Abstract
Five Rad51-like proteins, referred to as Rad51 paralogs, have been described in vertebrates. We show that two of them, Rad51B and Rad51C, are associated in a stable complex. Rad51B–Rad51C complex has ssDNA binding and ssDNA-stimulated ATPase activities. We also examined the functional interaction of Rad51B–Rad51C with Rad51 and RPA. Even though RPA enhances Rad51-catalyzed DNA joint formation via removal of secondary structure in the ssDNA substrate, it can also compete with Rad51 for binding to the substrate, leading to suppressed reaction efficiency. The competition by RPA for substrate binding can be partially alleviated by Rad51B–Rad51C. This recombination mediator function of Rad51B–Rad51C is likely required for the assembly of the Rad51-ssDNA nucleoprotein filament in vivo.
Keywords: DNA double-strand break repair, tumor suppression
Studies in Saccharomyces cerevisiae have identified a large number of genetic loci required for mitotic and meiotic recombination. These genes, comprising RAD50,RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, RDH54/TID1, MRE11, and XRS2 are collectively known as the RAD52 epistasis group. The RAD52 group of genes are also intimately involved in the repair of DNA double-strand breaks induced by exogenous agents such as ionizing radiation (Paques and Haber 1999; Sung et al. 2000) and for telomere maintenance in the absence of telomerase.
Cloning, genetic, and biochemical studies have indicated that the structure and function of the RAD52 group genes are highly conserved among eukaryotes, from yeast to humans (Sung et al. 2000; Thompson and Schild 2001). Interestingly, in mammals, the efficiency of recombination and DNA double-strand break repair is contingent upon the integrity of the tumor suppressors BRCA1 and BRCA2 (Dasika et al. 1999; Moynahan et al. 1999, 2001; Thompson and Schild 2001), underscoring the importance for deciphering the mechanistic basis of the recombination machinery.
In recombination processes that involve the formation of a DNA double-strand break, the ends of the DNA break are processed to yield single-stranded DNA tails. These DNA tails are utilized by the RAD52 group recombination factors for the formation of DNA joints with a homologous DNA template, contained within the sister chromatid or the chromosomal homolog. The nascent DNA joints are then extended in length by branch migration, followed by resolution of DNA intermediates to complete the recombination process (Paques and Haber 1999; Sung et al. 2000).
The RAD51 encoded product is the functional homolog of Escherichia coli RecA protein, and like RecA, possesses the ability to promote the homologous DNA pairing and strand exchange reaction that forms heteroduplex DNA joints. In mediating homologous DNA pairing and strand exchange, Rad51 must first assemble onto ssDNA as a nucleoprotein filament, in which the DNA is held in a highly extended conformation (Ogawa et al. 1993; Benson et al. 1994; Sung and Robberson 1995). Assembly of the Rad51-ssDNA nucleoprotein filament is rate-limiting and strongly inhibited by secondary structure in the ssDNA template. The removal of secondary structure can be effected by the single-strand DNA binding protein RPA, which has proved to be indispensable for homologous DNA pairing and strand exchange efficiency, especially when a plasmid-length ssDNA template is used as the initiating substrate (Sung et al. 2000; Sigurdsson et al. 2001).
Even though RPA is an important accessory factor for Rad51-mediated homologous DNA pairing and strand exchange, it can also compete with Rad51 for binding sites on the ssDNA template, which, when allowed to occur, suppresses pairing and strand exchange efficiency markedly (Sung et al. 2000). Here we demonstrate that the stoichiometric complex of the human Rad51B and Rad51C proteins, homologs of the S. cerevisiae Rad55 and Rad57 proteins (Sung et al. 2000), can partially overcome the suppressive effect of hRPA on hRad51-catalyzed DNA pairing and strand exchange, thus identifying the Rad51B–Rad51C complex as a mediator of recombination.
Results
Rad51B and Rad51C are associated in a stable complex
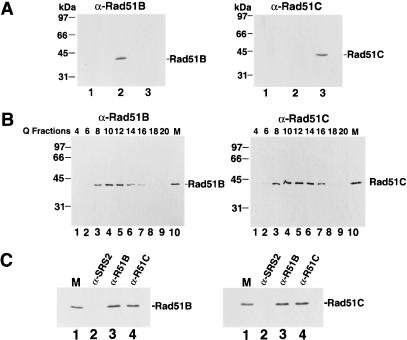
Because Rad51B and Rad51C interact in two-hybrid studies (Schild et al. 2000), we wished to address whether they are associated in human cells. For detecting Rad51B and Rad51C, we raised antibodies against these proteins expressed in E. coli and purified from inclusion bodies by preparative SDS-PAGE. The specificity of the anti-Rad51B and anti-Rad51C antibodies is shown in Figure 1A, in the left and right panels, in which extracts from yeast cells harboring the empty protein expression vector and plasmids expressing Rad51B and Rad51C were probed with the antibodies. A single 40-kD Rad51B and 42-kD Rad51C species was detected. The observed sizes of Rad51B and Rad51C are in good agreement with the predicted values of 39 kD for Rad51B and 42 kD for Rad51C. Furthermore, Rad51B and Rad51C endogenous to human HeLa cells have the same gel sizes as proteins expressed in yeast cells (see below). In both the immunoblot analysis (Fig. 1A) and immunoprecipitation (data not shown), the anti-Rad51B antibodies did not cross-react with Rad51C protein, nor did the anti-Rad51C antibodies cross-react with Rad51B protein.
Figure 1.
