Abstract

Decades of genetics research comparing mouse strains has identified many regions of the genome associated with quantitative traits. Microarrays have been used to identify which genes in those regions are differentially expressed and are therefore potentially causal; however, genetic variants that affect probe hybridization lead to many false conclusions. Here we used spectral counting to compare brain striata between two mouse strains. Using strain-specific protein databases, we concluded that proteomics was more robust to sequence differences than microarrays; however, some proteins were still significantly affected. To generate strain-specific databases, we used a complete database that contained all of the putative genetic isoforms for each protein. While the increased proteome coverage in the databases led to a 6.8% gain in peptide assignments compared to a nonredundant database, it also necessitated the development of a strategy for grouping similar proteins due to a large number of shared peptides. Of the 4563 identified proteins (2.1% FDR), there were 1807 quantifiable proteins/groups that exceeded minimum count cutoffs. With four pooled biological replicates per strain, we used quantile normalization, ComBat (a package that adjusts for batch effects), and edgeR (a package for differential expression analysis of count data) to identify 101 differentially expressed proteins/groups, 84 of which had a coding region within one of the genomic regions of interest identified by the Portland Alcohol Research Center.
Keywords: spectral counting, genetics, quantitative trait loci (QTL), single nucleotide polymorphisms (SNPs), substitutions, shared peptides, protein groups, protein database, batch effects, normalization
Short abstract
This study used spectral counting to compare two strains of mice used in genetics research. Similar studies using microarrays suffered from experimental artifacts due to sequence variants between strains. This work evaluated the impact of sequence variants on proteomics by using strain-specific protein databases. These highly redundant databases necessitated an approach for grouping similar proteins. Several grouping strategies are compared as well as several normalization and differential expression analysis approaches.
Introduction
Differences between individuals in a population are caused by genetic and environmental factors. Determining the influence of genomic variants on phenotypic traits in humans is challenging and requires very large sample sizes due to genetic complexity and environmental confounders. One alternative approach is to use model organisms where environment and breeding can be controlled. Genetic research in mice began in 1902, and successive generations of inbreeding have led to many genetically stable strains where tightly controlled housing and diet conditions reduce environmental noise.
One way to identify genes of interest for a quantitative trait is to cross two inbred strains that are widely divergent for the trait, measure the trait in the F2 offspring mice, and genotype the F2 mice to determine which genomic regions are associated with the trait. These regions are referred to as Quantitative Trait Loci (QTLs). The Portland Alcohol Research Center (PARC) has identified many QTLs that are responsible for differences in alcohol-drinking-related behaviors(1) between the two mouse strains investigated in this study.
QTL regions are often very broad and contain many genes. It is difficult to determine which gene, termed “quantitative trait gene”, is actually influencing the trait. An approach that the PARC has taken is to measure mRNA expression levels in regions of the brain that are expected to participate in alcohol-related decisions. Genes with coding regions that lie within the QTL regions and that are differentially expressed between the strains are suspect quantitative trait genes. However, searching for differentially expressed mRNAs between two mouse strains using microarrays is problematic. Genetic differences between the strains cause many false positives and negatives when a probe consistently hybridizes in one strain and does not in the other. In these strains, 16% of the Affymetrix mouse array has affected probes leading to a false positive rate of 22% and a false negative rate of 12%.(2) Similar issues have been found with human arrays.(3)
In this study, we compared these strains using quantitative proteomics. To our knowledge, this is the first time these strains have been compared using quantitative proteomics. Protein expression is important in searches for quantitative trait genes because studies have shown that protein levels generally do not correlate well with mRNA levels.4−10 Proteins that have coding regions that lie within QTL regions and that are differentially expressed between the strains would be putative “quantitative trait proteins”. We also investigated the influence of genetic differences on proteomic methodologies. If a genetic difference changes a protein sequence, then the peptide containing the substitution will likely not be identified. Using genome sequence data, we built strain-specific protein databases to evaluate the effect of genetic variants on peptide identification and protein quantification. This necessitated the use of complete protein databases constructed to contain all of the known gene duplication and alternative splicing isoforms for all of the proteins. We evaluated several protein grouping approaches to reduce spectral counting errors when proteins share large fractions of their identified peptides. These cases occur more frequently in complete databases with high levels of sequence similarity. We also compared our approach to results obtained when searching a database with little sequence redundancy.
