Abstract
Objective
Uncovering the dominant molecular deregulation among the multitude of pathways implicated in aggressive prostate cancer is essential to intelligently developing targeted therapies. Paradoxically, published prostate cancer gene expression signatures of poor prognosis share little overlap and thus do not reveal shared mechanisms. The authors hypothesize that, by analyzing gene signatures with quantitative models of protein–protein interactions, key pathways will be elucidated and shown to be shared.
Design
The authors statistically prioritized common interactors between established cancer genes and genes from each prostate cancer signature of poor prognosis independently via a previously validated single protein analysis of network (SPAN) methodology. Additionally, they computationally identified pathways among the aggregated interactors across signatures and validated them using a similarity metric and patient survival.
Measurement
Using an information-theoretic metric, the authors assessed the mechanistic similarity of the interactor signature. Its prognostic ability was assessed in an independent cohort of 198 patients with high-Gleason prostate cancer using Kaplan–Meier analysis.
Results
Of the 13 prostate cancer signatures that were evaluated, eight interacted significantly with established cancer genes (false discovery rate <5%) and generated a 42-gene interactor signature that showed the highest mechanistic similarity (p<0.0001). Via parameter-free unsupervised classification, the interactor signature dichotomized the independent prostate cancer cohort with a significant survival difference (p=0.009). Interpretation of the network not only recapitulated phosphatidylinositol-3 kinase/NF-κB signaling, but also highlighted less well established relevant pathways such as the Janus kinase 2 cascade.
Conclusions
SPAN methodolgy provides a robust means of abstracting disparate prostate cancer gene expression signatures into clinically useful, prioritized pathways as well as useful mechanistic pathways.
Keywords: Prostate cancer, protein networks, systems biology, information theory, network modeling, Simulation of complex systems (at all levels: molecules to work groups to organizations), knowledge representations, Uncertain reasoning and decision theory, languages and computational methods, statistical analysis of large datasets, advanced algorithms, discovery and text and data mining methods, Natural-language processing, Automated learning, Ontologies
Introduction
Gene signatures provide a glimpse into critical molecular pathways, as they essentially serve as a bridge between clinical phenotypes and genomics. As defined by Richard Simon, ‘a multigene expression signature classifier is a function that provides a classification of a tumor based on the expression levels of the component genes. The classes are often good-risk or poor-risk, but classifiers can be defined to distinguish any set of classes for which a training set of cases exist for each class.1’ These signatures have traditionally been derived by examining the differential expression of mRNA from discrete cancer states such as tumor versus normal tissue or high-grade versus low-grade tumors. Beginning over a decade ago with the identification of poor-risk breast cancer gene sets,2 3 these gene signatures have rapidly proliferated to the point where nearly 1000 entries exist in a gene signature database established to catalog them.4 Surprisingly, despite their proliferation, few of these signatures have been commercialized and adopted by the medical community. In the USA, only one product in breast cancer, OncotypeDX, has achieved widespread adoption5; however, newer tests such as a ‘tumor of origin’ assay6 for cancers of unknown primary may gain in popularity. In contrast, biomarkers such as prostate-specific antigen (PSA) in prostate cancer, HER2/Neu in breast cancer, and epidermal growth factor receptor (EGFR) in colon cancer have enjoyed rapid usage among practitioners with a multitude of clinical trials.
Indeed, the vast majority of biomarkers are functionally and biologically understood, in stark contrast with gene signatures. Moreover, biomarkers tend to be single-pathway-specific, whereas gene signatures may span multiple mechanisms. To add to the confusion, genes constituting distinct signatures are rarely shared among gene signatures even though they paradoxically occupy a common prognostic space.7 Their similar efficiency in predicting poor clinical outcomes in new cohorts has led some observers such as Joan Massague in his 2007 New England Journal of Medicine editorial to call for research into ‘sorting out’ these gene signatures and elucidating their common overlap.8 Thus a critical problem for those in oncology has been determining whether these disjointed genetic signatures can ‘jointly’ provide a unified mechanistic rationale bridging both gene expression and clinical outcomes.
To address this challenge, we have previously demonstrated that, by aggregating different, published genetic signatures of poor prognosis, we can reveal shared molecular pathways—for example, excess direct interactions with oncogenes and tumor suppressors—through the application of a network modeling technique termed single protein analysis of networks (SPAN).9 SPAN, previously validated,10 takes advantage of protein–protein interaction networks that have been used to generate robust clinical predictions in other tumor types.9 11 In essence, SPAN uses as input a set of uncategorized protein interactions; as output, SPAN returns proteins that are more connected than can be expected by chance. The advantage of SPAN over purely expression- or literature-based methods of prioritization is that it will detect important proteins even if they are not overtly modified or amplified.9 Thus SPAN provides critical information that may not be accessible through expression data alone.
In this paper, we turn our attention to prostate cancer, as it faces a similar data prioritization problem. The treatment of prostate cancer has historically been centered around deregulation of the androgen receptor (AR) to effectively eliminate the effects of testosterone, the ligand for the AR. However, despite AR-specific targeted therapy, most patients eventually develop resistance to these agents. Consequently, multiple alternative pathways of ‘poor prognosis’ have been studied for therapeutic targeting, as many molecular mechanisms have been implicated in AR cross-talk, such as the Janus kinase (JAK)/STAT12 and platelet-derived growth factor (PDGF) receptor pathways.13 There has been no integrative approach to elucidating the key regulatory pathways. Importantly, we believe that, not only can we uncover key molecular pathways, but we can also generate gene signatures that are mechanistically coherent—or, in other words, enriched for the same molecular pathways.
While past computational approaches in prostate cancer have focused on ranking single gene targets among multiple diseases,14 we hypothesized that, using protein interactions, we could take advantage of the richness that gene signatures have to offer in the selection of molecular pathways that play essential roles in prostate cancer progression. To this end, we extracted a broad representation of poor-prognosis gene expression prostate cancer signatures from the literature (seed signatures). We then evaluated their individual protein interactions with known cancer genes curated by the Wellcome Trust Sanger Institute via SPAN. We further assembled the significant interactions of each signature. The result is what we term an ‘interactor signature’—a prioritized list of genes relating independent prostate cancer gene signatures. We evaluated this interactor signature in two ways: its internal mechanistic coherence using a novel application of information theory similarity and then its intrinsic ability to predict survival in a cohort independent of the seed signatures' cohorts. Finally, we added a qualitative evaluation of the signature against known prostate cancer pathways and current therapies. Taken together, we show that, through an extensive network analysis, prostate cancer gene expression signatures can be transformed into a set of prioritized pathways that ultimately provide a useful guide for therapeutic development.