Rad51B and Rad51C form a stable complex. (A) Specificity of antibodies. Yeast cells harboring the empty expression vector pPM231 (2μ, GAL-PGK; lane 1), the Rad51B expression plasmid pR51B.1 (2μ, GAL-PGK-RAD51B; lane 2), and the Rad51C expression plasmid pR51C.1 (2μ, PGK-RAD51C; lane 3) were run in an 11% polyacrylamide gel and then subjected to immunoblot analysis with either anti-Rad51B (αRad51B; left panel) or anti-Rad51C antibodies (αRad51C; right panel). (B) Rad51B and Rad51C are associated in a complex in human cells. HeLa cell extract was fractionated in a Q Sepharose column, and the indicated fractions were run in an 11% gel and then subjected to immunoblot analysis with anti-Rad51B antibodies (α-Rad51B; left panel) or anti-Rad51C antibodies (α-Rad51C; right panel). Yeast extracts containing Rad51B (lane 10 in left panel, marked M) or Rad51C (lane 10 in right panel, marked M) were used to help identify these proteins in the Q column fractions. (C) Coimmunoprecipitation of Rad51B and Rad51C from the Q column fractions. The Q Sepharose pool (fractions 8–16) was subjected to immunoprecipitation with protein A beads containing anti-ySrs2 antibodies (α-SRS2), anti-Rad51B antibodies (α-R51B) and anti-Rad51C antibodies (α-R51C). Proteins bound to the various immunobeads were eluted by SDS treatment and analyzed for their content of Rad51B (left panel) and Rad51C (right panel).
To identify Rad51B and Rad51C endogenous to human cells, an extract from HeLa cells was fractionated in a Q Sepharose column, and the column fractions were subjected to immunoblotting analysis. In this analysis, we used extracts from yeast cells expressing Rad51B and Rad51C (Fig. 1B, lane 10 in both panels) to aid in the identification of the endogenous proteins. The results (Fig. 1B) showed that Rad51B and Rad51C coeluted from Q Sepharose precisely, from fractions 8 to 16. To investigate whether Rad51B and Rad51C in the Q Sepharose fractions were stably associated, we examined whether they could be coimmunoprecipitated. As shown in Figure 1C, anti-Rad51B antibodies precipitated not only Rad51B, but also Rad51C. Similarly, Rad51B coprecipitated with Rad51C in the anti-Rad51C immunoprecipitation (Fig. 1C). Importantly, the quantity of Rad51B and Rad51C that coprecipitated with the other protein was similar to the amount of these proteins precipitated by their cognate antibodies, suggesting that Rad51B and Rad51C in the Q column fractions were associated as a stable complex. Neither Rad51B nor Rad51C was precipitated by control antibodies raised against yeast Srs2 protein (Fig. 1C). Rad51C also forms a complex with XRCC3 (Schild et al. 2000; Kurumizaka et al. 2001; Masson et al. 2001), and it remains possible that a portion of Rad51C in the Q Sepharose fractions was bound to XRCC3 or that the XRCC3-Rad51C complex was not retained on the Q column.
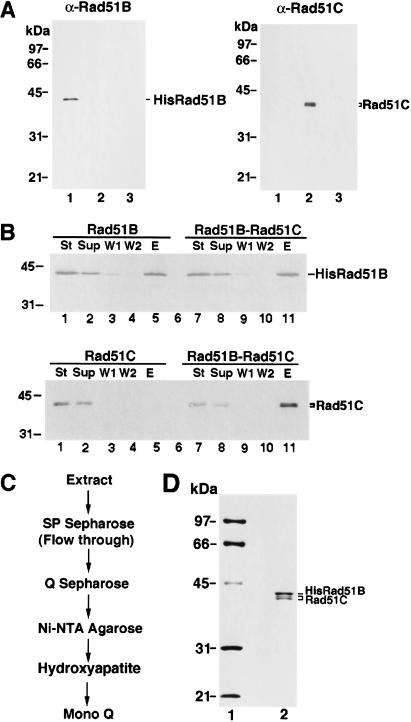
The results above indicated that Rad51B and Rad51C are associated in a stable complex in HeLa cell extract, but they could not address whether association of these two proteins was due to direct interaction between them or whether an intermediary is required. To determine whether Rad51B and Rad51C interact directly, we carried out immunoprecipitation using extracts from yeast cells that expressed these two factors. As expected, Rad51B and Rad51C were precipitated by their cognate antibodies but not by antibodies specific for the other protein (data not shown). Importantly, upon mixing of the extracts containing Rad51B and Rad51C, coprecipitation of the two proteins occurred (data not shown). Direct interaction between Rad51B and Rad51C was demonstrated another way. In this case, we constructed recombinant baculoviruses that encoded a six-histidine-tagged form of Rad51B and an untagged form of Rad51C. The expression of Rad51B and Rad51C in insect cells infected separately with these baculoviruses was verified by immunoblotting (Fig. 2A). Because Rad51B was tagged with a six-histidine sequence, we could in this instance use affinity binding of the six-histidine tag to nickel-NTA agarose as criterion for protein–protein interaction when extracts were mixed. As summarized in Figure 2B, untagged Rad51C alone did not bind the nickel matrix, whereas a significant portion of it was retained on the affinity matrix in the presence of six-histidine-tagged Rad51B. Taken together, these results made it clear that Rad51B and Rad51C form a stable complex via direct interaction. The results presented below further indicated that the complex of Rad51B and Rad51C is highly stable, and contains stoichiometric amounts of the two proteins.
Figure 2.
Purification of Rad51B and Rad51C from insect cells. (A) Expression of six-histidine-tagged Rad51B and Rad51C in insect cells. Nitrocellulose blots containing extracts from insect cells without any baculovirus (lane 3 of both panels) and infected with the recombinant 6His-tagged Rad51B baculovirus (lane 1 of both panels) or Rad51C baculovirus (lane 2 of both panels) were probed with either anti-Rad51B antibodies (αRad51B; left panel) or anti-Rad51C antibodies (αRad51C; right panel). (B) Complex formation between 6His-tagged Rad51B and Rad51C. Extracts from insect cells expressing 6His-tagged Rad51B (Rad51B), Rad51C (Rad51C), and the mixture of these extracts (Rad51B–Rad51C) were incubated with nickel-NTA agarose beads, which were washed with 10 mM, 20 mM, and then with 150 mM imadazole. The starting fractions (St), the supernatants containing unbound proteins (Sup), the 10 mM (W1) and 20 mM (W2) imidazole washes, and the 150 mM imidazole eluate (E) were subjected to immunoblotting to determine their content of Rad51B (upper panel) and Rad51C (lower panel). (C) Purification scheme for Rad51B–Rad51C complex. (D) Purity analysis. The purified Rad51B–Rad51C complex, 1.5 μg in lane 2, was run alongside molecular size markers (lane 1) in an 11% denaturing polyacrylamide gel and stained with Coomassie blue.