Experimental Methods
Sample Collection and Processing
All animal handling procedures were done in accordance with federal guidelines and approved by the OHSU IACUC. Adult 10-week-old male ethanol-naïve mice (C57BL/6 (B6) and DBA/2 (D2)) were sacrificed and whole striata were immediately dissected from their brains and snap frozen until further processing. Four biological replicates from each strain were analyzed where each biological replicate consisted of a pool of tissue from six mice to reduce within-strain variation and provide sufficient protein (Figure 1). The experiments were performed in two batches approximately seven months apart. Two replicates from each strain were analyzed in each batch. A protocol developed by Smit et al.11,12 was used to deplete mitochondrial and structural proteins and to aid in the identification and quantification of less-abundant synaptic proteins. Following suspension of the final synaptosome pellet in 0.5 mL 5 mM Hepes (pH 7.4) buffer, a protein assay was performed (BCA assay kit, Pierce, Rockville, IL), and 500 μg portions of protein were dried by vacuum centrifugation.
Figure 1.
Experimental Design and Spearman-Rank clustering of samples before normalization, after normalization, and after batch adjustment. Striata from six mice were pooled for each sample to reduce within-strain variation and to obtain enough protein. Batch 1 contained samples B61, B62, D21, and D22. Batch 2 contained samples B63, B64, D23, and D24. The strains formed B6 and D2 clusters only after applying batch corrections.
Protein Digestion, Peptide Separation, Mass Spectrometry
The 500 μg portions of synaptosome proteins were suspended in 100 μL of 100 mM ammonium bicarbonate buffer containing 4 mg/mL RapiGest SF detergent (Waters, Milford, MA), reduced by addition of 10 μL of 100 mM dithioerythritol, and incubated at 60 °C for 30 min. Alkylation of free cysteines was then performed by addition of 30 μL of 100 mM iodoacetamide and incubation at room temperature for 30 min. Sixty microliters of 0.3 mg/mL trypsin (Proteomics grade, Sigma, St Louis, MO) was then added and the samples digested overnight at 37 °C with shaking. Detergent was then removed by addition of 200 μL of 2% trifluoroacetic acid, incubation at 37 °C for 45 min, centrifugation at 8000× g for 15 min, and removal of the supernatant. Digests were then solid phase extracted (Sep Pak Light Cartridges, Waters Corp) and peptides were separated by cation exchange chromatography into 35 fractions using a polysulfethyl A column (PolyLC Inc., Columbia MD) as previously described.(13) Of each cation exchange fraction, 40% was then separated by reverse phase chromatography and 100 min of tandem mass spectrometry data was collected for each of the 35 fractions using an LTQ linear ion trap (Thermo Scientific, San Jose, CA) as previously described.(14)
Database Searches
Peptide identification was performed using SEQUEST (Version 28, rev. 12, Thermo Fisher). Parent ion average mass tolerance was 2.5 Da and monoisotopic fragment ion tolerance was 1.0 Da. Tryptic cleavage was specified with a static modification of +57 Da on cysteine residues and a variable modification of +16 Da on methionines. A pipeline developed in-house was used to identify peptides and proteins with carefully controlled false discovery rates estimated using sequence-reversed databases.(15) Protein identification criteria were two distinct, fully tryptic peptides per protein per sample. All samples were searched against three different protein databases: UniProtKB/Swiss-Prot (release 57.8; 16 191 entries; reviewed canonical sequences) and two versions of the Ensembl protein database (release 57; 35 412 entries; ab initio predicted proteins were not included), one representing the B6 strain and one representing the D2 strain. Protein sequences that were exact duplicates or exact subsets of another protein sequence from the same gene were removed from the Ensembl databases before searching. The Ensembl genome is based on the B6 strain, so the reference Mus musculus protein database was used as the B6 database. To generate a D2-specific database, the D2 pileup file (dated 12/9/2009) containing over 5 million genomic single nucleotide polymorphisms (SNPs) and short insertions and deletions (InDels) was downloaded from the Wellcome Trust Sanger Institute Mouse Genomes Project ftp site. Using the Ensembl Perl API, the SNPs and InDels were inserted into the correct locations in the transcripts and the proteins were retranslated. Approximately 20% of the proteins had altered sequences and 0.25% had premature stop codons. Unless otherwise noted, the quantitative results in this paper were calculated using counts from the B6 (reference) Ensembl database for the B6 samples and the D2 Ensembl database for the D2 samples.
