged strains (Fig. 2A; data not shown). In addition to Set3–TAP, the other six identified proteins were Snt1 (proposed name for YCR033w on the basis of the presence of two SANT domains; Aasland et al. 1996), YIL112w, Sif2 (SIR4 interacting factor 2; Cockell et al. 1998), Hos2 (Rundlett et al. 1996), Hst1 (Brachmann et al. 1995), and Cpr1 (cyclophilin A; for review, see Dolinski and Heitman 1997) as indicated in Figure 2A. The heat-shock proteins Ssa1 and Ssb1 were also present as minor bands, however, we consistently find these highly abundant proteins in wild-type control and unrelated purifications under the conditions used in this study, indicating that they are unspecific contaminants (data not shown). On the basis of Coomassie staining (Fig. 2A), we estimate relative stoicheiometries (indicated in brackets) as follows: Snt1 (2); YIL112w (1); Set3 (1); Sif2 (2); Hos2 (1); Hst1 (<1/4); and Cpr1 (1).
Figure 2.
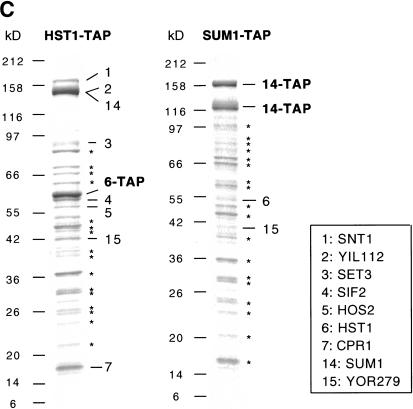
The Set3 protein complex. (A) Set3–TAP purification. Coomassie-stained 7%–25% polyacrylamide SDS gel showing the purified complex (Set3–TAP). Cartoons of MS-identified proteins are shown with full-length protein sizes in amino acids (aa). Protein domains are also indicated in amino acids. The tagged protein, Set3, is indicated in bold together with its C-terminal remnant of the TAP tag. Protein size markers (broad range, NEB) are indicated in kilodaltons. Ssa1 and Ssb1 are heat-shock proteins also found in mock purifications, and therefore represent unspecific contaminants. (B) Verification of the complex. As in A, showing the complexes obtained by TAP purification of Snt1, YIL112w, Sif2, or Hos2, as indicated. The tagged proteins with remaining TAP fusions are indicated in bold. TRiC complex in Hos2–TAP: 8, Cct6; 9, Cct8; 10, Cct2; 11, Cct3; 12, Cct4; 13, Cct7. (C) Defining a proteomic border of Set3C. As in A and B, showing TAP purification of Hst1 and Sum1, as indicated. The presence of many bands, marked by asterisks, was later found to be due to the batches of calmodulin beads used. All the bands marked by an asterisk were identified by MS, and are nonspecific contaminants, also visible in Figure 3. (They are, from top to bottom, in the Hst1–TAP panel: J04692; Ssa2: Ssb1: HSP60; ef1-α; enolase2; phosphoglycerate kinase; 60s ribosomal protein [rp] l4-B; fructose-biphosphate aldolase; glyceraldehyde 3-phosphate dehydrogenase; 60s rp l2; mixture of 40s rp s1-B, 60s rp l8-A, 40s rp s4; mixture of 60s rp l1, triosephosphate isomerase; mixture of 60 ns rp l7-A, 60 s rp l10; mixture of 40s rp s7-A, 60s rp l13-A; 60 s rp l20; and in the SUM1–TAP panel: mixture of sum1, ef2; hsp82; jo4692; sum1; ssa2; ssb2; pyruvate kinase 1; mixture of sum1, pyruvate decarboxylase isozyme; ef1α; mixture of enolase 2, 60s rp l3; 60s rp l4-A; glyceraldehyde3-phosphate dehydrogenase; 60s rpl2; mixture of 40s rp s1-B, 40s rp s4, 60s rp l8-B; mixture of 60s rp l7-B, 60s rp l10, 60s rp l1; mixture of 60s l13-B, 40s rp s7-A; 60s rp l20; mixture of 40s rp s24, 40s rp s17-A, 60s rpl25, 40s rp s19-B).