Methods
Datasets used
Prostate cancer signatures
We evaluated 12 previously published prostate gene signatures of poor prognosis15–27 and a previously unpublished prostate cancer gene signature derived from a Mayo Clinic dataset22 listed in table 1. Thus a total of 13 gene signatures were evaluated. Signatures were deliberately chosen to span various phenotypes (eg, high-grade tumor, stem cell nature) but unified in their ability to prognosticate either decreased overall survival or early disease relapse in prostate cancer datasets. These distinct specific phenotypic conditions are well-established biological or clinical indicators of aggressive malignancy. A full listing of the genes from the included prostate cancer signatures and their translation are available at http://lussierlab.org/publications/ProstateSignature.
Table 1.
Prostate cancer gene signatures evaluated
| Signature No | Phenotype | Samples used to generate signature | Author | Available genes | In Network* |
| 1 | Aggressive disease | Divided 66 microdissected prostate cancer specimens into two groups based on clinical aggressiveness defined as prostate-specific antigen (PSA) relapse following radical retropubic prostatectomy (RRP), distant metastasis, or cancer invasion into adjacent organs | Yu et al28 | 26 | Yes |
| 2 | Benign versus cancerous prostate tissue | Proteomic screen of microdissected prostate tissue embedded in tissue microarray for genes that best discriminated between benign, localized prostate cancer and metastatic disease | Bismar et al18 | 12 | Yes |
| 3 | High-grade tumor | Examined 12 microdissected RRP Gleason pattern 3 specimens compared with that of 12 Gleason pattern 4 and eight Gleason pattern 5 | True et al27 | 85 | Yes |
| 4 | PTEN pathway/poor prognosis | Comparison of 35 phosphatase and tensin homolog (PTEN) negative and 70 PTEN positive based on immunohistochemistry from stage II estrogen receptor status-matched breast cancer specimens. Signature subsequently validated in a historic dataset of 79 prostate cancer specimens19 described above in the Sun et al signature | Saal et al24 | 184 | Yes |
| 5 | Recurrence signature in solid tumors | Comparison of gene expression signature from 12 metastatic adenocarcinoma nodules from prostate and five other tissue types compared to 64 primary adenocarcinomas from primary tumors | Ramaswamy et al23 | 17 | Yes |
| 6 | Recurrent/aggressive disease | Evaluated 62 primary prostate tumors, 41 normal prostate specimens and nine lymph node metastases to develop a two-gene model of recurrence | Lapointe et al21 | 2 | No |
| 7 | Recurrent disease | 79 patient RRP specimens from patients with clinically localized prostate cancer. 39 cases with recurrence defined as three consecutive elevations in PSA for at least 5 years | Sun and Goodison26 | 11 | No |
| 8 | Recurrent disease | Using 21 prostate cancer samples, five genes using k-nn clustering were identified. | Singh et al25 | 5 | No |
| 9 | Recurrent disease /High-Gleason score | 512 candidate genes were analyzed for correlation with Gleason score from 71 patient RRP specimens (16 patients with relapsed disease defined as two consecutive PSA elevations over 84 months) | Bibikova et al16 | 16 | Yes |
| 10 | Relapse-free survival | Using 21 prostate cancer samples from Singh et al, the authors identified three signatures of recurrence and subsequently validated these signatures on a set of 79 tumors | Glinsky et al19 | 4/4/5 | No |
| 11 | Stem cell nature | Comparison of CD133+/α2β1hi cell culture specimens from 12 human prostate cancers compared with eight CD133–/α2β1low specimens. | Birnie et al17 | 22 | Yes |
| 12 | Systemic disease after relapse, Sig 1 | 213 patients with prostate cancer PSA relapse and no evidence of systemic disease (defined as a positive bone scan or CT scan) were compared with 213 patients with prostate cancer with PSA relapse | Nakagawa et al22 | 17 | No |
| 13 | Systemic disease after relapse, Sig 2 | Reanalysis of the above Nagakawa et al (Mayo Clinic dataset) as described in the Methods section | 133 | Yes |
In Network indicates that the listed gene signature connects to the Sanger cancer genes via SPAN and composes part of the interactor signature.
Cancer mechanism genes
The Sanger Cancer Gene Census is a database maintained by the Wellcome Trust Cancer Genome Project, which contains a catalog of genes for which mutations have been causally implicated in cancer, acquired and updated through literature-based methods. We downloaded the Cancer Gene Census on October 9, 2009 from http://www.sanger.ac.uk/genetics/CGP.
Protein–protein interaction network
In brief, the protein interactions were downloaded from the Search Tool for the Retrieval of Interacting Genes version 8.0 on December 19, 2008 (STRING; http://string.embl.de).29 STRING is a repository maintained by a European consortium of genomics facilities which contains known and predicted protein–protein interactions derived from such sources as high-throughput experiments, co-expression data, and literature. We extracted all human protein–protein interactions and retained those with a combined score of >900 (highly reliable score) that also had gene fusion, experimental, or database evidence. Thus text mining results of STRING were filtered out. A total of 72 617 distinct interactions between 7681 distinct proteins were retained.11 Proteins were considered to be nodes, and interactions between proteins are links.
Signature generation from the Mayo Clinic prostate cancer data
The original Mayo Clinic signature was significantly smaller than other comparable signatures, and we thus recalculated a broader signature as follows. Gene Expression Omnibus (GSE10645) was downloaded and analyzed using R/Bioconductor. We compared men who had biochemical relapse with systemic disease (bone or visceral disease) with those who did not. Genes with little to no change in expression levels were filtered using a covariance of expression parameter of 0.3. Significance analysis of microarrays30 was then performed to obtain a gene signature with a false discovery rate (FDR) <5%.