Protein Group Summarization and Database Comparison
Sequence similarities in the Ensembl protein databases resulted in large numbers of ambiguous (shared) peptides that were assigned to multiple proteins. Methods for splitting shared peptides using unique peptide information have been proposed and have been shown to provide more accurate protein total counts.16,17 Splitting peptides on the basis of relative unique peptide counts, however, fails when unique counts are too low. To avoid these errors, we evaluated two methods to identify and group similar proteins before applying peptide splitting.
The first method grouped proteins that belong to the same Ensembl protein family. Ensembl provides protein family annotations for each of its proteins. Proteins were clustered into protein families based on sequence similarity (for more details see http://www.ensembl.org/info/docs/compara/family.html).
In the second grouping method, all pairwise comparisons of proteins were performed, and proteins A and B were merged into one group if both proteins had fewer than X exclusive peptides with a total of Y exclusive peptide counts (spectra) to distinguish between them. Several values of X and Y were evaluated to cover the spectrum of stringency. The baseline (least aggressive) grouping approach (where X = 1 and Y = 1) merged two proteins unless they each had at least one exclusive peptide. This is similar to previously published parsimony methods that group proteins with redundant peptide sets and remove proteins with subset peptide sets.18,19 Our method was slightly more aggressive, however, because if proteins A and B were grouped together and B and C were grouped together, then A and C were also grouped together. Increasing the values for X and Y made the algorithm group more aggressively because more exclusive peptide data were required in order for two proteins to remain independent. It should be noted that many groups contained single proteins independent of grouping method or values of X and Y. After grouping the proteins, any peptides that were found in multiple groups were split using protein group unique peptide evidence similar to previous methods.16,17
For some species, such as mouse, curated databases are available that have the advantages of smaller sizes, reduced instances of shared peptides, and higher quality annotations. These advantages reduce search times and reduce the risk of incorrectly counting shared peptides. We compared searching the data using the more complete Ensembl databases to the manually reviewed canonical Swiss-Prot database (without expanded isoform entries) to see if increased peptide identifications justified the increased difficulty in interpreting protein results. To compare the databases, we mapped the Ensembl families to the Swiss-Prot proteins by comparing the sets of identified peptides. A Swiss-Prot protein mapped to an Ensembl protein family if they shared one or more peptides. Swiss-Prot proteins that mapped to multiple families and families that mapped to multiple Swiss-Prot proteins were discarded for the analysis comparing the two databases (see Supporting Information Part 1.)