To verify the Set3 complex (Set3C) and to analyze further the interacting proteins of the other six Set3C components, the TAP tag was fused to the coding regions of each and the same protocols of purification and MS analysis were applied. The purifications of Snt1, YIL112, Sif2, and Hos2 yielded very similar results as the Set3–TAP purification (Fig. 2B). Along with additional observations, the results shown in Figure 2B are summarized as follows and establish that Set3C is composed of seven proteins: (1) Snt1–TAP purification yielded MS identifications of all proteins of Set3C. Relative stoicheiometries were similar as in Set3–TAP, and no new copurifying proteins could be detected, showing that endogenous Snt1 is present in Set3C and not in other complexes or as free protein. (2) YIL112w–TAP yielded all seven proteins of Set3C without any new copurified protein. Thus, YIL112w protein–protein interactions are limited to Set3C by this experimental criterion. Whereas the stoicheiometry of Set3:Sif2:Hos2:Hst1:Cpr1 was again estimated as 1:2:1:<1/4:1, comigration of YIL112w–TAP with Snt1 obscured the relative stoicheiometry of these two Set3C components. (3) TAP purification of Sif2 yielded the entire Set3C without any new copurifying proteins, but in this case, Sif2–TAP was present in clear excess over other members of Set3C. This indicates that endogenous Sif2 occurs as free protein as well as in the Set3C. Sif2 has been identified as a Sir4-interacting factor because of an interaction in a yeast two-hybrid assay (Cockell et al. 1998). We failed to detect the presence of Sir4 in Sif2–TAP purifications (Fig. 2B) or the presence of Sif2 in Sir4–TAP purifications (data not shown), suggesting that Sif2–Sir4 interaction might be transient and/or too lowly abundant to be detected by this method. (4) Hos2–TAP yielded the entire Set3C. In addition, the complete TRiC complex (Leroux and Hartl 2000) was identified. This may indicate that Hos2 is present in two complexes, Set3C and TRiC. More likely, the chaperone function of the TRiC complex may be involved in folding Hos2 or the Hos2–TAP fusion protein. (5) Importantly, TAP purification of the substoicheiometric component Hst1 yielded all seven proteins of Set3C (Fig. 2C), confirming the presence of Hst1 in Set3C. However, about fourfold more Hst1–TAP was retrieved than Set3C components and two additional proteins were copurified. Sum1 was copurified in approximately equal abundance to Hst1 along with the pioneer ORF protein, YOR279. Physical interaction between Hst1 and Sum1 has been proposed on the basis of similar behavior in a genetic screen for repressors of mid sporulation genes (Xie et al. 1999), and Hst1 and Sum1 have recently been coimmunoprecipitated (Rusché and Rine 2001). Consequently, we knocked the TAP tag into sum1, and retrieved Hst1 and YOR279, but not any other member of Set3C, or any other specific copurifying proteins (Fig. 2C). As opposed to approximately equal quantities of Sum1 and Hst1–TAP, Sum1–TAP was retrieved in greater abundance than Hst1. Also a specific shorter, 120-kD version of Sum1–TAP was observed that was not observed in the Hst1–TAP purification. These observations indicate that Sum1 exists partly complexed with Hst1 and partly as free protein, and suggests that an N-terminally shorter version of Sum1 does not associate with Hst1. We also conclude that Hst1 is present in two distinct complexes, Set3C and the Hst1–Sum1 complex. (6) TAP purification of Cpr1 yielded very high levels of Cpr1–TAP without apparent copurifying proteins (data not shown). As opposed to other members of Set3C, Cpr1 is a highly abundant protein. Despite the absence of other Set3C members in Cpr1–TAP purifications, the consistent presence of Cpr1 in TAP purifications of SET3, Snt1, YIL112w, Sif2, Hos2, and Hst1, but not wild-type control, Sum1, or unrelated purifications (Fig. 2B,C; data not shown) strongly implicates Cpr1 as a bona fide member of Set3C. Furthermore, Cpr1 is required for function of the Hos2-related histone deacetylase, Rpd3 (Arevalo-Rodriguez et al. 2000), suggesting a functional relationship between cyclophilins and Rpd3/Hos2-type histone deacetylases.
In summary, sequential TAP purification identified (1) the Set3C consisting of the following seven proteins: Snt1, YIL112, Set3, Sif2, Hos2, Hst1, and Cpr1; with all available data indicating an approximate stoicheiometric compostion of 2:1:1:2:1:<1/4:1, respectively; (2) the Hst1–Sum1 complex consisting of Hst1, Sum1, and YOR279; (3) free, uncomplexed Sif2; (4) free, uncomplexed Sum1; (5) free, uncomplexed Cpr1.
Requirements for Set3C integrity
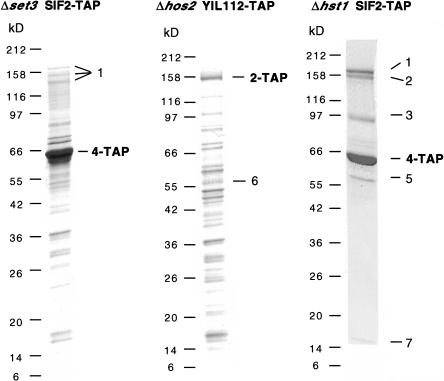
To dissect protein–protein interactions within Set3C, we constructed strains in which one complex member carried the TAP tag, whereas a part of the complex was deleted. Sif2–TAP purification in a Δset3 strain yielded Sif2–TAP and Snt1 without the other Set3C members (Fig. 3, left). Snt1 appeared partially degraded, indicating instability outside intact Set3C. This shows that Sif2 and Snt1 interact directly and require Set3 to interact with Hos2, YIL112w, Hst1, and Cpr1. Furthermore, TAP purification of a Set3–TAP allele in which the SET domain had been precisely deleted from the rest of the protein failed to retrieve anything. However, Western detection of the TAP tag on this Set3–ΔSET domain–TAP protein in total cell lysates yielded detectable but very low levels of expression (data not shown).