SPAN analysis of the prostate cancer gene signatures
The SPAN method has been extensively described previously,9 11 and expanded details can be found in the online supplemental methods. In brief, each prostate cancer gene signature was compared with the Sanger Cancer Gene set using SPAN. The observed number of interactions between the prostate cancer signatures and the Sanger cancer genes were derived and compared with an expected distribution through permutation resampling. The unadjusted p value of each signature gene's connectivity was further adjusted for multiplicity using Bonferroni-type methods. A converse calculation was performed where each single Sanger cancer gene was analyzed for its total number of interactions with each independent, unique gene in the amalgamated prostate cancer signatures independently and assigned a p value and a Benjamini–Hochberg FDR. Prioritized genes and their interactors that had a FDR <5% were retained. The resulting statistically significant genes were then aggregated to form an ‘interactor signature’. As each SPAN protein keeps an equal number of partners in the empirical distribution (constant node degree), ‘hub proteins’ are statistically prioritized using conservative controls. See figure 1 and the online supplemental methods for details of interactor signature assembly from seed signatures. The resulting network was then displayed in Cytoscape31 where Sanger cancer genes that are also members of the expression signature gene lists are clearly represented. Further, when SPAN-prioritized Sanger cancer genes from individual signature genes also overlap between these signatures, these shared known cancer interactions common among signatures serve as a ‘quasi-gold standard’ because of the very high statistical and biological significance of such an occurrence. Visualization of statistically significant pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) provided a further unbiased evaluation of reference gold-standard pathway genes and their associated networks.32
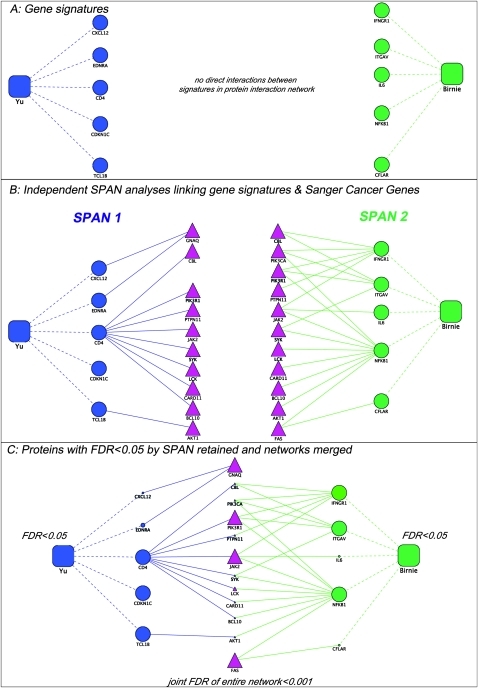
Figure 1.
Representative assembly of the protein network from disparate gene signatures. As shown in (A), signature 1 gene/proteins (blue circles) do not connect directly with signature 2 proteins (green circles). Protein interaction networks can independently link (solid line) each gene signature to a common set of cancer genes from the Sanger database (red triangles in B). Subsequently, using single protein analysis of network (SPAN), only protein–protein interactions with a false discovery rate (FDR) <0.05 are retained in each signature and an aggregate of interactors is assembled from all SPAN analyses of each signature, thus generating a composite network with a FDR <1% ((C) large shapes=FDR <5%, small shapes=FDR >5%). Prognostic gene expression signatures are represented as squares, and their respective genes are related with dotted lines.
Gene Ontology enrichment of the interactor signature
Using FuncAssociate 2.0 software,33 we evaluated the resulting interactor signature genes for common molecular processes and biological functions from annotations found in Gene Ontology (GO). GO annotations that were statistically over-represented with a FDR <1% were noted. A local minimum algorithm was then used to identify more informative GO terms.11
Evaluating pathway similarity among sets of gene signatures
In order to determine if sets of genes comprised related or unrelated molecular pathways, we calculated a metric of information theoretic similarity (ITS) applied to GO that we and others have previously validated.10 Among the approaches used to estimate similarity between gene functions, those derived from information theory and GO are considered robust and state of the art,34 and we have previously demonstrated their utility in calculating the similarity between breast cancer expression signatures.9 We performed two evaluations using this metric. First, we examined the similarity of annotations within each seed signature geneset (lists of its genes) and the interactor signature in the context of GO to derive an ITS score by examining each gene–gene distance using the information theoretic distance. The information theoretic distance of each gene–gene distance was based on their respective annotations in GO. We then took the summation of all the scores of the unique gene pairs. We divided this total score by the number of genes in the signature. These scores were not further normalized, as we used an information theoretic distance, not the gene expression level, between the genes. Information theoretic distances are calculated as a continuous variable between 0 and 1. Therefore all measurements are within the same scale, and in our estimation did not require further normalization. Scores were calculated for the interactor signature and for the original gene signatures. To control the ITS for length of signature, we then generated an empiric distribution by using a bootstrap, resampling without replacement, of genes from the protein network for each signature; we selected the same number of signature genes, calculated the ITS, and repeated this procedure 10 000 times. We then observed the rank of the original gene signature ITS score within the individual empirical distributions and calculated a p value (reported in table 1).
Generation of a prioritized phenotype–pathway map
We examined the connectivity of the interactor signature to itself using SPAN. Significant proteins with a FDR <0.05 were retained. We then overlaid KEGG pathway data on to the resultant protein network using the DAVID tool35 (adjusted p value <0.05). The final annotated protein network is what is considered to be the phenotype–pathway map. This pathway map has been thoroughly reviewed in the literature for its biomolecular mechanistic relevance to prostate cancer progression and prognosis.
Survival analysis
To test the clinical relevance of the interactor signature, we examined its ability to find a survival difference in a large and independent retrospective dataset of 281 Swedish men who underwent a course of ‘watchful waiting’ after being diagnosed with prostate cancer (GSE10645).22 This survival analysis using a separate dataset serves as a type of clinical evaluation of the interactor signature. This set of 281 only included patients who were alive or had died from prostate-cancer-specific causes. For each patient, gene expression levels from the interactor signature were totaled to develop a per-patient score. Patients were placed in one of two groups on the basis of whether or not they were above or below the mean score. Kaplan–Meier analysis was then performed using time from diagnosis until death. In a second analysis, only patients with undisputed disease (Gleason scores of 7, 8, or 9) were included for analysis.
Qualitative validation of interactor genes, connections, and network
To establish whether the genes, interactions, and network prioritized via our analyses were relevant in prostate cancer, multiple reference sources were queried. (i) PubMed literature searches restricted from 2000 to 2010 were entered with the target genes and their interactions of interest and the keyword ‘prostate’. Relevant and high priority pathways were then identified and reported. (ii) Genes were analyzed by Ingenuity Pathway Analysis (Ingenuity Systems; http://www.ingenuity.com) to observe literature-based connections among the genes and canonical sub-networks. (iii) ClinicalTrials.gov website from December 1, 2010 was queried for each SPAN prioritized gene in the second interactor signature to observe whether relevant clinical trials were ongoing or planned.