Rad51C expressed in insect cells consists of two closely spaced species (Fig. 2A,B), with the top band having the same gel mobility as Rad51C seen in extract from HeLa cells or yeast cells expressing this protein (data not shown). The slower migrating species of the two Rad51C immunoreactive bands is likely full-size Rad51C, whereas the faster migrating form, which represents ∼60% of the total Rad51C amount, could be a proteolytic product or the result of an aberrant expression in insect cells.
Expression and purification of the Rad51B–Rad51C complex
We used insect cells as the medium for the purification of the Rad51B–Rad51C complex for the following two reasons: (1) much larger amounts of Rad51B–Rad51C complex can be obtained from insect cells infected with the recombinant baculoviruses than from HeLa cell extract or yeast cells harboring the Rad51B and Rad51C expression plasmids, and (2) the six-histidine tag engineered in the recombinant Rad51B baculovirus allowed us to use nickel-NTA agarose as an affinity step to facilitate the purification of this complex.
We initially attempted to purify Rad51B and Rad51C individually, but our efforts were hampered by the complications that a significant portion (>75%) of these two proteins was insoluble and the soluble portion gave broad peaks during chromatographic fractionation procedures. To determine whether the Rad51B–Rad51C complex might be more amenable to purification than the individual components, we coinfected the Rad51B and Rad51C recombinant baculoviruses into insect cells. Interestingly, coexpression of Rad51B and Rad51C improved the solubility of these two proteins, even though the overall protein amounts in the infected insect cells remained relatively unchanged (data not shown). Importantly, the Rad51B–Rad51C complex eluted from various chromatographic matrices as relatively well defined peaks, thus enabling us to obtain substantial purification of the complex. Through many small-scale trials, a procedure was devised to encompass fractionation of insect cell extract containing the Rad51B–Rad51C complex in SP Sepharose, Q Sepharose, Hydroxyapatite, Mono Q, and affinity chromatography on nickel-NTA agarose (Fig. 2C) to purify this complex to near homogeneity (Fig. 2D). Stoichiometric amounts of Rad51B and Rad51C cofractionated during the entire purification procedure, indicating a high degree of stability of the complex. Using the aforementioned purification protocol, we could obtain about 100 μg of Rad51B–Rad51C complex from one liter of insect cell culture. Three independent preparations of Rad51B–Rad51C complex gave similar results in the experiments described below.
Rad51B–Rad51C complex binds DNA and hydrolyzes ATP
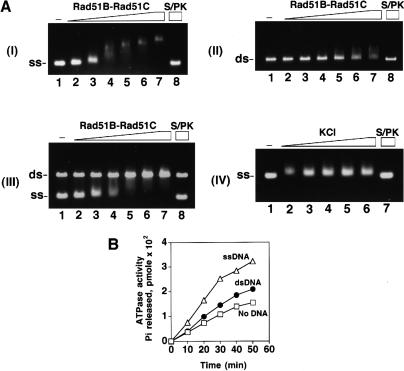
Because Rad51B and Rad51C are involved in recombination, we tested the purified Rad51B–Rad51C complex for binding to DNA. For this, an increasing amount of Rad51B–Rad51C complex was incubated with either φX ssDNA or dsDNA. The reaction mixtures were run in agarose gels, followed by staining with ethidium bromide to detect shifting of the DNA species. As shown in Figure 3A, while clear shifting of the ssDNA occurred at the lowest Rad51B–Rad51C concentration of 0.15 μM (DNA to protein ratio of 80 nucleotides/protein complex; see lane 2 of panel I), no shifting of the dsDNA was seen until the Rad51B–Rad51C concentration reached 0.6 μM (7 base pairs/protein complex; see lane 5 of panel II). These observations suggested that Rad51B–Rad51C complex binds ssDNA readily but has a lower affinity for dsDNA. To validate this conclusion, we repeated the DNA binding experiment by coincubating the ssDNA and dsDNA with the same concentration range of Rad51B–Rad51C used before. The results (Fig. 3A, panel III) revealed that Rad51B–Rad51C complex preferentially shifted the ssDNA without binding significantly to the dsDNA. In these experiments, ATP was included in the buffer used for the binding reaction, but the exclusion of ATP or its substitution with the nonhydrolyzable ATP-γ-S did not affect the binding results appreciably (data not shown). Taken together, these findings led us to conclude that the Rad51B–Rad51C complex binds ssDNA preferentially. We also examined ssDNA binding by Rad51B–Rad51C as a function of the ionic strength. To do this, a fixed quantity of Rad51B–Rad51C complex (0.3 μM) was incubated with the ssDNA (12 μM nucleotides) in the presence of increasing concentrations (50, 100, 150, 200, and 250 mM) of KCl. The results (Fig. 3A, panel IV) showed that ssDNA binding was not diminished significantly by even the highest concentration of KCl (250 mM), indicating a high degree of avidity of Rad51B–Rad51C complex for the DNA.
Figure 3.
Rad51B–Rad51C binds DNA and hydrolyzes ATP. (A) Rad51B–Rad51C complex (0.15, 0.3, 0.45, 0.6, 0.75, and 0.9 μM in lanes 2–7, respectively) was incubated with φX ssDNA (12 μM nucleotides in panel I; designated as ss), φX dsDNA (4 μM base pair in panel II; designated as ds), or with both the ssDNA and dsDNA (panel III) for 10 min at 37°C and then run in a 0.9% agarose gel. The DNA species were stained with ethidium bromide. In lane 8 of all three panels, the nucleoprotein complex formed with 0.9 μM of Rad51B–Rad51C complex was treated with 0.5% SDS and 500 μg/mL proteinase K at 37°C for 5 min before loading onto the agarose gel. In lane 1 of all three panels, DNA was incubated in buffer without protein. In panel IV, Rad51B–Rad51C complex (0.3 μM) was incubated with ssDNA (12 μM nucleotides) in the presence of increasing concentrations (50, 100, 150, 200, and 250 mM in lanes 2–6, respectively) of KCl at 37°C for 10 min and then analyzed. (B) Rad51B–Rad51C, 1.8 μM, was incubated with 1 mM ATP in the absence of DNA (designated by the squares) and in the presence of ssDNA (20 μM nucleotides; designated by the triangles) or dsDNA (20 μM base pairs; designated by the closed circles) for the indicated times at 37°C.