Normalization and Differential Expression Analysis
We compared three normalization approaches (sum total, sum total with protein length, and quantile(20)) and four differential expression analysis approaches (ANOVA with factors for strain and batch, Significance Analysis of Microarrays(21) blocked on batch, Quasi-Poisson Generalized Linear Model(22) with factors for strain and batch, and edgeR(23)) (see Supporting Information Part 2 for a comparison of results). As this is a biological data set rather than an experimental mixture with known amounts of spiked-in proteins, an analysis of the sensitivity and specificity of these methods could not be performed. Proteins having small spectral count numbers and many missing observations across replicates violate most expression analysis method assumptions, so we imposed a minimum protein spectral count total sum of 10 across all samples for quantification. We chose to use a combination of quantile normalization, batch adjustment,(24) and edgeR for differential expression analysis. Batch adjustments were deemed necessary as batch effects that changed protein ranks remained after normalization (Figure 1, also see Supporting Information and ref (25)). To adjust for batch effects, we used nonparametric adjustments in the ComBat package.(24) The software package edgeR was designed to analyze count data that measures expression across many genes, such as SAGE, RNA-seq, and MS/MS spectral counting. It uses shared information across genes to estimate dispersion, and we used the common dispersion option. All packages are open source and were used in the R statistical programming environment.
Mapping to Portland Alcohol Research Center Quantitative Trait Loci (QTL)
QTL genomic regions were obtained from the Portland Alcohol Research Center (http://www.ohsu.edu/parc/by_phen.shtml). Genome coordinates given in cM were converted to bases using the Jackson Laboratory Mouse Map Converter (http://cgd.jax.org/mousemapconverter). For QTLs that did not have ranges given, the peak ±20Mb (1/2 of the median of the observed ranges) was used. A family mapped to a QTL if: 1. It contained a protein that had a coding region within the QTL range, and 2. There was peptide evidence that the protein within the QTL was present in the samples. A list of which families overlap with QTLs can be found in Supplemental Table 5 (Supporting Information).
Results and Discussion
Treatment of Shared Peptide Artifacts with Protein Grouping
When we searched the 4 049 668 spectra data set against the Ensembl protein databases, 423 376 MS2 spectra passed the thresholds with 5650 reversed-sequence matches (1.33% peptide FDR). We identified 33 297 unique peptides belonging to 6602 different proteins. When a standard peptide subset removal parsimony analysis was performed (equivalent to DTASelect with Occam’s razor filter(26)), the protein identifications were reduced to 4593 redundant target matches (3284 nonredundant) with 98 decoy matches (2.1% protein FDR), excluding common contaminants (Supplemental Tables 1–4, Supporting Information). To evaluate alternative grouping approaches and to avoid the loss of annotation, we retained the redundant protein identifiers.
The algorithm we used to split shared peptide spectral counts was based on the fraction of unique peptide counts found for each protein containing the shared peptide.16,17 We determined this approach to be problematic for some proteins when GAPDH, a highly abundant housekeeping protein that is known to vary little between samples, appeared to be highly differentially expressed. The gene for GAPDH is duplicated many times in the genome, which led to multiple similar GAPDH Ensembl entries. A small number of unique peptides prevented the parsimony analysis from collapsing all of the GAPDH entries into one group. A single amino acid substitution in one of the protein isoforms led to an increase in unique peptide counts which led the splitting algorithm to assign many of the spectral counts to this one isoform. However, this only occurred in two samples, both of which belonged to the B6 strain. This led to the isoform appearing to be differentially expressed between strains. Small unique count numbers for protein families with high sequence homology (e.g., GAPDH, actins, tubulins), where the bulk of their spectral counts come from shared peptides, can produce large fluctuations in protein total spectral counts after splitting. This led us to investigate strategies for grouping such similar proteins prior to the splitting algorithm.
Ensembl Family and Peptide-Based Grouping Strategies
In one approach, we grouped similar proteins into Ensembl-defined protein families and then counted the spectral counts found per family. After grouping similar proteins into families, only 0.59% of the peptides were ambiguously assigned to multiple families. After filtering out families with a sum of fewer than 10 counts across all 8 samples and 1 family with severe batch effects, 1807 families remained for further analysis.
An alternative grouping approach was to group two similar proteins if they each had fewer than X exclusive observed peptides with a total of Y exclusive peptide counts (spectra) to distinguish between them. We compared grouping by Ensembl protein family to five versions of this peptide-based grouping strategy: 1. No grouping, 2. Baseline grouping (requires each protein to have at least one exclusive peptide), 3. Light grouping (requires at least one exclusive peptide with a total of five exclusive peptide counts), 4. Moderate grouping (requires at least two exclusive peptides with a total of 10 exclusive peptide counts), and 5. Aggressive grouping (proteins are grouped if they share any peptides) (Table 1).