Results and discussion
Prostate cancer gene signatures are tightly interwoven and have greater interacting partners than expected by chance
We began with 13 prostate cancer signatures that all had statistically significant worsened survival outcomes in independent cohorts of patients with prostate cancer. Eight gene signatures among the 13 met the compound connectivity criteria of (i) FDR <5% and (ii) having two or more interactors between the gene expression signature and the Sanger cancer genes (Methods). Of note, traditional measures of prostate cancer aggressiveness are based on the tumor morphology or grade, and thus four of the signatures examined this specifically: (i, ii) benign versus cancerous prostate tissue,18 (iii) high-Gleason score16 and (iv) high-grade tumor.27 Recurrent disease is by definition already more aggressive, and multiple gene expression profiles were derived from tumors with this phenotype: (v, vi, vii) recurrent disease,19 25 26 (viii) recurrence signature in solid tumors,23 (ix, x) systemic disease after relapse22 and recalculated systemic disease after relapse (Mayo Clinic dataset), and (xi) aggressive disease—which included patients who relapsed after primary therapy.28 The last two signatures are based on principles of the cancer biology of aggressiveness—namely more primitive appearing cancers—(xii) stem cell in nature17 or cancers that have a known phosphatase and tensin homolog (PTEN) deregulation (xiii) PTEN pathway.24 Please refer to table 1 for full details.
Using SPAN methodology, we evaluated whether any genes from the prostate cancer gene signatures that were significantly connected to the Sanger cancer genes curated by the Wellcome Trust Cancer Genome. We also examined whether there were Sanger genes that were significantly connected to each gene signature. In total, 42 genes were statistically significant with a FDR of 5% and met criteria for having at least two interacting partners (table 1, online supplemental table 1). We call these 42 genes the ‘interactor signature’. Eight of the 13 gene signatures were connected via SPAN.
We also examined the interactor signature genes' connectivity to other genes. As a check of our prioritization method, we believed that our interactor genes would have importance within a network context. To confirm this, we relied on work published by the Gerstein laboratory,36 who had identified specific network proteins as having biologically significant properties. They defined ‘hubs’ as proteins that have the 20% highest number of neighbors, and ‘bottlenecks’ as the proteins that are in the top 20% in terms of betweenness (connecting groups of proteins). In our network, 29 (69%) proteins were bottlenecks, 25 (59%) were hubs, and 24 (57%) were both bottleneck and hubs (online supplemental table 1). SPAN analyses are conservatively controlled for hubness. Each protein keeps its node degree (number of protein interactions) constant in each permutation, while its interactors are resampled. The fact that 57% were both hub and bottleneck proteins is in far excess of the baseline 10.14% in a random distribution of proteins from the network (p<0.0001, Fisher exact test).36 This confirmed to us that, at a network structure level, our interactor signature identified critical players in poor-prognosis prostate cancer. The tightly interwoven nature of our interactor signature is readily evident in our graphical representation of its relationships (figure 2).
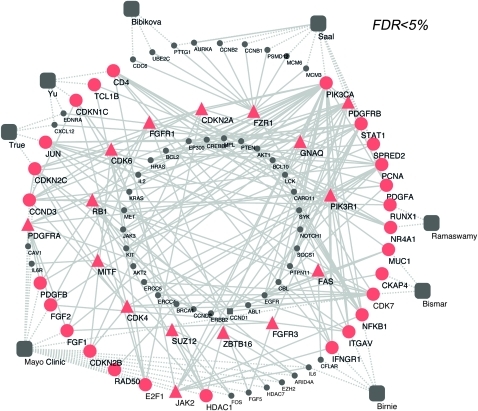
Figure 2.
Combined network of prioritized signature genes and cancer proteins derived from single protein analysis of network (SPAN) protein interaction analysis conducted over each expression signature. Prostate cancer gene signatures of poor prognosis (large grey squares) were evaluated for their protein–protein interaction connectivity to the Sanger cancer genes curated by the Wellcome Trust Cancer Gene Atlas through SPAN methodology. Squares represent prostate cancer gene signatures, circles indicate network genes, and triangles indicate Wellcome Trust Sanger cancer genes. Red indicates statistically significant proteins (false discovery rate (FDR) <5%) with at least two interacting partners, and grey indicates non-prioritized proteins. Nodes on the outer circle indicate prostate cancer signature genes, and nodes in the innermost circle indicate proteins contributing to prioritize the statistically significant ones but for which the FDR >5%. Dashed lines indicate linkages between signature genes and their respective signatures, and solid lines indicate a protein interaction.
Interactor signature genes are involved in cell cycle, PDGF and fibroblast growth factor (FGF) signaling, and phosphorylation
We sought to characterize the predominant biomolecular functions of the selected 42 genes. To do this, we evaluated the functional annotations found in GO of this interactor signature. GO is essentially a hierarchical lexicon of terms used to describe genes. We determined whether these descriptors of biomolecular functions were enriched in our gene set. Highly significant (adjusted p value <0.0001) descriptors that were associated with this set of genes were terms related to several pathways, namely PDGF and FGF signaling. Also notable were annotations related to cell cycle regulation and phosphorylation. Full results of this GO enrichment are listed in online supplemental table 2.
The 42-gene interactor signature prioritizes key pathways better than other prostate gene signatures
To evaluate whether the genes in our interactor signature were more related to one another (ie, involved in the same molecular pathway or performed the same molecular function) than genes in other prostate cancer gene signatures, we extended a method of evaluating the similarity of genes based on their shared annotation in GO.11 We computed an ITS score, which evaluates the average similarity of a set of genes. Using this algorithm, we systematically evaluated the ITS between each pair of signatures including the 42-gene interactor signature and the 13 original prostate cancer gene signatures. Next, to correct for gene signature length and to calculate an empiric p value, we generated 10 000 bootstraps of a similar length gene signature derived from the protein network and then examined the rank of each gene signature among the bootstraps. The interactor signature ranked first in 10 000 bootstraps (p≤0.0001) of a similar number of genes as demonstrated by table 2. The only other signature that was statistically significant was Bibikova's high-Gleason signature, which resulted in a significant but lower p value of 0.017.
Table 2.