Figure 3.
Set3 and Hos2 are central components of the Set3C but Hst1 is not. Coomassie-stained polyacrylamide SDS gels showing TAP purifications of Sif2–TAP in a Δset3 background (left), YIL112–TAP in a Δhos2 background (middle), and Sif2–TAP in Δhst1 background (right). The tagged proteins with remaining TAP fusion are indicated in bold.
YIL112w–TAP purification in a Δhos2 background yielded YIL112w–TAP and Hst1, but no other Set3C members (Fig. 3, middle). This shows that YIL112w and Hst1 interact directly and require Hos2 to interact with Set3, Sif2, Snt1, and Cpr1.
In contrast, deletion of hst1 did not disturb Set3C purified from Sif2–TAP (Fig. 3, right). Thus, Hst1 is peripheral to Set3C. These results define Set3 and Hos2 as crucial members of Set3C required for complex integrity.
Enzyme activities in Set3C
The presence of a SET domain in Set3C suggested that it may include a histone methyltransferase activity. We failed to detect any such activity in Set3C in several assays, although TAP–Clr4 (expressed from a CEN plasmid and purified by the same methodology) and Set1, in the context of its complex, Set1C, proved to be very active (Roguev et al. 2001). Nevertheless, Set3C may be a methyltransferase when tested under different conditions.
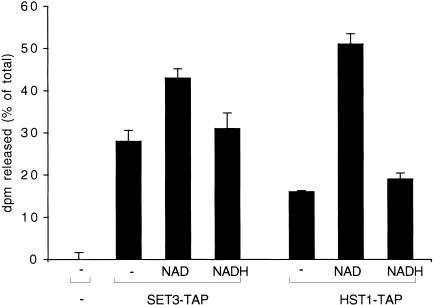
Set3C includes two predicted histone deacetylase enzyme activities. Hos2 is one of the five S. cerevisiae members of the first identified histone deacetylase family that includes Rpd3 and Hda1 (Rundlett et al. 1996). Hst1 is one of the five S. cerevisiae members of the second, NAD-dependent, histone deacetylase family that includes Sir2 (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). To determine whether Set3C contains histone deacetylase activity, we tested deacetylation of partially purified Set3–TAP or Hst1–TAP complexes with 3H-acetylated histone H4 peptide with and without NAD. Consistent with the relative stoicheiometries in Set3C (indicated in brackets) of Hos2 (1) and Hst1 (<1/4), the predominant deacetylase activity in the Set3–TAP eluate was NAD independent, and was moderately stimulated by NAD (Fig. 4).
Figure 4.
The Set3 and Hst1 complexes contain both NAD-independent and NAD-dependent histone deacetylase activities. IgG Sepharose eluates from the strains indicated were assayed for histone deacetylase activity as described in Materials and Methods. The presence or absence of 0.5 mM NAD or NADH in the reaction is also indicated. Data represent 3H release as a percentage of input, and are means ± SEM.
Application of the same assay to Hst1–TAP eluate, which contains both Set3C and the Hst1–Sum1 complex, showed, as expected, lower levels of NAD-independent and higher levels of NAD-stimulated histone deacetylase activity (Fig. 4). These results indicate that Hos2 in Set3C and Hst1 in both Set3C and the Hst1–Sum1 complexes are active histone deacetylases.
Set3C is a repressor of meiosis
Diploid Δset3 and Δhos2 strains were tested for meiotic landmarks during sporulation. Figure 5A shows that kinetics of premeiotic DNA replication, as measured by FACS (fluorescence activated cell sorter) analysis, were unchanged in Δset3 and Δhos2 strains as compared with wild type. Therefore, Set3C is unlikely to be involved in the regulation of early meiotic events such as initiation of meiosis or premeiotic DNA synthesis. In contrast, both Δset3 and Δhos2 strains underwent faster progression through meiosis I, meiosis II, and ascus formation than wild type, and resulted in reduced levels of viable ascus formation, as determined by morphological and tetrad analyses (Fig. 5B; data not shown). Thus, Δset3 and Δhos2 phenotypes are very similar, consistent with the proteomic analysis of Set3C, and indicate a role for Set3C in middle sporulation.
Figure 5.