Both Rad51B and Rad51C contain Walker ATP binding motifs, suggestive of an ability to bind and hydrolyze ATP (Sung et al. 2000; Thompson and Schild 2001). For this reason, we examined the purified Rad51B–Rad51C complex for ATPase activity. As shown in Figure 3B, Rad51B–Rad51C complex possesses an ATP hydrolytic activity that is stimulated by DNA. Reproducibly, ssDNA was more effective at stimulating ATP hydrolysis than was dsDNA. The kcat values for the Rad51B–Rad51C ATPase were found to be 0.15/min in the absence of DNA, and 0.19/min and 0.3/min in the presence of dsDNA and ssDNA, respectively.
Effects of RPA on Rad51-mediated DNA joint formation
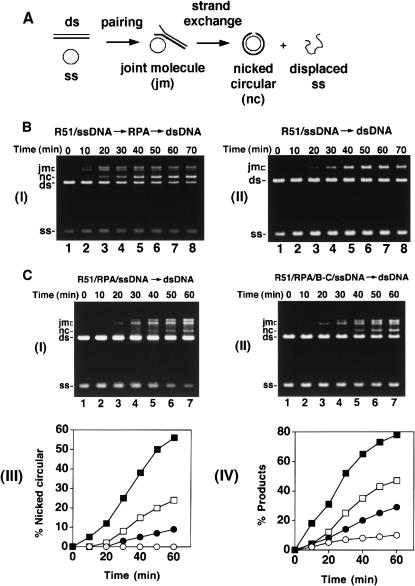
Human Rad51 (hRad51) can make joints between homologous single-stranded and double-stranded DNA molecules, but was thought to have only limited DNA strand exchange activity (Baumann and West 1997, 1999; Gupta et al. 1997). We recently described a DNA strand exchange system (Fig. 4A) wherein hRad51 mediates a substantial amount of DNA strand exchange (Sigurdsson et al. 2001). In this new system, the efficiency of homologous DNA pairing and strand exchange is strongly dependent on the heterotrimeric ssDNA binding factor RPA. To assemble the reaction, hRad51 is preincubated with circular ssDNA at the optimal ratio of 4 nucleotides per protein monomer, followed by the addition of hRPA at 20 nucleotides per protein monomer and the homologous linear duplex substrate (Fig. 4A). The reaction products, that is, joint molecules and nicked circular duplex (Fig. 4A), are separated from the input substrates by agarose gel electrophoresis and visualized by staining with ethidium bromide. An example of this reaction is shown in Figure 4B, panel I. As reported earlier (Sigurdsson et al. 2001) and reiterated here, omission of RPA from the reaction reduced the amount of products markedly (Fig. 4B, panel II). Importantly, little or no nicked circular duplex, the product of full strand exchange, was generated in the reaction that did not contain RPA (Fig. 4B, panel II).
Figure 4.
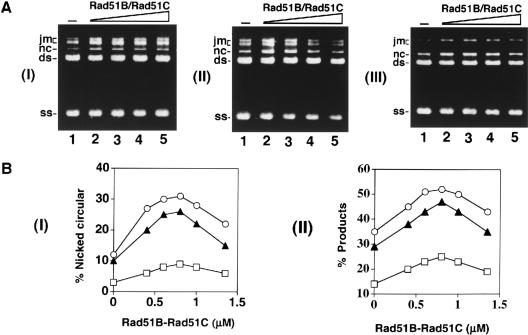
Mediator function of Rad51B–Rad51C. (A) Schematic of the homologous DNA pairing and strand exchange reaction using φX174 DNA substrates. Linear duplex is paired with the homologous ssDNA circle to yield a joint molecule. DNA strand exchange, if successful over the length (5.4 kb) of the DNA molecules, results in the formation of the nicked circular duplex. (B) Rad51-mediated DNA pairing and strand exchange was carried out with RPA (panel I) or without it (panel II). In panel I, the ssDNA was preincubated with Rad51 (R51) before RPA was added. The concentrations of the reaction components were: Rad51, 7.5 μM; RPA, 1.5 μM; ssDNA, 30 μM nucleotides; linear duplex, 15 μM base pairs. (C) In the DNA strand exchange reaction in panel I, the ssDNA was incubated with both Rad51 (R51) and RPA simultaneously, and in the reaction in panel II, the ssDNA was incubated with Rad51, RPA and Rad51B–Rad51C (B–C) simultaneously. The concentration of Rad51B–Rad51C was 0.8 μM, while the concentrations of the other components were exactly as those in B. In panel III, the amounts of nicked circular duplex in the reactions represented in B panel I (filled squares) and panel II (open circles) and in C panel I (filled circles) and panel II (open squares) are plotted. In panel IV, the amounts of total reaction products (sum of joint molecules and nicked circular duplex) in the reactions represented in B panel I (filled squares) and panel II (open circles) and in C panel I (filled circles) and panel II (open squares) are plotted.
Interestingly, coaddition of RPA with Rad51 to the ssDNA to mimic what may be expected to occur in vivo resulted in a marked reduction in reaction efficiency (Fig. 4C, panels I, III, and IV). Specifically, while ∼38% and ∼56% of the input linear duplex substrate had been converted into nicked circular duplex after 40 min and 60 min in the standard reaction (Fig. 4B, panel I and 4C, panel III), coaddition of hRPA with hRad51 to the ssDNA yielded only 5% and 9% of nicked circular duplex after these reaction time (Fig. 4C, panels I and III). In the homologous DNA pairing and strand exchange reaction mediated by E. coli RecA or yeast Rad51, SSB/yRPA added to the ssDNA template before or at the same time as the recombinase causes a notable suppression of the reaction as well (Umezu et al. 1993; Sung 1997a; New et al. 1998; Shinohara and Ogawa 1998). In these cases, suppression of the reaction efficiency by SSB/yRPA is due to competition of these ssDNA binding factors with RecA/yRad51 for sites on the ssDNA template. Based on the paradigm established with the E. coli and yeast recombination systems (Bianco et al. 1998; Sung et al. 2000), we also attribute the suppression of hRad51-mediated DNA pairing and strand exchange by hRPA to the exclusion of hRad51 from the ssDNA template.