Table 1. Comparison of Strategies for Grouping Similar Proteinsa.
| grouping strategy | percent of peptides shared | total number of groups | number of groups with >10 counts | percent of groups containing any shared peptides | percent of groups containing only one protein | number of groups differentially expressed (p < 0.05/q < 0.05) |
|---|---|---|---|---|---|---|
| No grouping | 31.16% | 4593b | 2583 | 52.03% | 100.00% | 116/16 |
| Baseline grouping (1/1) | 11.94% | 3264 | 2405 | 33.76% | 77.51% | 120/17 |
| Light grouping (1/5) | 6.84% | 2998 | 2329 | 26.66% | 70.92% | 119/17 |
| Swiss-Prot search with no grouping | 4.78% | 2976 | 2201 | 27.21% | 100.00% | 110/16 |
| Moderate grouping (2/10) | 4.62% | 2885 | 2259 | 22.13% | 69.06% | 123/16 |
| Ensembl family grouping | 0.59% | 2343 | 1808 | 4.54% | 55.65% | 101/19 |
| Aggressive grouping | 0.00% | 2579 | 1958 | 0.00% | 63.31% | 111/14 |
Grouping label (2/10) indicates that two proteins with any shared peptides are merged unless they each have 2 exclusive peptides with a total of 10 exclusive peptide counts to distinguish between them.
The “no grouping” protein set includes redundant proteins.
Due to the relatively low similarity threshold set by Ensembl when they constructed the protein families, we found grouping by Ensembl family to be on the aggressive end of the spectrum. We mapped groups formed using Ensembl families to groups formed using the moderate (2/10) grouping criteria. We found that only 3.7% of the groups formed using peptide-based criteria contained proteins belonging to multiple Ensembl families. This indicated that grouping using moderate peptide criteria rarely groups two proteins that belong to different families and are therefore most likely functionally distinct. Conversely, 19.0% of Ensembl families mapped to multiple groups in the moderate grouping scheme. This suggests that grouping by Ensembl family may be overly aggressive in some cases because there may be sufficient peptide data to quantify some members of the families individually. We decided to use the Ensembl family grouping for further analyses because of the family level annotation provided by Ensembl.
Increase in Peptide and Spectral Counts When Using a Complete vs a Nonredundant Database
To avoid the problems associated with shared peptides, proteomics data can be searched against databases with minimal sequence redundancy, such as Swiss-Prot. When a protein has multiple isoforms, Swiss-Prot usually has one canonical sequence to represent the set. Ensembl, as well as other more complete databases, include separate entries for gene duplications and splice isoforms, leading to higher sequence redundancy within the databases. For our data set, 31.1% of Ensembl peptides were ambiguous before protein grouping whereas only 4.8% of Swiss-Prot peptides were. We searched our data set against both the Ensembl (reference/B6) and Swiss-Prot databases so that we could determine if the additional information content in a complete database would significantly increase peptide identifications and spectral counts. Using Ensembl, we observed a 6.8% increase in successful spectrum-to-peptide assignments. Using a standard parsimony analysis, an average of 3336 (SD = 732) additional peptides and 176 (SD = 22) proteins were identified per sample when searching Ensembl compared to Swiss-Prot. Complete results are in Supporting Information Part 1. A comparison between other more complete databases (e.g., NCBI RefSeq, UniProtKB/TrEMBL, and IPI) was not attempted but similar increases in peptide identifications would be expected.