Set3C is a repressor of meiosis. Wild type, Δset3, or Δhos2 diploid strains were sporulated and assayed for meiotic landmarks at the times indicated (in hours). (A) Premeiotic DNA synthesis as measured by FACS analysis. X-axis, fluorescence intensity; Y-axis, number of cells. (Left peaks) 2n DNA; (right peaks) 4n DNA (as confirmed by microscopic analysis of sorted cells). (Blue graphs) wild type; (orange graphs) Δset3; (green graphs) Δhos2. (B) Cells completing meiosis I, meiosis II, and ascus formation (Asci) were counted using a fluorescence microscope. At least 300 cells per time point per strain were counted. (C) Northern blot analysis of meiotic genes during sporulation of wild-type (wt) or Δset3 cells. Data were obtained from a separate experiment as in A and B, and the absolute time of sporulation onset is therefore slightly different. Relative differences between wild type and Δset3 are similar to those in B. Genes examined are indicated next to the expression data, as are corresponding gene clusters (Primig et al. 2000) and gene functions. The mRNA coding for the ribosomal protein Tcm1 was used as control for RNA. Decreased expression of this gene very early during sporulation, followed by stable expression levels, is as reported (Chu and Herskowitz 1998). Expression of asc1 and dal7 confirmed the quality of the RNA at these early time points. (D) As in C, except RNA was harvested from wild-type, Δhst1, and Δhst1,Δhos2 strains.
DNA microarray analysis has led to the clustering of genes induced during sporulation based on the timing of their expression (Chu et al. 1998; Primig et al. 2000). To determine whether disruption of Set3C affected sporulation-specific gene expression, we performed Northern blot analysis of representative members of these clusters and of known key regulators of sporulation during a sporulation time course (Fig. 5C). Genes in clusters 2 (acs1) and 3 (dal7), which have amino acid and other metabolic functions, had unchanged expression levels in Δset3 cells. Expression of the cluster 4 gene rfa1, which functions in DNA synthesis, repair and recombination, was only slightly increased in Δset3 cells. In contrast, expression of spo11, which also belongs to cluster 4 but functions in chromosome synapsis and segregation, was precociously induced in Δset3, reaching higher levels than in wild-type cells. The cluster 5 genes spr3, sps2, and sps18, which function in cytokinesis and differentiation, were all expressed earlier and at higher levels in Δset3.
Analysis of key regulators of sporulation revealed that ime1, which is required for early meiotic gene expression including ime2 and spo11 (Mitchell et al. 1990), was expressed at slightly higher levels in Δset3 cells. In contrast, ime2, which is involved in both early and middle/late gene expression including spo11 and sps2 (Mitchell et al. 1990), was expressed earlier and at higher levels in Δset3, as was ndt80, a regulator of middle/late genes including itself, spr3, sps2, and sps18 (Chu and Herskowitz 1998). Notably, none of the genes repressed by Set3C in sporulation were up-regulated at the zero time point, which is representative of gene expression during vegetative growth. Hence, we identify Set3C as a meiotic specific repressor of cluster 4 and 5 genes of the sporulation program.
Deletion of hst1 did not relieve repression of ime2 or ndt80, whereas deletion of hos2 in the absence of or presence of hst1 did (Fig. 5D; data not shown). These results parallel the effects on Set3C of set3, hos2, and hst1 deletions. Both Set3 and Hos2 are required for Set3C integrity, whereas Hst1 is not (Fig. 3). Thus, in early sporulation, Set3C represses two key regulators of the sporulation gene program, ime2 and ndt80, and Hst1 is not required for Set3C formation or function.
Discussion
Protein interactions of Set3
A proteomic approach based on sequential TAP tagging and purification was used to define the protein interactions of Set3 and all associated factors to define the proteomic environment of Set3C (Figs. 1–3; schematized in Fig. 6). Set3 occurs in a single complex, Set3C, consisting of Snt1 (2), YIL112w (1), Set3 (1), Sif2 (2), Hos2 (1), Hst1 (<1/4), and Cpr1 (1) (estimated stoicheiometries in brackets). The relatively low amount of Hst1 in this complex suggests that it is transiently interacting with Set3C. Furthermore, Hst1 is also present in a complex with Sum1 and YOR279 (Hst1–Sum1 complex). TAP purification of Sum1 did not copurify Set3C. Vice versa, Sum1 was not detected in TAP purifications of core Set3C members. Thus, Set3C and the Hst1–Sum1 complex are distinct entities. However, we cannot exclude the possibility that these complexes interact, and that a Sum1–Set3C interaction is too weak to be detected by the approach used. We also conclude that Snt1, YIL112w, Set3, and Hos2 are predominantly included in Set3C and are not present as free proteins or in other stable protein complexes, whereas Sif2, Cpr1, and Sum1 do exist as free proteins in addition to their involvement in the observed complexes.
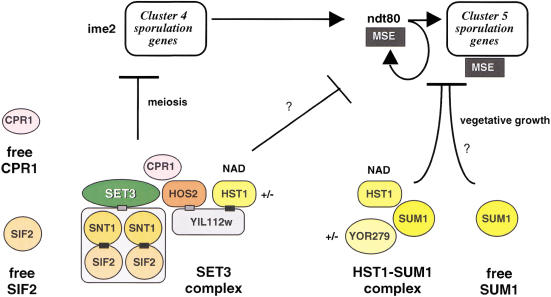
Figure 6.