Rad51B–Rad51C complex has a presynaptic mediator function
Rad51B and Rad51C are both required for recombination and DNA double-strand break repair in vivo. In chicken DT40 cells deleted for either Rad51B or Rad51C, the assembly of Rad51 nuclear foci in response to DNA damage is compromised (Takata et al. 2000, 2001). Based on these results, we wished to test whether Rad51B–Rad51C could promote homologous DNA pairing and strand exchange by Rad51 with RPA competing for binding sites on the initiating ssDNA substrate. To do this, we added Rad51B–Rad51C complex (0.8 μM) with Rad51 (7.5 μM) and RPA (1.5 μM) during the preincubation with ssDNA (30 μM), and then completed the reaction mixture by adding the homologous duplex. Importantly, upon inclusion of the Rad51B–Rad51C complex, significantly higher amounts of the reaction products were seen (Fig. 4C, panels II, III, and IV). In particular, the level of the nicked circular duplex, product of complete strand exchange (see Fig. 4A), was formed at a significantly higher rate than in the absence of Rad51B–Rad51C (Fig. 4C, panels II and III). We also examined whether amounts of Rad51B–Rad51C below and above that used before would lead to different levels of homologous DNA pairing and strand exchange. As shown in Figure 5, the optimal concentration of Rad51B–Rad51C was from 0.4 to 1.0 μM, and amounts of Rad51B–Rad51C above the optimal level in fact led to gradual inhibition of the reaction (Fig. 5; data not shown).
Figure 5.
Mediator activity as a function of Rad51B–Rad51C concentration. (A) The φX174 ssDNA template (30 μM nucleotides) was incubated with Rad51 (7.5 μM), RPA (1.5 μM), and increasing concentrations of Rad51B–Rad51C (0, 0.6, 0.8, 1.0, and 1.4 μM in lanes 1–5, respectively) for 10 min before the φX174 linear duplex (15 μM base pairs) was incorporated to complete the reaction mixtures. Portions of the reaction mixtures were withdrawn at 30 min (panel I), 60 min (panel II), and 80 min (panel III) and then processed for agarose gel electrophoresis. (B) The results from A and from two other independent experiments performed under the same reaction conditions were compiled and graphed. Symbols: results from the 30 min timepoint (squares), the 60 min timepoint (filled triangles), and the 80 min timepoint (circles). Panel I shows the levels of nicked circular duplex formed, and panel II shows the amounts of total reaction products (joint molecules and nicked circular duplex).
One possible explanation for the stimulatory effect of Rad51B–Rad51C complex (Fig. 5) is that this protein complex promotes homologous DNA pairing and strand exchange regardless of the order of addition of RPA. To test this possibility, we used the same amount of Rad51B–Rad51C complex (0.8 μM) that afforded the maximal restoration of DNA pairing and strand exchange (see Fig. 5A,B) in reactions wherein the protein complex was (1) added with Rad51 to the ssDNA substrate, followed by RPA; (2) added with RPA after Rad51 had already nucleated onto the ssDNA substrate; and (3) added after the ssDNA had first been incubated with Rad51 and then with RPA. No measurable effect of the Rad51B–Rad51C complex on the rate of formation of joint molecules and nicked circular duplex was recorded in any of these experiments (data not shown). We also tested whether the inclusion of Rad51B–Rad51C complex would alter the concentration of Rad51 needed for optimal pairing and strand exchange, determined previously to be from 3 to 4 nucleotides per Rad51 monomer (Baumann and West 1997, 1999; Gupta et al. 1997; Sigurdsson et al. 2001). However, Rad51B–Rad51C did not change the concentration of Rad51 needed for optimal reaction efficiency (data not shown).
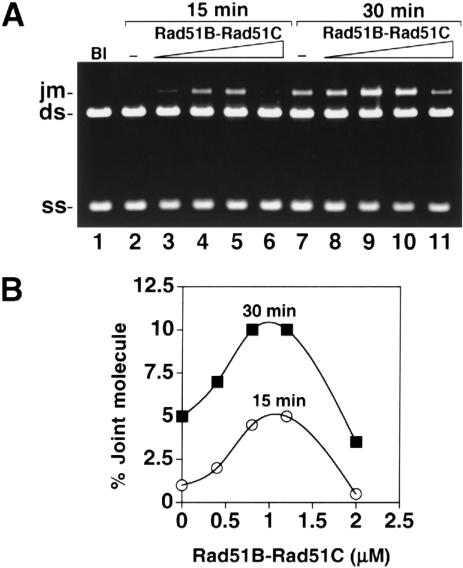
To examine whether the Rad51B–Rad51C complex can substitute for RPA in the DNA pairing and strand exchange reaction, we used a range of Rad51B–Rad51C concentrations (0.4, 0.8, 1.2, and 2.0 μM) with a fixed concentration of Rad51 (7.5 μM) and ssDNA (30 μM) without RPA. Rad51B–Rad51C did not promote the formation of nicked circular duplex (Fig. 6) even after 60 min of reaction (data not shown), showing that it cannot substitute for RPA, which is highly effective in enabling Rad51 to make nicked circular duplex (Fig. 4; Sigurdsson et al. 2001). Interestingly, we observed an increase in joint molecules at 0.8 and 1.2 μM of Rad51B–Rad51C complex (Fig. 6). Specifically, after 30 min, the level of DNA joint molecules increased from ∼5% in the absence of Rad51B–Rad51C to ∼10% upon the inclusion of 1.2 μM Rad51B–Rad51C. As with the earlier strand exchange restoration experiment (Fig. 5), suppression of DNA joint molecule formation was seen at higher concentrations of the Rad51B–Rad51C complex (Fig. 6A,B; data not shown). In other experiments, over the range of Rad51B–Rad51C concentration from 3 to 45 nucleotides/protein complex, we did not detect any DNA joint molecule with the φX substrates, regardless of whether RPA was present (data not shown). These observations suggested that the Rad51B–Rad51C complex is devoid of homologous DNA pairing activity.
Figure 6.
Rad51B–Rad51C stimulates DNA joint formation but does not replace RPA in DNA strand exchange. (A) Rad51 and Rad51B–Rad51C were used in the homologous DNA pairing and strand exchange reaction without RPA. The concentrations of the reaction components were: Rad51, 7.5 μM; Rad51B–Rad51C, 0.4 to 2 μM, as indicated; ssDNA, 30 μM nucleotides; linear duplex, 15 μM base pairs. (B) Graphic representation of the results from the experiment in A.