Significance of pathway similarity among sets of gene signatures
| Signature number | Signature name | p Value | Rank among 10 000 gene length identical bootstraps | Phenotype |
| Na | Interactor signature | ≤0.0001 | 1 | Na |
| 9 | Bibikova | 0.017 | 174 | Recurrent disease/high-Gleason |
| 10 | Glinsky 3 | 0.070 | 701 | Relapse-free survival |
| 12 | Nakagawa | 0.086 | 858 | Systemic disease after relapse |
| 6 | Lapointe | 0.114 | 1146 | Recurrent/aggressive disease |
| 5 | Ramaswamy | 0.173 | 1728 | Recurrence signature in solid tumors |
| 13 | Mayo Clinic dataset | 0.309 | 3091 | Relapse-free survival |
| 1 | Yu | 0.344 | 3440 | Aggressive disease |
| 4 | Saal | 0.452 | 4520 | PTEN pathway/poor prognosis |
| 7 | Sun | 0.452 | 4520 | Recurrent disease |
| 10 | Glinksy 2 | 0.463 | 4632 | Relapse-free survival |
| 3 | True | 0.519 | 5186 | High-grade tumor |
| 8 | Singh | 0.523 | 5233 | Recurrent disease |
| 10 | Glinsky 1 | 0.538 | 5382 | Relapse-free survival |
| 2 | Bismar | 0.652 | 6615 | Benign versus cancerous prostate tissue |
| 11 | Birnie | 0.887 | 8874 | Stem cell nature |
The SPAN-generated interactor signature has prognostic significance in newly diagnosed prostate cancer
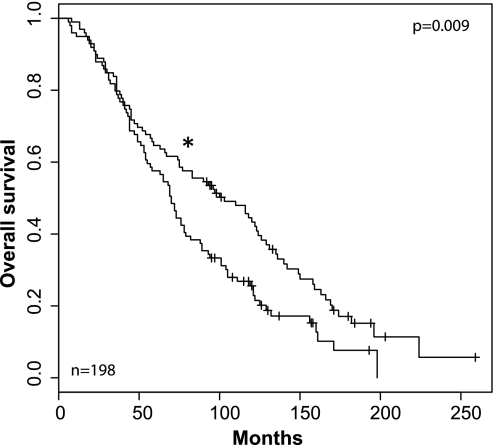
To evaluate the clinical relevance of the interactor signature, we performed our evaluation in a completely independent dataset, the Swedish Watchful Waiting Cohort.37 In this study, 281 men underwent a course of watchful waiting after diagnosis of prostate cancer. We asked whether interactor signature overexpression was able to distinguish a group with poorer survival. Of the genes in the interactor signature, 35 were available for analysis. We divided the patients into two groups on the basis of whether their mean gene expression was higher than the average of the entire cohort. Kaplan–Meier survival analysis of the two groups from the date of their diagnosis was performed. The log rank test gave a p value that approached significance at 0.052. Importantly, given the heterogeneity of prostate cancer, we were able to detect an even greater significance (p=0.009) when we only evaluated a subset of 198 patients with high-grade prostate cancer (Gleason 7–10) (figure 3).
Figure 3.
Kaplan–Meier analysis of the 42-gene interactor signature revealed a clinically significant signal. Genes from the interactor signature that were available for analysis (35 genes total) from an active surveillance study of prostate cancer were used for analysis. 198 patients with high-grade (Gleason 7–10) disease were used, and overall survival from time of diagnosis was determined. The log rank test showed a significant survival difference in patients who had higher average expression levels of the genes of interest versus those who had lower average expression (p=0.009; Kaplan–Meier analysis). Asterix (*) indicates lower expression of interactor signature.
SPAN analysis of the interactor signature emphasizes pathways of prostate cancer progression
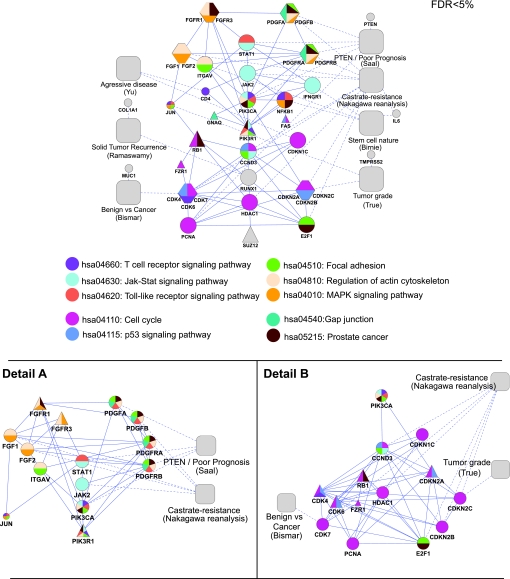
Our first SPAN analysis generated a set of highly connected genes (interactor signature) related to prostate cancer. A second SPAN analysis over the interactor signature allows us to prioritize molecular pathways vis-à-vis their protein interactions with one another. In other words, the first SPAN allowed us to identify disparate expression signatures interacting with common cancer proteins of the gold standard Sanger cancer genes (Methods). For the 42 key protein interactors thus generated, we then further annotated the most central ones in the network. The determination of centrality was performed via a second SPAN analysis over the interactor signature proteins that resulted in their prioritization (node size) and clarification of their interactions as shown in figure 4. To highlight established pathways, we overlaid canonical pathway information from the KEGG32 after calculating which of the pathways were represented at a statistically significant level (p<0.05). The result, when graphed, is what we call a phenotype–pathway map (figure 4). In this prostate cancer phenotype–pathway map of poor prognosis, seven of the original prostate cancer gene signatures form coherent subgroups that are consistent with established pathways.
Figure 4.
Prostate phenotype–pathway map. A second single protein analysis of network (SPAN) was conducted over the network presented in figure 2 to prioritize a subset of the 42-gene interactor prostate signature of poor prognosis which revealed a tightly interwoven network (top panel; Methods). Proteins with an empiric false discovery rate (FDR) <0.05 were retained and are indicated by the larger size shape. Significant KEGG pathways (FDR <0.05) were overlaid on to the network and colorized as indicated. Detail A and Detail B expand areas in the top panel that were simplified. Square shapes denote prognostic expression signatures with dotted lines to their associated gene; hexagons represent several proteins that are closely associated with one another and combined for purposes of simplicity of representation; triangles denote Sanger genes as compared with circle shapes which denote the protein products of signature genes.
Our second SPAN and resulting prostate phenotype–pathway map allows us to better understand the biological meaning of the interactor signature. By looking for dominant molecular mechanisms and highly connected genes, we can begin to untangle, and conjecture about, the key pathways of poor-prognosis prostate cancer.