The proteomic environment of Set3. The proteomic environment of Set3, associated factors, and their regulation of part of the sporulation program is presented. The proteins involved are shown as colored objects below the regulatory interactions. Question marks indicate uncertainties in the regulation circuitry. Note that Set3C repression applies in meiosis and Hst1–Sum1 repression in vegetative growth. (MSE) Middle sporulation element. The Set3C includes two molecules of each Snt1 and Sif2 in a tetramer. Protein interactions in Set3C are shown as black boxes, probable interactions as gray boxes. The +/− beside Hst1 and YOR279 indicates the likelihood that these proteins are not always present in their respective complexes. Those proteins that are also found free are indicated, as is the relationship between Hst1 and NAD.
In addition to the deacetylase domains and the SET domain and PHD finger of Set3, Set3C includes several other identifiable protein motifs. The SANT domain was identified by its occurrence in chromatin proteins including Swi3, Ada2, NcoR, and TFIIIB“ (Aasland et al. 1996). It is distantly related to the DNA-binding domain of c-myb and may act in nonsequence-specific DNA binding by TFIIIB“ (Kumar et al. 1997). Thus, Snt1 may be a DNA-binding component of Set3C. Sif2 is a seven WD40-repeat propeller protein. Such proteins participate in many cellular functions, however, TBL1 [transducin (β)-like I], Groucho, and TUP1 have been shown to bind histones (Edmonson et al. 1996; Palaparti et al. 1997; Guenther et al. 2000). Hence, Sif2 may contribute nucleosomal binding activity to Set3C. It is interesting to note that Snt1 and Sif2 interact (Fig. 3) and are the two members of Set3C that are present twice each. Hence, they may present a tetrameric chromatin interaction module for Set3C (indicated as a box in Fig. 6). YIL112w contains four ankyrin repeats, which have been shown previously to participate in a variety of protein–protein interactions (Sedgwick and Smerdon 1999). The ankyrin repeats of YIL112w and/or its coiled-coil region may be involved in the direct interaction with Hst1 and/or the implied interaction with Hos2.
Biochemical activities of Set3C and probable conservation in eukaryotes
Although certain SET domain proteins including Su(var)3–9 have histone methyltransferase activity (Rea et al. 2000), we were unable to detect this activity in Set3C. However, Set3 is required for Set3C (Fig. 3), indicating a structural role in complex integrity.
Set3C combines both NAD-dependent and independent histone deacetylase activities (Fig. 4). In concordance with other studies on histone deacetylases (Rundlett et al. 1996; Hassig et al. 1998), we find that the activities associated with Set3C and Hst1–Sum1 complex deacetylate several histone substrates in vitro with no apparent substrate specificity (data not shown). Furthermore, Hst1 is dispensible for Set3C integrity and function in meiotic repression of early/middle sporulation genes (Figs. 3C, 5D). Consequently, the functional significance of combining the two types of histone deacetylase activities in the same complex remains to be determined.
Set3C shows interesting similarities to the partially characterized mammalian HDAC3 complexes (Guenther et al. 2000; Li et al. 2000). In addition to the fact that Hos2 is the closest yeast homolog of HDAC3, complexes from HeLa cells include the corepressors SMRT and NCoR, which like Snt1 are SANT domain proteins. The HeLa complexes also include the seven WD40-repeat protein, TBL1. Among the many seven WD40-repeat proteins in mammals and yeast, Sif2 is clearly the yeast homolog of TBL1. We speculate that further characterizations of HDAC3 complexes may present more similarities, including, possibly, the inclusion in HDAC3 complexes of an NAD-dependent histone deacetylase and the uncharacterized SET and PHD domain protein predicted by EST 095038, which is the closest human homolog of Set3.
Function of the Set3C and the Hst1–Sum1 complex
Examination of the kinetics of sporulation landmarks revealed that Δhos2 or Δset3 strains underwent normal premeiotic DNA synthesis but showed a faster progression through meiosis. Deletion of hos2 has been reported to reduce ascus formation dramatically (Bisland et al. 1998). We found only a slight reduction in Δhos2 strains, both in the strain of Bisland et al. (1998) and the strain used throughout our studies under various conditions (Fig. 5; data not shown). Presumably, loss of hos2 has a more profound impact on ascus production under other conditions. In concordance with the fast meiotic phenotypes shown by Δhos2 or Δset3, loss of either protein disrupts Set3C, leaving subcomponents (Fig. 3) that did not retain Set3C function. Hence, repression by Set3C may require both the essential components and the integrity of the complex.
Northern analysis showed that Set3C function is directed toward repression of clusters 4 and 5 of the sporulation gene program, including the key regulators ime2 and ndt80. Set3C repression of ndt80 could be indirect, as ime2 is required for expression of ndt80, but not vice versa (Hepworth et al. 1998). Hence, the fast meiotic phenotypes shown by Δhos2 or Δset3, and the precocious induction of ndt80 and other later genes, could be solely due to earlier Set3C repression of ime2 and other cluster 4 genes.