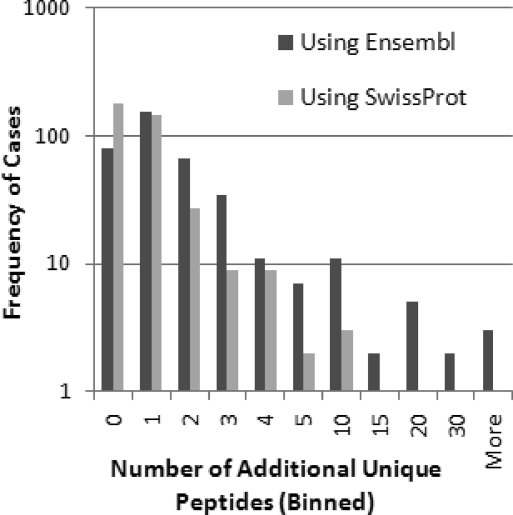
To make a fair protein-level comparison, we selected only the 749 cases where there were one-to-one matches between Swiss-Prot proteins and Ensembl families that contained multiple isoforms. In 376 of those cases, additional peptides were found using Ensembl or Swiss-Prot (Figure 2). Of these, 296 (78.1%) gained additional peptides when all of the isoforms in the Ensembl family were considered. A total of 30 proteins gained five or more additional distinct peptides. In all of these 30 cases, there is peptide evidence that multiple isoforms are present in the samples. Spectral counts increased dramatically in some cases. Several specific examples are given in Supporting Information Part 1.
Figure 2.
Histogram of the number of additional unique peptides identified when using Ensembl vs Swiss-Prot. Only the cases where one Swiss-Prot protein mapped to one Ensembl family that represented two or more isoforms (and where additional peptides were found using Ensembl or Swiss-Prot) are shown.
Additional distinct peptides were also found using Swiss-Prot, which suggests that either Ensembl does not contain all of the sequences that are used in Swiss-Prot or that searching a larger database reduces search sensitivity for low-scoring peptides. Of the 30 539 Swiss-Prot peptides identified by SEQUEST, 620 (2.0%) were not found in the reference Ensembl database search. Of these, 69 were found in the D2 Ensembl database search because Swiss-Prot contained the D2 version of the peptide. This could be because there are multiple versions of the peptide across the strains and that Swiss-Prot contains the version found in the D2 strain, or it could be due to an error in the Ensembl genome. Of the remaining 551 peptides not found in either Ensembl search, 341 were in fact present in the Ensembl database but were not found in the SEQUEST search due to reduced sensitivity when searching a larger database. The remaining 210 could not be found at all in Ensembl, indicating missing sequence data or annotation. (Additional details can be found in Supporting Information Part 1.)
Differential Expression Results
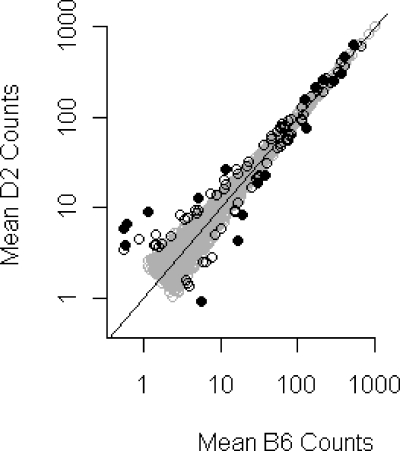
Striatal protein expression was very similar between B6 and D2 (Figure 3, Pearson r = 0.997, p < 2e-16). Of the 1,807 families exceeding minimum count cutoffs that we were able to quantify, 101 were significantly different between strains (p < 0.05). After a False Discovery Rate (FDR) adjustment for multiple comparisons, 19 remained significant (q < 0.05) (Figure 3). Ten of the 19 had p-values of less than 0.05 even when no batch adjustment was performed.
Figure 3.
Protein families found to be significantly different between strains. Gray circles represent all of the data. Black open circles represent a p-value of less than 0.05. Black closed circles represent an FDR-adjusted q-value of less than 0.05. Quantile normalized and batch adjusted data is shown, but a plot of the raw data was similar.