The phenotype–pathway map recapitulates phosphatidylinositol-3 kinase (PI3K)/NF-κB centrality to prostate cancer progression and highlights driver pathways
Examining figure 4 in detail, we can see that the PI3K/NF-κ B signaling cascade is central to this phenotype–pathway map, as it is a common end point for the various upstream signaling cascades. The role of the PI3K/NF-κB in prostate cancer progression is well established and is believed to be a mechanism for cross-talk with the androgen receptor and thus is implicated in androgen independence.38 This finding has been noted in prostate cancer by multiple observers.39 As in the following qualitative discussion, we can recapitulate and prioritize major drivers of poor-prognosis prostate cancer, as well as describe under-reported findings. As shown below in our review of the literature, the Janus kinase 2 (JAK2) and STAT1 stories were perhaps the most novel and under-reported.
Feeding into the PIK3/NF-κB pathway are driver pathways that include1 the PDGF signaling cascade,2 FGF signaling,3 interferon (IFN)γ signaling, and4 the JAK/STAT pathway. When we consider the KEGG annotations of the pathways, we observe that the pathway ‘regulation of actin cytoskeleton’ (hsa:04010) encompasses FGF and PDGF signaling through the PI3K/NF-κB cascade. A second KEGG pathway, JAK/STAT (hsa: 04630), captures IFN signaling. The importance of the JAK/STAT pathways is consistent with conclusions in a separate paper analyzing molecular profiling of prostate cancer stem cells.17
Key regulators of cell cycle derangements constitute a substantial portion of the phenotype–pathway map
Consistent with the established role of PI3K/NF-κB in mitogenic activation, downstream proteins were nearly all associated with the cell cycle (hsa: 04110) (figure 4B). Cell cycle kinases, regulatory proteins and proliferating cell nuclear antigen, a known marker of proliferation,40 constitute the majority of the identified proteins. Cyclin D3 (CCND3) and its ligand, the tumor suppressor protein retinoblastoma 1 (RB1), were prioritized as part of cell cycle regulation. RUNX1—normally associated with acute myeloid leukemia (AML)—was tightly associated with this sub-network as well. Previous work has demonstrated that RUNX1 cooperates with E-twenty six transcription to activate transcription in the setting of androgen deprivation.41
JAK2 is uniquely positioned in the phenotype–pathway map as an activator of the PI3K/NF-κB cascade
Perhaps most interesting from a translational medicine perspective is the utility of the phenotype–pathway map in helping identify key genetic lynchpins. JAK2 is involved in cytokine receptor signaling and has been experimentally confirmed in prostate cancer.12 Examination of figure 4A reveals that JAK2 is connected either directly or indirectly to nearly all the proteins that are upstream of the PI3K/NF-κB signaling cascade. We note the interplay of JAK2 with the FGF, PDGF and IFN pathways. Indeed, phosphorylation of the Stat3 oncogene via the FGF pathway is dependent on JAK2.42 The Stat3 oncogene in turn is believed to be downstream of PDGF and also activated via JAK2.43 PDGF activation can then proceed through the PI3K/NF-κB pathway44 to activate proliferation. Similarly, the proinflammatory cytokine, IFNγ, is traditionally thought to bind to the IFNγ receptor (partly encoded by IFNGR1) and then act via the JAK2/STAT1 pathway in a tumor suppressor role in prostate cancer.45 In fact, STAT1 activation may be a marker of derangement in the sensitivity in IFN signaling and has been associated with chemoresistance in the castrate setting.46 In other work, Gu et al have published a series of papers characterizing the behavior of STAT3 and STAT5a/b transcription factors using in vivo and in vitro models, and recently posited whether JAK2 is the ‘common denominator’ for their dual activation in clinical prostate cancers.12 Thus the prostate phenotype–pathway map highlights the centrality of JAK2 as a mediator of the prioritized pathways. Whether JAK2 inhibition alone is sufficient to prevent the recurrence of prostate cancer or is a bona fide therapeutic target for advanced disease remains to be determined in clinical trials.
Bioinformatics can provide an ‘executive synopsis’ of relevant molecular pathways and points to potential drug targets
This is the second study that confirms mechanistic overlap in relatively disjointed but prognostically congruent gene signatures, a paradox noted by Joan Massague in 2007,8 that was solved with our previous publication.9 We initially demonstrated that cancer genes, both oncogenes and tumor suppressors, were interacting with signature genes more than expected by empirical distribution in our network modeling of protein interactions. Although our previous study was conducted in the breast domain, this study corroborates our previous findings in the prostate arena but differs in two ways. First, we ensure the biological significance of our findings by introducing an information theory-based metric. Second, we provide a completely unbiased patient cohort to confirm the clinical relevance of our study.
In this paper, we demonstrate, through established bioinformatics methods, that we can transform heterogeneous and complex prostate cancer gene signatures of poor prognosis into a clinically meaningful ‘executive synopsis’ of the most relevant pathways and critical genes such as JAK2. There are undoubtedly many such genes. Table 3 provides a summary of the members of the phenotype–pathway and their exploration as potential therapies. A comparison list of drugs and drug targets derived from KEGG can be found in online supplemental table 3. As shown, these in silico results mirror relevant prostate cancer pathways that have been found and verified experimentally in vitro/in vivo.
Table 3.
Phenotype–pathway map genes and their stage of clinical drug development within prostate cancer (source: http://ClinicalTrials.gov and PubMed data as of December 2010)
| Prioritized gene | Relation to prostate cancer | Signature or cancer gene | Clinical drug development |
| CCND3 | Established47 | Both | No data |
| CDK(4/6/7) | Well-established48 | Cancer/cancer/signature | Yes |
| CDKN1C | Well-established49 | Signature | Tumor suppressor |
| CDKN(2A/2B/2C) | Established50 | Cancer/signature/signature | Tumor suppressor |
| E2F1 | Well-established51 | Signature | No data |
| FGF(1/2), FGFR(1/3) | Well-established42 | Signature/signature/both/cancer | Yes |
| HDAC1 | Well-established52 | Signature | Yes |
| IFNGR1 | Established53 | Signature | No data |
| ITGAV | Established54 | Signature | No data |
| JAK2 | Well-established55 | Both | Yes |
| NFKB1 | Well-established56 | Signature | Yes |
| PCNA | Well-established40 | Signature | No data |
| PDGF(A/B), PDGFR(A/B) | Well-established13 | Signature/both/both/both | Yes |
| PI3KR1/PI3KCA | Well-established38 | Both/cancer | Yes |
| RB1 | Well-established57 | Cancer | Tumor suppressor |
| RUNX1 | Established58 | Both | No data |
| STAT1 | Role unclear46 | Signature | No data |
| SUZ12 | No data | Cancer | Yes |
PCNA, proliferating cell nuclear antigen.