Discussion
Paradoxical effects of RPA on homologous DNA pairing and strand exchange
Like other members of the RecA/Rad51 class of recombinases (Ogawa et al. 1993; Sung and Robberson 1995; Roca and Cox 1997; Bianco et al. 1998), hRad51 assembles onto ssDNA to form a nucleoprotein filament in an ATP-dependent manner (Benson et al. 1994). Extensive biochemical studies with RecA have indicated that the search for DNA homology, DNA joint formation, and DNA strand exchange all occur within the confines of the RecA-ssDNA nucleoprotein filament. The assembly of the recombinase-ssDNA nucleoprotein filament is therefore the critical first step in the homologous DNA pairing and strand exchange reaction, and is generally referred to as the presynaptic phase of this reaction (Roca and Cox 1997; Bianco et al. 1998; Sung et al. 2000).
For RecA and yeast Rad51, the assembly of the presynaptic filament on plasmid-length DNA molecules is dependent on the cognate single-strand binding protein, E. coli SSB or yeast RPA, which acts to minimize the secondary structure in the DNA template and hence renders extension of the nascent nucleoprotein filament facile. In most experimental systems, the single-strand DNA binding factor is added subsequent to the recombinase, that is, after nucleation of the recombinase onto the ssDNA substrate has already commenced. Interestingly, precoating of the ssDNA template with the single-strand binding protein (Umezu et al. 1993; Sugiyama et al. 1997; New et al. 1998; Shinohara and Ogawa 1998) or coincubation of RPA, Rad51, and the ssDNA substrate (Sung 1997a,b) results in pronounced suppression of the reaction efficiency. We have shown in the present study that the hRad51 recombinase activity is similarly suppressed by RPA during the presynaptic phase. Taken together, the results indicate that the single-strand binding protein, while important for secondary structure removal in the ssDNA template, can interfere with the nucleation of the recombinase onto the DNA template and can thus prevent the formation of a contiguous presynaptic filament.
Mediator function in the Rad51B–Rad51C complex
Exploiting the paradoxical behavior of the single-strand DNA binding protein in the assembly of the presynaptic recombinase filament, various recombination mediator proteins capable of overcoming the suppressive effect of the single-strand binding protein have been identified in prokaryotes and yeast cells (Umezu et al. 1993; Sung 1997a,b; New et al. 1998; Shinohara and Ogawa 1998; Beernink and Morrical 1999). In yeast, two such mediators—Rad52 and the Rad55–Rad57 complex—have been described. Rad55 and Rad57 are similar to Rad51B and Rad51C in that they too exhibit homology to Rad51 and form a stable stoichiometric complex via direct interaction (Sung et al. 2000). Importantly, both Rad51B–Rad51C and Rad55–Rad57 complexes are needed for the assembly of Rad51 nuclear foci in response to DNA damaging treatment (Gasior et al. 1998, 2001; Takata et al. 2000, 2001), which are thought to correspond to the sites of ongoing DNA damage repair.
We show here that purified Rad51B–Rad51C complex has single-strand DNA binding and ssDNA-stimulated ATPase activities. Importantly, Rad51B–Rad51C can promote the Rad51-mediated homologous DNA pairing and strand exchange reaction under conditions wherein Rad51 must contend with the competition by RPA for binding sites on the initiating ssDNA substrate. In addition, DNA joint formation by Rad51 in the absence of RPA is also enhanced by Rad51B–Rad51C. We asked whether Rad51B–Rad51C could stimulate the rate of DNA pairing and strand exchange, but did not uncover a postsynaptic role (Roca and Cox 1997; Bianco et al. 1998; Sung et al. 2000) in this protein complex. Rad51B–Rad51C complex did not pair the φX DNA substrates used, nor did it lower the concentration of Rad51 needed to attain optimal reaction efficiency. Thus, the results of our study suggest a specific role of Rad51B–Rad51C in promoting the assembly of the presynaptic Rad51 filament, identifying it as a mediator of recombination, in a manner analogous to what has been described for the yeast Rad55–Rad57 complex (Sung et al. 2000). Given that both Rad51B and Rad51C are indispensable for recombination and DNA repair and needed for the normal assembly of Rad51 nuclear foci after DNA damaging treatment, we surmise that the Rad51B–Rad51C mediator function likely contributes to the recruitment of hRad51 to sites of recombination and DNA damage repair.
Role of other Rad51 paralogs in recombination processes
Aside from Rad51B and Rad51C, three additional Rad51 paralogs, namely, XRCC2, XRCC3, and Rad51D, have been described (Thompson and Schild 2001). XRCC3 and Rad51C also form a stable complex that has DNA binding activity (Kurumizaka et al. 2001; Masson et al. 2001). Rad51D (also called Rad51L3) has DNA binding and ssDNA-stimulated ATPase activities, and it forms a stable complex with XRCC2 (Braybrooke et al. 2000). Whether the XRCC2–Rad51D and XRCC3–Rad51C complexes also have a recombination mediator function remains to be determined. However, that the formation of DNA damage-induced Rad51 nuclear foci is impaired in mutants of all five of the Rad51 paralogs would seem to suggest that they all play a role in the delivery of Rad51 to DNA lesions in vivo. It thus seems possible that some combination of complexes of the Rad51 paralogs could have an enhanced mediator function in the Rad51-catalyzed homologous DNA pairing and strand exchange reaction. In fact, our observation that Rad51B–Rad51C only partially overcomes the suppressive effect of RPA is consistent with such a scenario. It remains to be seen whether the other two Rad51 paralog pairs, either by themselves or in combination with each other and with Rad51B–Rad51C, may also function in the postsynaptic phase of the pairing and strand exchange reaction by enhancing the efficiency of DNA joint formation and promoting DNA branch migration.