Eighty-four (83%) of the significantly differentially expressed families had coding regions that fell within one of the genomic regions of interest identified by the Portland Alcohol Research Center (Table 2). This is significantly more than expected by chance as these regions cover only 64% of the genome (p = 0.00002) and only 73% of all of the families identified overlapped with these regions (p = 0.01). Differentially expressed proteins that overlap with these regions are suspect “quantitative trait proteins” and are listed in Supplemental Table 5 (Supporting Information).
Table 2. Number of Significantly Differentially Expressed Protein Families that have at least One Protein that was Identified in the Data Set and that Lie within a Region of the Genome Found to be Associated with the Given Phenotype.
| quantitative phenotype | p < 0.05 | q < 0.05 |
|---|---|---|
| Acute Alcohol Withdrawal | 13 | 3 |
| Alcohol Acceptance | 5 | 2 |
| Alcohol Metabolism | 14 | 3 |
| Alcohol Preference Drinking | 54 | 10 |
| Alcohol Response Conditioning | 21 | 1 |
| Alcohol Stimulated Activity | 65 | 13 |
| Chronic Alcohol Withdrawal | 24 | 3 |
| Hypothermia | 6 | 2 |
| Loss of Righting Reflex | 12 | 3 |
Influence of Strain-Specific Databases on Spectral Counts and Differential Expression Analysis
We compared quantitative results obtained from searching the D2 samples on the reference Ensembl database vs an Ensembl database adapted to match the D2 genome sequence. On average, we identified an additional 239 peptides per sample when using the D2 database, which represents an increase of 0.44%. Only 62 (3.4%) of the protein families had spectral count differences of greater than 5%. Of those 62, just 7 went from differentially expressed to not or vice versa.
If we assume true counts are obtained using the D2 database on the D2 samples, we obtained 91 true positives (the protein family was determined to be significantly differentially expressed using either database), 11 false positives, 10 false negatives, and 1695 true negatives. These led to a false positive rate of 0.64% and a false negative rate of 9.9%. Six of the false positives and 7 of the false negatives had only a small change in their p-value, which led to a change in differential expression status due to the arbitrary cutoff of 0.05. Five false positives and two false negatives had significantly altered p-values due to low peptides counts for the D2 strain when searched on the reference database. In these cases, at least one D2 peptide was absent in the reference database but was present in the D2 database. This led to an increase in peptide counts in the D2 samples and a change in differential expression status when the appropriate database was used. An example peptide containing an amino acid substitution is shown in Table 3. Spectra for the two peptide forms confirming the amino acid substitution are provided in Supporting Information Part 3.
Table 3. Effect of a Single Amino Acid Substitution on Protein Family ENSFM00250000001899a.
| protein ID: ENSMUSP00000068260 | reference DB |
D2 DB |
||||||
|---|---|---|---|---|---|---|---|---|
| peptide sequence | B6–1 | B6–2 | B6–3 | B6–4 | D2–1 | D2–2 | D2–3 | D2–4 |
| ELSGLPSGPSVGSGPPPPPPGPPPPPIPTSSGSDDSASR | 0 | 0 | 0 | 0 | 10 | 6 | 10 | 6 |
| ELSGLPSGPSVGSGPPPPPPGPPPPPISTSSGSDDSASR | 5 | 8 | 8 | 10 | 0 | 0 | 0 | 0 |
Using the Ensembl reference database, this family was considered differentially expressed with a total of 185 counts in the B6 strain and 148 counts in the D2 strain (edgeR, p = 0.0077). Using the D2 database on the D2 samples increased the D2 counts to 180, making the family no longer significant (p = 0.20). This change is due to the single amino acid substitution S242P in protein ENSMUSP00000068260.
The remaining false negative was a low count protein family that appeared to have missing counts in both strains when the D2 database was used. This suggested an error in the D2 database. Because we searched both strains on both Ensembl databases, we were able to identify cases where discrepancies likely arose due to sequence errors in the reference or D2 databases. For example, we found 29 peptides that were present in both strains when using the D2 database, but were absent when using the Ensembl reference database. This suggests there is an error in the Ensembl reference sequence. Conversely, there were 37 peptides that were found in both strains when using the reference database and were absent when using the D2 database. This suggests there is an error in the D2 genome sequence or the Ensembl transcript coordinates used to insert the polymorphism and retranslate the protein.