Consequently, an attractive explanation of repression by Set3C action involves Hos2 deacetylation of nucleosomes near the ime2 gene. This presumes that a change early after induction of sporulation, which could include altered acetylation of these nucleosomes, potentially activates ime2 gene expression. However, action by Set3C delays the onset of expression so that the regulatory cascade controlled by Ime2 is delayed. This delay may serve to improve sporulation efficiency, and/or to integrate a further regulatory checkpoint. This model requires a mechanism to abrogate Set3C repression in middle sporulation. Studies to test aspects of this model are underway, however, biochemical analysis of Set3C during sporulation showed no apparent changes in Set3C composition between vegetative and sporulation conditions, indicating that regulation of Set3C activity does not involve loss or gain of proteins bound in Set3C (data not shown).
Although Hst1 is a part of Set3C, it is not required for the function of Set3C described here. Rather, Hst1 represses middle sporulation genes in vegetative growth, probably through the Hst1–Sum1 complex (Xie et al. 1999; Fig. 2C). Hst1/Sum1 repression operates through Sum1 binding the MSE (middle sporulation element) DNA sequence element, which is present in the cluster 5 genes spr3, sps2, sps18, and the key regulator ndt80 (Chu and Herskowitz 1998). Why Hst1 is also present in Set3C is not clear. Because both Set3C and the Hst1–Sum1 complex act as repressors, appear to include histone deacetylase activities (Fig. 4), and repress overlapping sets of sporulation-specific genes, it is very likely that the presence of Hst1 in Set3C has functional significance. As with the questions regarding targets and mechanism of Set3C repression discussed above, further work is required to define the events involved in regulation of Set3C and Hst1/Sum1 repression.
Methodological approaches in proteomics
Ongoing discussions about proteomics have focused on the methodological options available (Rigaut et al. 1999; Shevchenko et al. 1999; Vidal 2001), with global yeast two-hybrid approaches assuming prominence. We scrutinized all available two-hybrid data, including global analyses (Uetz et al. 2000; Ito et al. 2001) for interactions between any of the factors encompassed here. Only an interaction between Sif2 and YIL112w was reported (in both studies; Uetz et al. 2000; Ito et al. 2001). This interaction was not detected by us with either TAP-tagged Sif2 or YIL112 in the absence of Set3 or Hos2, respectively (Fig. 3), suggesting that the published two-hybrid results relied on the presence of endongenous Set3 and Hos2. Ito et al. (2001) also reported interaction between Sif2–YJR141w, which could not be confirmed by our biochemical analysis. Furthermore, we failed to detect any evidence for the yeast two-hybrid interaction between Sif2 and Sir4 (Cockell et al. 1998). The lack of concurrence between our results and yeast two-hybrid results provokes questions about the fidelity and utility of two-hybrid analyses (Seol et al. 2001). We think that coherent documentation and understanding of proteomes must be based on core data sets composed of strong, biochemically documented, interactions. Mapping of two-hybrid data onto these core data sets may sort out true from false positives and point to higher-order regulatory circuits.
Materials and methods
Sequence analysis
The yeast genome was searched for SET domains and PHD fingers by BLASTP searches (Altschul et al. 1997) and Profilesearch (as implemented on the Bioccelerator facility at EMBL; http://eta.embl-heidelberg.de:8000/) using profiles based on alignments of previously identified SET domains and PHD fingers (http://www.uib.no/aasland/set/).
Strains
Strains are listed in Table 1. Yeast transformations were performed as described (Soni et al. 1993). All haploid strains were derived from MGD353-13D (Puig et al. 1998). The TAP tag was introduced at the endogenous locus of the gene of interest as an in-frame C-terminal fusion with the coding sequence as described (Puig et al. 1998). Gene disruptions were performed as described (Puig et al. 1998). The diploid strains Δset3 and Δhos2 were obtained by crossing to BSY 323 (Matα genotype) followed by tetrad analysis. Resulting haploids carrying the desired disruption were mated. Correct modifications were confirmed by PCR and Western blot analysis. The double mutant Δhst1 Δhos2 (DS140) was obtained by integrating a K.l. ura3 disruption cassette flanked by loxP sites. Transient expression of Cre-recombinase resulted in cassette excision. The hos2 ORF was then disrupted by inserting a cassette, again using K.l. ura3 as selectable marker.
Table 1.