We also learned that the best discriminatory genes for an expression signature do not necessarily make the best input into the SPAN network. The 17-gene Nakagawa signature, which was developed using a non-parametric supervised learning method, ultimately did not connect to the network. In contrast, a 132-gene signature derived in an unbiased manner from the same dataset connected to nearly all the other signatures via SPAN. Thus genes that have the greatest degree of discriminatory ability may indeed be ‘passenger’ genes rather than ‘driver’ genes. For purposes of SPAN analysis, we believe it is better to pursue the most unbiased gene signature to identify a larger grouping of statistically relevant genes and allow the protein network to perform the filtering. In other words, a larger set of genes is more likely to comprise the ‘drivers’ of cancer mechanisms from which SPAN can assert protein interactions, rather than the ‘passenger’ genes that simply correlate with outcome and thus cannot contribute mechanisms in network models.
Limitations
By design, we used a simple, single protein network interaction model to calculate p values, as it is easier to interpret significant results by clinicians and biologist. However, more sensitive and powerful network modeling is likely to yield additional insight, such as diffusion kernels.59 Furthermore, we conducted the analysis using STRING version 8.0, which contains a limited number of interactions and thus limited the study to this subset of proteins. While the computational controls show that the observed network signature is highly statistically significant, the qualitative evaluation of the results rely on previously published data. We are therefore beholden to the different methodologies and the multitude of oligonucleotide arrays used to derive the gene signatures. We have attempted to overcome this by carefully incorporating multiple independent gene signatures and using stringent statistical cut-offs to ensure a conservative evaluation. Indeed, each seed signature SPAN analysis required an FDR <0.05 and more than one interactor; this suggests that the relevant interactors of this network signature (spanning multiple seed signatures) more likely have a significance of FDR <<0.05. Additionally, the Kaplan–Meier analysis was performed in a completely independent dataset of all the signatures and therefore provided an unbiased evaluation.
The protein-interaction network and the Sanger cancer genes are not static but vibrant growing entities. As we learn more about factors contributing to prostate cancer, undoubtedly there will be additions and variations to the phenotype–pathway map. Nevertheless, the intent of this study was to provide and explore a tool for understanding gene expression signatures quickly at this moment in time. Going forward, we do intend to rerun these analyses with updated and expanded lists of gene signatures. Furthermore, as we have shown that different interactors prioritized in distinct seed signatures may be related to the same oncogene by interactions, this fact suggests that novel methods should be developed to produce expression classifiers where the interaction is investigated ab initio rather than a posteriori. Such an approach could be designed to identify, across samples, significant, yet distinct, interactors to an oncogene, thus promoting within the principles of personal genomics a fundamental paradigm shift from the current cohort-wide requirements.
In summary, the phenotype–pathway map provides an excellent starting point for developing rational clinical trial designs, as it can inform researchers about what therapy should be attempted first that may be helpful for the largest number of patients. To this end, we are working on translating our prioritized pathway findings into the clinical setting; simultaneously, we are extending this technique to other tumor types. As more knowledge accumulates about oncogenes and gene signatures, reanalysis by this technique may reveal new pathways and interconnections that were heretofore unknown or understudied.
Conclusion
By analyzing multiple prostate cancer signatures of poor prognosis, we have uncovered seven highly connected cancer genes that were not among our original gene signatures of poor prognosis. This further confirms our hypothesis that, while multiple genes in a high-throughput analysis may change along with the activity of oncogenes or tumor suppressors, the critical information contained in direct physical interactions among proteins is not accessible via expression arrays. As a result, a multiscale approach incorporating gene expression data and protein interaction networks can elucidate otherwise neglected targets and underlying molecular sub-networks underpinning the phenotypic concordance of genetically disparate gene signatures. At the gene expression signature level, the pathways are not apparent. However, the interactor signature not only prioritizes biological mechanisms underpinning multiple signatures, it also recapitulates in good part known pathways involved in prostate cancer oncogenesis. Indeed, the phenotype–pathway map generated by our interactor signature truly recapitulates and underscores the centrality of the PI3K/NF-κB pathway and other known mechanisms for prostate cancer progression. Moreover, through a systems biology approach, we are able to prioritize less well-established pathways, such as JAK2, that may ultimately serve as attractive drug targets. From seed signatures generated at the cohort level, we have demonstrated a posteriori that expression changes in direct, yet distinct, interactors to oncogenes correlate with prognosis. Thus we propose that ab initio design of mechanistically anchored gene expression classifiers are more likely than current cohort-level classifier approaches to be sensitive to individual variation in personal genomics.
Supplementary Material
Footnotes
Funding: NIH/NNLM, Grant Number 1U54CA121852-01A1.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J Clin Oncol 2005;23:7332–41 [DOI] [PubMed] [Google Scholar]
- 2.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999–2009 [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52 [DOI] [PubMed] [Google Scholar]
- 4.Culhane AC, Schwarzl T, Sultana R, et al. GeneSigDB–a curated database of gene expression signatures. Nucleic Acids Res 2010;38(Database issue):D716–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 2006;24:3726–34 [DOI] [PubMed] [Google Scholar]
- 6.Monzon FA, Lyons-Weiler M, Buturovic LJ, et al. Multicenter validation of a 1,550-gene expression profile for identification of tumor tissue of origin. J Clin Oncol 2009;27:2503–8 [DOI] [PubMed] [Google Scholar]
- 7.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 2006;355:560–9 [DOI] [PubMed] [Google Scholar]
- 8.Massague J. Sorting out breast-cancer gene signatures. N Engl J Med 2007;356:294–7 [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Sam L, Huang Y, et al. Protein interaction network underpins concordant prognosis among heterogeneous breast cancer signatures. J Biomed Inform 2010;43:385–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao Y, Sam L, Li J, et al. Information theory applied to the sparse gene ontology annotation network to predict novel gene function. Bioinformatics 2007;23:i529–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee Y, Yang X, Huang Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol 2010;6:e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu L, Dagvadorj A, Lutz J, et al. Transcription factor Stat3 stimulates metastatic behavior of human prostate cancer cells in vivo, whereas Stat5b has a preferential role in the promotion of prostate cancer cell viability and tumor growth. Am J Pathol 2010;176:1959–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Kong D, Li Y, et al. PDGF-D signaling: a novel target in cancer therapy. Curr Drug Targets 2009;10:38–41 [DOI] [PubMed] [Google Scholar]