Materials and methods
Antibodies
Rad51B protein was expressed in E. coli BL21 (DE3) using the T7 promoter in the vector pET11a. The insoluble Rad51B protein was purified by preparative SDS-PAGE, dialyzed into phosphate-buffered saline (10 mM NaH2PO4 at pH 7.2, 150 mM NaCl), and then used as antigen for production of antisera in rabbits. A GST–Rad51C fusion protein was expressed in E. coli BL21 (DE3) using the vector pGEX-2T. The GST–Rad51C fusion protein is also insoluble and was purified for antibody production in rabbits as described for Rad51B. The antigens were covalently conjugated to cyanogen bromide activated Sepharose 4B (Pharmacia-LKB) for use as affinity matrices to purify monospecific antibodies from rabbit antisera (Sung et al. 1987).
DNA substrates
φX174 viral (+) strand was purchased from New England Biolabs, and the φX174 replicative form I DNA was from GIBCO BRL. The replicative form I DNA was linearized with ApaLI. All of the DNA substrates were stored in TE (10 mM Tris-HCl at pH 7.5, 0.5 mM EDTA).
Fractionation of HeLa cell extract
To make extract, 10.4 g of HeLa S3 cells (National Cell Culture Center) was suspended in 15 mL of cell breakage buffer (50 mM Tris/HCl at pH 7.5, 2 mM EDTA, 10% sucrose, 100 mM KCl, 1 mM dithiothreitol and the following protease inhibitors: aprotinin, chymostatin, leupeptin, pepstatin, all at 3 μg/mL, and 1 mM phenylmethylsulfonyl fluoride) and passed through a French Press at 20,000 psi. After centrifugation (100,000g for 1 h), the clarified extract (20 mL) was loaded onto a column of Q-Sepharose (4 mL), which was fractionated with a 45 mL gradient of KCl from 100 to 600 mM in K buffer (20 mM KH2PO4 at pH 7.4, 0.5 mM EDTA, 1 mM DTT, and 10% glycerol) collecting 30 fractions. To identify the Rad51B and Rad51C proteins, the Q-Sepharose fractions (8 μL) were subjected to immunoblot analyses with anti-Rad51B and anti-Rad51C antibodies after SDS-PAGE in 11% gels.
Expression of Rad51B and Rad51C in yeast cells
The RAD51B gene was placed under the control of the GAL-PGK promoter in vector pPM231 (2μ, GAL-PGK, LEU2d) to yield pRad51B.1 (2μ, GAL-PGK-RAD51B, LEU2d) and RAD51C were placed under the control of the PGK promoter in vector pPM255 (2μ, PGK, URA3) to yield pRad51C.1 (2μ, PGK-RAD51C, URA3). The empty expression vector, pR51B.1, and pR51C.1 were introduced into the protease-deficient yeast strain BJ5464 (MATα, ura3-52, trp-1, leu2Δ1, his3Δ200, pep4∷HIS3, prbΔ1.6R). Induction of Rad51B and Rad51C followed the protocol of Petukhova et al. (2000). Extracts (2 mL of buffer per gram of yeast cells) were prepared using a French press and clarified by centrifugation, as described above.
Purification of the Rad51B–Rad51C complex
Six-histidine (6His)-tagged Rad51B and untagged Rad51C recombinant baculoviruses were constructed by cloning the cDNAs for these proteins into the baculovirus transfer vector pVL1393 (Invitrogen). Amplification of the recombinant viruses was carried out in Sf9 cells and for protein expression, High-Five insect cells (Invitrogen) were infected with the 6His-tagged Rad51B and Rad51C baculoviruses at an MOI of 5. The insect cells were harvested 36–48 h postinfection. For the purification of the Rad51B–Rad51C complex, High-Five cells were coinfected with the Rad51B and Rad51C baculoviruses at an MOI of 5 for both viruses. The insect cells were harvested 36–48 h postinfection.
All the protein purification steps were carried out at 0 to 4°C. Cell lysate was prepared from 1 L of insect cell culture (15 g) using a French press in 60 mL of cell breakage buffer. The lysate was clarified by centrifugation (100,000g for 1 h) and the supernatant was applied on a 10 mL SP-Sepharose column. The flow-through from SP-Sepharose was then fractionated in a column of Q-Sepharose (10 mL of matrix) with a 30 mL, 100 to 600 mM KCl gradient in buffer K. Rad51B–Rad51C eluted from 200–300 mM KCl, and the peak fractions were pooled and mixed with 0.6 mL of Ni-NTA agarose for 2 h, followed by fractionation with a 15 mL, 0 to 250 mM imidazole in buffer K. The Rad51B–Rad51C peak fractions were pooled and further fractionated in a column of Macro-hydroxyapatite (0.5 mL matrix) with a 15 mL, 0 to 180 mM KH2PO4 gradient in buffer K. The peak fractions were pooled and dialyzed against buffer K with 50 mM KCl and applied onto a Mono Q column (HR5/5), which was developed with a 15 mL gradient from 100 to 800 mM KCl in buffer K. Rad51B–Rad51C from the Mono Q step was concentrated in a Centricon-30 microconcentrator to 2 mg/mL and stored at −70°C.
Immunoprecipitation
Affinity-purified anti-Rad51B, anti-Rad51C, and anti-ySrs2 antibodies, 1.0 mg each, were coupled to 300 μL protein A agarose beads as described previously (Sung 1997a). In Figure 1C, 0.3 mL of the Q-Sepharose pool (fractions 8 to 16) containing the Rad51B–Rad51C peak was mixed with 10 μL of protein A beads containing anti-Rad51B, anti-Rad51C, or anti-ySrs2 antibodies at 4°C for 5 h. The beads were washed twice with 200 μL of buffer K containing 1 M KCl and once with 200 μL of buffer K, before being eluted with 20 μL of 2% SDS at 37°C for 10 min. The SDS eluates, 2 μL each, were analyzed by immunoblotting to reveal their content of Rad51B and Rad51C.
Affinity binding of the Rad51B–Rad51C complex to nickel-NTA agarose
Extract from 0.5 mL packed cell volume of High-Five insect cells harboring either the six-histidine-tagged Rad51B or Rad51C recombinant baculovirus was prepared as described above, using 3 mL of cell breakage buffer. After ultracentrifugation (100,000g for 1 h), 0.25 mL of the Rad51B and Rad51C containing extracts were mixed either with each other or with 0.25 mL of extract from insect cells that did not contain any recombinant baculovirus. After incubation on ice for 1 h, the various mixtures were subjected to ultracentrifugation (100,000g for 1h), and then rocked gently with 50 μL of nickel-NTA agarose beads (QIAGEN) at 4°C for 3 h. The beads were washed sequentially with 0.2 mL of 10 mM and 20 mM imidazole, and then with 0.1 mL of 150 mM imidazole. The starting fractions, supernatants after nickel-binding, and the three imidazole washes, 5 μL each, were subjected to immunoblot analysis to determine their content of Rad51B and Rad51C.