S. cerevisiae strains used in this study
| Name
|
Genotype G
|
Strains
|
Reference
|
|---|---|---|---|
| MGD-35313D | Mata | ade2; arg4 (RV−), leu2-3, 112; trp1-289l ura3-52 | Puig et al. 1998 |
| BSY320 | Mata/α | ade2/ade2; arg4 (RV−)/arg4 (RV−); leu2-3,112/leu2-3, 112; trp1-289; ura3-52/ura3-52 | Puig et al. 1998 |
| BSY323 | Mat α | ade2; leu2-3,112; lys2;trp1-289; ura3-52 | This study |
| BSY792 | Mata | set3 C-term. TAP tagged introducing K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS4 | Mata | yill 12w C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS6 | Mata | snt1 C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS8 | Mata | sif2 C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS9 | Mata | hos2 C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS22 | Mata | cpr1 C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| AR5 | Mata | hst1 C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS101 | Mata | sum1 C-term. TAP K.l. TRP1 ade2; arg4 (RV−); leu2-3,112; ura3-52 | This study |
| DS16 | Mata | set3∷ K.l. URA3 ade2; arg4 (RV−); leu2-3,112; trp1-289 | This study |
| DS18 | Mata | hos2∷ K.l. URA3 ade2; arg4 (RV−); leu2-3,112; trp1-289 | This study |
| DS19 | Mata | yil112w C-term. TAP K.l. TRP1; hos2∷K.l.URA3 ade2; arg4 (RV−); leu2-3,112 | This study |
| DS21 | Mata | sif2 C-term. TAP K.l. TRP1; set3∷K.l.URA3 ade2; arg4 (RV−); leu2-3,112 | This study |
| DS169 | Mata | sif2 C-term. TAP K.l. TRP1; hst1∷K.l.URA3 ade2; arg4 (RV−); leu2-3,112 | This study |
| DS52-1 | Mata/α | set3∷ K.l. URA3/set3∷K.l.URA3 ade2/ade2; ARG4/arg4 (RV−); leu2-3,112/leu2-3,112; lys2/LYS2; trp1-289/trp1-289 | This study |
| DS53-3 | Mata/α | hos2∷ K.l. URA3/hos2∷K.l.URA3 ade2/ade2; ARG4/arg4 (RV−); leu2-3,112/leu2-3,112; lys2/LYS2; trp1-289/trp1-289 | This study |
| DS129 | Mata/α | Hst1∷floxed K.l. URA3/hst1∷floxedK.l.URA3 ade2/ade2; ARG4/arg4 (RV−); leu2-3,112/leu2-3,112; lys2/LYS2; trp1-289/trp1-289 | This study |
| DS140 | Mata/α | hst1∷loxP/hst1∷loxP hos2∷K.l. URA3/hos2∷K.l.URA3 ade2/ade2; ARG4/arg4 (RV−); leu2-3,112/leu2-3,112; lys2/LYS2; trp1-289/trp1-289 | This study |
TAP purification and mass spectrometry
The extraction of yeast cells was performed as described for the yeast Swi/Snf complex (Logie and Peterson 1999). The TAP tag consists of a calmodulin binding peptide (CBP), a TEV protease cleavage site, and two IgG-binding units of protein A, respectively, as described (Rigaut et al. 1999). TAP purification was performed according to Rigaut et al. (1999), with the following modifications: 10 mL of supernatant of the 43,000 rpm centrifugation (Logie and Peterson 1999) was allowed to bind to 200 μL of IgG Sepharose (Pharmacia), equilibrated in buffer E (Logie and Peterson 1999) for 2 h at 4°C by use of a Bio-Rad disposable column. Two to three columns (the equivalent of 4–6 L yeast culture at OD600 2–3) were used per purification shown. The IgG Sepharose column was washed with 35 mL of buffer E lacking proteinase inhibitors, followed by 10 mL of TEV cleavage buffer (Rigaut et al. 1999). TEV cleavage was performed using 10 μL of rTEV (GIBCO) in 1 mL of TEV cleavage buffer for 2 h at 16°C. Calmodulin Sepharose (Stratagene) purification was performed as described (Rigaut et al. 1999). Purified proteins were concentrated as described by Wessel and Flügge (1984). After separation on 7%–25% SDS–polyacrylamide gels, proteins were stained with Coomassie, in-gel digested with trypsin, and identified by MS as described (Shevchenko et al. 1996a,b).
Histone deacetylase assay
Yeast Histone H4 peptide (amino acids 2–20, Upstate) was chemically acetylated by use of sodium [3H]acetate (NEN, 2.8 Ci/mmole) as described (Taunton et al. 1996), and purified by gel filtration over G15 Sepharose. IgG Sepharose eluates were prepared as above, and were added to 30,000 dpm 3H H4 peptide in a final volume of 30 μL 10 mM Tris (pH 8.0), 150 mM NaCl, 0.1 μg/μL BSA, and 0.5 mM NAD or NADH (Sigma) as indicated. After incubation at 30°C for 90 min, the reaction was spotted on P81 phosphocellulose paper (Whatmann). Filters were washed four times for 15 min in 50 mM NaHCO3 (pH 9), dried completely, and counted in 2 mL of scintillation cocktail.
Sporulation assays
Diploid strains were grown in YPD to OD600 of 2.0. Cells were harvested by centrifugation, and resuspended in sporulation medium (1.5% KAc at pH 7.5, dilution 1:3) supplemented with required amino acids. Cultures were incubated at 23°C, and samples were taken at the times indicated. DNA was stained with propidium iodide as described (Haase and Lew 1997). Cells were sonicated three times for 2 sec with 5-sec intervals at 20% power to resolve cell clumps. DNA synthesis was measured by FACS analysis. Microscopic analysis showed that this treatment did not disrupt individual cells. Progression into meiosis was determined by counting at least 300 cells per point using a fluorescent microscope. Northern blot analysis was performed by use of standard procedures. Probes were prepared by random prime labeling of PCR fragments of the genes indicated.