- 14.Franke L, van Bakel H, Fokkens L, et al. Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet 2006;78:1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Best CJ, Gillespie JW, Yi Y, et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res 2005;11:6823–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibikova M, Chudin E, Arsanjani A, et al. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics 2007;89:666–72 [DOI] [PubMed] [Google Scholar]
- 17.Birnie R, Bryce SD, Roome C, et al. Gene expression profiling of human prostate cancer stem cells reveals a pro-inflammatory phenotype and the importance of extracellular matrix interactions. Genome Biol 2008;9:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bismar TA, Demichelis F, Riva A, et al. Defining aggressive prostate cancer using a 12-gene model. Neoplasia 2006;8:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glinsky GV, Glinskii AB, Stephenson AJ, et al. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 2004;113:913–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henshall SM, Afar DE, Hiller J, et al. Survival analysis of genome-wide gene expression profiles of prostate cancers identifies new prognostic targets of disease relapse. Cancer Res 2003;63:4196–203 [PubMed] [Google Scholar]
- 21.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A 2004;101:811–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One 2008;3:e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet 2003;33:49–54 [DOI] [PubMed] [Google Scholar]
- 24.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A 2007;104:7564–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002;1:203–9 [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Goodison S. Optimizing molecular signatures for predicting prostate cancer recurrence. Prostate 2009;69:1119–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.True L, Coleman I, Hawley S, et al. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci U S A 2006;103:10991–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 2004;22:2790–9 [DOI] [PubMed] [Google Scholar]
- 29.Jensen LJ, Kuhn M, Stark M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 2009;37(Database issue):D412–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001;98:5116–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2010;38(Database issue):D355–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berriz GF, King OD, Bryant B, et al. Characterizing gene sets with FuncAssociate. Bioinformatics 2003;19:2502–4 [DOI] [PubMed] [Google Scholar]
- 34.Alterovitz G, Xiang M, Hill DP, et al. Ontology engineering. Nat Biotechnol 2010;28:128–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:P3. [PubMed] [Google Scholar]
- 36.Yu H, Kim PM, Sprecher E, et al. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 2007;3:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sboner A, Demichelis F, Calza S, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics 2010;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dubrovska A, Kim S, Salamone RJ, et al. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci U S A 2009;106:268–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer 2010;10:561–74 [DOI] [PubMed] [Google Scholar]
- 40.McNeal JE, Haillot O, Yemoto C. Cell proliferation in dysplasia of the prostate: analysis by PCNA immunostaining. Prostate 1995;27:258–68 [DOI] [PubMed] [Google Scholar]
- 41.Banach-Petrosky W, Jessen WJ, Ouyang X, et al. Prolonged exposure to reduced levels of androgen accelerates prostate cancer progression in Nkx3.1; Pten mutant mice. Cancer Res 2007;67:9089–96 [DOI] [PubMed] [Google Scholar]
- 42.Dudka AA, Sweet SM, Heath JK. Signal transducers and activators of transcription-3 binding to the fibroblast growth factor receptor is activated by receptor amplification. Cancer Res 2010;70:3391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowman T, Garcia R, Turkson J, et al. STATs in oncogenesis. Oncogene 2000;19:2474–88 [DOI] [PubMed] [Google Scholar]
- 44.Uehara H, Kim SJ, Karashima T, et al. Effects of blocking platelet-derived growth factor-receptor signaling in a mouse model of experimental prostate cancer bone metastases. J Natl Cancer Inst 2003;95:458–70 [DOI] [PubMed] [Google Scholar]
- 45.Shou J, Soriano R, Hayward SW, et al. Expression profiling of a human cell line model of prostatic cancer reveals a direct involvement of interferon signaling in prostate tumor progression. Proc Natl Acad Sci U S A 2002;99:2830–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patterson SG, Wei S, Chen X, et al. Novel role of Stat1 in the development of docetaxel resistance in prostate tumor cells. Oncogene 2006;25:6113–22 [DOI] [PubMed] [Google Scholar]
- 47.Olshavsky NA, Groh EM, Comstock CE, et al. Cyclin D3 action in androgen receptor regulation and prostate cancer. Oncogene 2008;27:3111–21 [DOI] [PubMed] [Google Scholar]
- 48.Rigas AC, Robson CN, Curtin NJ. Therapeutic potential of CDK inhibitor NU2058 in androgen-independent prostate cancer. Oncogene 2007;26:7611–19 [DOI] [PubMed] [Google Scholar]
- 49.Cote RJ, Shi Y, Groshen S, et al. Association of p27Kip1 levels with recurrence and survival in patients with stage C prostate carcinoma. J Natl Cancer Inst 1998;90:916–20 [DOI] [PubMed] [Google Scholar]
- 50.Ashida S, Nakagawa H, Katagiri T, et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res 2004;64:5963–72 [DOI] [PubMed] [Google Scholar]
- 51.Zheng C, Ren Z, Wang H, et al. E2F1 Induces tumor cell survival via nuclear factor-kappaB-dependent induction of EGR1 transcription in prostate cancer cells. Cancer Res 2009;69:2324–31 [DOI] [PubMed] [Google Scholar]
- 52.Noonan EJ, Place RF, Pookot D, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009;28:1714–24 [DOI] [PubMed] [Google Scholar]
- 53.Dunn GP, Sheehan KC, Old LJ, et al. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res 2005;65:3447–53 [DOI] [PubMed] [Google Scholar]
- 54.Cooper CR, Chay CH, Pienta KJ. The role of alpha(v)beta(3) in prostate cancer progression. Neoplasia 2002;4:191–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hedvat M, Huszar D, Herrmann A, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell 2009;16:487–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L, Charron M, Wright WW, et al. Nuclear factor-kappaB activates transcription of the androgen receptor gene in Sertoli cells isolated from testes of adult rats. Endocrinology 2004;145:781–9 [DOI] [PubMed] [Google Scholar]
- 57.Mack PC, Chi SG, Meyers FJ, et al. Increased RB1 abnormalities in human primary prostate cancer following combined androgen blockade. Prostate 1998;34:145–51 [DOI] [PubMed] [Google Scholar]
- 58.Fowler M, Borazanci E, McGhee L, et al. RUNX1 (AML-1) and RUNX2 (AML-3) cooperate with prostate-derived Ets factor to activate transcription from the PSA upstream regulatory region. J Cell Biochem 2006;97:1–17 [DOI] [PubMed] [Google Scholar]
- 59.Tsuda K, Noble WS. Learning kernels from biological networks by maximizing entropy. Bioinformatics 2004;20(Suppl 1):i326–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.