Other recombination proteins
Human Rad51 (hRad51 Lys313) was expressed in and purified from E. coli as described (Sigurdsson et al. 2001). Human RPA was purified from E. coli cells transformed with a plasmid that co-overexpresses the three subunits of this factor (Henricksen et al. 1994), as described (Sigurdsson et al. 2001). Both hRad51 and hRPA were nearly homogeneous.
Homologous DNA pairing and strand exchange reactions
All the reaction steps were carried out at 37°C. The reaction buffer was 40 mM Tris-HCl (pH 7.8), 2 mM ATP, 1 mM MgCl2, and 1 mM DTT, and contained an ATP regenerating system consisting of 8 mM creatine phosphate and 28 μg/mL creatine kinase. The standard reaction had a final volume of 12.5 μL, and was assembled by first incubating hRad51 (7.5 μM) added in 0.5 μL of storage buffer and φX174 viral (+) strand (30 μM nucleotides) added in 1 μL for 5 min. After this, hRPA (1.5 μM) in 0.5 μL of storage buffer was added and following a 5 min incubation, 1.25 μL of ammonium sulfate (1 M stock, final concentration of 100 mM) was incorporated. Immediately afterward, linear φX174 replicative form I DNA (15 μM base pairs) in 1 μL was added to complete the reaction. At the indicated times, 5 μL portions were withdrawn, mixed with 7.5 μL of 0.8% SDS and 800 μg/mL proteinase K, incubated for 15 min at 37°C, before being run in 0.9% agarose gels in TAE buffer (40 mM Tris acetate at pH 7.4, 0.5 mM EDTA). The gels were stained in ethidium bromide (2 μg/mL in H2O) for 1 h, destained for 12 to 18 h in a large volume of water at 4°C, and then subjected to image analysis in a NucleoTech gel documentation station equipped with a CCD camera. In some experiments, the reaction was scaled up appropriately to accommodate the increased number of timepoints used. In Figure 4B, panel II, storage buffer was added instead of RPA, but otherwise the reaction was assembled in exactly the same manner as the standard reaction. In Figure 5A, panel I, Rad51 and RPA were added together to ssDNA at the beginning of the reaction and incubated for 10 min with the latter, but otherwise the additions of the ammonium sulfate and linear φX duplex followed the procedure described for the standard reaction. In Figure 5A, panel II and in Figure 5B, Rad51, RPA, and the indicated amounts of Rad51B–Rad51C were incubated with the ssDNA for 10 min, but otherwise the additions of the ammonium sulfate and linear φX duplex followed the procedure described for the standard reaction. In Figure 6, Rad51 and the indicated amounts of Rad51B–Rad51C were incubated with the ssDNA for 10 min, but otherwise the additions of the ammonium sulfate and linear φX duplex followed the procedure described for the standard reaction.
DNA mobility shift and ATPase assays
The indicated amounts of Rad51B–Rad51C complex were incubated with φX ssDNA (12 μM nucleotides), dsDNA (4 μM base pairs), or both ssDNA (12 μM nucleotides) and dsDNA (4 μM base pairs) in reaction buffer (50 mM Tris-HCl at pH 7.8, 1 mM DTT, 100 μg/mL BSA, 1 mM MgCl2, 1 mM ATP, and 100 mM KCl) for 10 min at 37°C. After electrophoresis in 0.9% agarose gels in TAE buffer at 4°C, the gels were stained in ethidium bromide (2 μg/mL in H2O) for 1 h and destained at 4°C for 12 to 18 h, before being subjected to image analysis in the gel documentation station.
Rad51B–Rad51C (1.8 μM) was incubated in the absence or presence of ssDNA (20 μM nucleotides) or dsDNA (20 μM base pairs) in 10 μL of reaction buffer (50 mM Tris-HCl at pH 7.8, 1 mM DTT, 1 mM MgCl2) containing 1 mM [γ-32P]ATP for the indicated times at 37°C. The level of ATP hydrolysis was determined by thin layer chromatography as described previously (Petukhova et al. 2000).
Acknowledgments
We thank Susie Zhang for constructing the Rad51C recombinant baculovirus. This work was supported by Public Health Service grants PO1CA81020, GM57814, ES07061, GM30990, CA81019-01 and by California Breast Cancer Research Program grant 5KB-0123. S.S. was supported in part by U.S. Army Training Grant DAMD17-99-1-9402 and Predoctoral Fellowship DAMD-17-01-1-0412. S.V.K. was supported in part by U.S. Army Predoctoral Fellowship DAMD-17-01-1-0414.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sung@uthscsa.edu; FAX (210) 567-7277.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.935501.
References
- Baumann P, West SC. The human Rad51 protein: Polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Heteroduplex formation by human Rad51 protein: Effects of DNA end-structure, hRP-A and hRad52. J Mol Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- Beernink HT, Morrical SW. RMPs: Recombination/replication mediator proteins. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- Benson FE, Stasiak A, West SC. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;23:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Braybrooke JP, Spink KG, Thacker J, Hickson ID. The Rad51 family member, Rad51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J Biol Chem. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes & Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc Natl Acad Sci. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T. Homologous-pairing activity of the human DNA-repair proteins Xrcc3–Rad51C. Proc Natl Acad Sci. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human Rad51C and XRCC3 recombination repair proteins. Proc Natl Acad Sci. 2001;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;2:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes & Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca AI, Cox MM. RecA protein: Structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Schild D, Lio Y, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997a;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- ————— Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes & Dev. 1997b;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Sung P, Prakash L, Matson SW, Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci. 1987;84:8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Trujillo K, Van Komen S. Recombination factors of Saccharomyces cerevisiae. Mutat Res. 2000;451:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers MA, Kanaar R, Thompson LH, Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc Natl Acad Sci. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]