Acknowledgments
We thank Colin Logie, Henk Stunnenberg, Alexander Brehm, Peter Becker, Jeanette Rientjes, Elisabeth Bragado-Nilsson, Wolfgang Zachariae, Anne Atzberger, Susan Gasser, and Moira Cockell for reagents, advice, and discussions; Joep Muyrers and Julia Schaft for auxiliary experiments; and Michelle Meredyth for critical reading of the manuscript. W.P. was supported by an EU Biotechnology fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stewart@mpi-cbg.de; FAX 49-351-210-1409.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.207401.
References
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Aasland R, Stewart AF, Gibson T. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrand P-O, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- Arevalo-Rodriguez M, Cardenas ME, Wu X, Hanes SD, Heitman J. Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 2000;19:3739–3749. doi: 10.1093/emboj/19.14.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisland E, Dahlén M, Sunnerhagen P. Genomic disruption of six budding yeast genes gives one drastic example of phenotype strain-dependence. Yeast. 1998;14:655–664. doi: 10.1002/(SICI)1097-0061(199805)14:7<655::AID-YEA257>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Capili AD, Schultz DC, Rauscher IF, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Cockell M, Renauld H, Watt P, Gasser SM. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr Biol. 1998;8:787–790. doi: 10.1016/s0960-9822(98)70304-5. [DOI] [PubMed] [Google Scholar]
- Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- Dolinski K, Heitman J. Peptidyl-prolyl isomerases (PPIases) In: Tooze S, editor. Guidebook to molecular chaperones and protein folding catalysts. Oxford, UK: Oxford University Press; 1997. pp. 359–369. [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes & Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Lew DJ. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 1997;283:322–332. doi: 10.1016/s0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Trithorax and the regulation of homeotic gene expression in Drosophila: A historical perspective. Int J Dev Biol. 1998;42:423–429. [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kassavetis GA, Geiduschek EP, Hambalko M, Brent CJ. Functional dissection of the B‘ component of RNA polymerase III transcription factor IIIB: A scaffolding protein with multiple roles in assembly and initiation of transcription. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux MR, Hartl FU. Protein folding: Versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol. 2000;10:R260–R264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4250. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren A, Bungard D, Pierce M, Xie J, Vershon A, Winter E. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 2000;19:6489–6497. doi: 10.1093/emboj/19.23.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logie C, Peterson CL. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 1999;304:726–741. doi: 10.1016/s0076-6879(99)04044-6. [DOI] [PubMed] [Google Scholar]
- Mitchell AP, Driscoll SE, Smith HE. Positive control of sporulation genes by the Ime1 and Ime2 products in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2104–2110. doi: 10.1128/mcb.10.5.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Puig O, Rutz B, Luukkonen BG, Kandels-Lewis S, Bragado-Nilsson E, Seraphin B. New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast. 1998;14:1139–1146. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1139::AID-YEA306>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Roguev, A., Schaft, D., Shevchenko, A., Pijnappel, W.W.M., Wilm, M., Aasland, R. and Stewart, A.F. 2001. The S. cerevisiae Set1 complex includes an Ash2 homolog and methylates histone 3 lysine 4. EMBO J. (in press). [DOI] [PMC free article] [PubMed]
- Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce CM, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben-Simchon L, Croce CM, Mazo A, et al. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché LN, Rine J. Conversion of a gene-specific repressor to a regional silencer. Genes & Dev. 2001;15:955–967. doi: 10.1101/gad.873601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Zachariae W, Shevchenko A. A strategy for the characterization of protein interaction networks by mass spectrometry. Biochem Soc Trans. 1999;27:549–554. doi: 10.1042/bst0270549. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996a;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov V, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci. 1996b;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Murray JA. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr Genet. 1993;24:455–459. doi: 10.1007/BF00351857. [DOI] [PubMed] [Google Scholar]
- Stassen MJ, Bailey D, Nelson S, Chinwalla V, Harte PJ. The Drosophila trithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech Dev. 1995;52:209–223. doi: 10.1016/0925-4773(95)00402-m. [DOI] [PubMed] [Google Scholar]
- Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, Altherr MR, den Dunnen JT. WHSC1, a 90 kb SET domain-containing gene maps in the Wolf-Hirschorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripoulas N, LaJeunesse D, Gildea J, Shearn A. The Drosophila ash1 gene product, which is localized at specific sites on polytene chromosomes, contains a SET domain and a PHD finger. Genetics. 1996;143:913–928. doi: 10.1093/genetics/143.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Vidal M. A biological atlas of functional maps. Cell. 2001;104:333–339. doi: 10.1016/s0092-8674(01)00221-5. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]