Conclusions
Managing Sequence Redundancy
We identified 6.8% more peptides (approximately 27 000 matches) when we used the complete Ensembl database with explicit isoform entries rather than the nonredundant Swiss-Prot database. This illustrates the value of using a complete database that includes multiple isoforms as independent entries rather than a nonredundant database that uses a canonical sequence to represent a protein family. We found that 46% of the proteins with multiple isoforms had increased counts when their isoforms were considered. We chose the Ensembl database because of its straightforward mapping onto the mouse genome so that we could construct strain-specific databases. Although we did not evaluate other frequently used examples of complete databases such as UniProtKB/TrEMBL, NCBI RefSeq, and IPI (now discontinued), many of which include even more isoforms than Ensembl, we expect that utilizing these complete databases would also increase peptide identifications by similar or more amounts.
Although searching more complete databases increased peptide counts, additional analysis steps were necessary to address sequence redundancy in the databases. Extensive sequence redundancy can lead to many peptides being shared. If these shared peptides are not counted properly, inaccurate total protein counts may result and lead to erroneous differential expression candidates. Peptide splitting algorithms based on unique peptide counts become unreliable when unique counts are too small.
We explored two different ways to group similar proteins before peptide splitting was performed. One approach for grouping similar proteins was to compare the sets of peptides found for each protein. Most proteomics analysis pipelines group proteins that have identical peptide sets (redundant proteins) and remove peptide subsets (parsimony analysis).18,19 We extended these concepts by grouping proteins that shared most of their peptides and had few exclusive peptides to distinguish between them before applying a shared-peptide splitting calculation. A single unique peptide may suggest a protein’s presence in the sample, but it may not provide sufficient data to quantify the protein independent of its family members.(27) Grouping similar proteins, even if there was some limited unique peptide evidence, fixed the unreliable quantitative results we observed without grouping.
In analogy to definitions of “minimal identifiable protein sets”, we attempted to define the “minimal quantifiable protein set”. For example, if at least two distinct peptides and at least ten peptide counts were required to consider a protein quantifiable, then should not it logically follow that at least two unique peptides and at least ten unique peptide counts be required to separately quantify two similar isoforms? The exact definition of what is quantifiable depends on many factors and our definition is what made sense for our data and quantification technique. For most experiments, the number of identifiable proteins will exceed, sometimes greatly, the number of quantifiable proteins.
An alternative grouping approach was to use protein families based on sequence similarity. This is algorithmically complex to compute, but is conveniently provided for Ensembl proteins. We chose to utilize this grouping because of the useful family annotations provided by Ensembl. Managing annotations for proteins grouped on a by-experiment basis is a challenge that is typically overlooked. There is a downside to using Ensembl protein families, however. If one member of the family is significantly differentially expressed, and the others are not, that difference may no longer appear significant when the counts are summed into families. We observed this behavior for 25 proteins, 16 of which were confirmed using strictly unique peptide counts (p<0.05). Grouping related proteins using protein families may cause us to miss some significantly differentially expressed proteins; however this is preferable to keeping related proteins separate when there is insufficient unique peptide information to reliably split their shared peptides.
Normalization and Batch Corrections
Large-scale technologies such as microarrays and mass spectrometry often involve multiple samples processed at different times and require normalization to remove nonbiological variability. We compared several normalization methods (see Supporting Information Part 2) and found that quantile normalization—a powerful, nonlinear normalization method frequently used for microarrays(20)—performed the best. Quantile normalization makes the distribution of spectral count values nearly identical between samples, an assumption that is reasonable for this comparison of the same tissue between very similar mouse strains. There may be many other situations where quantile normalization would not be appropriate. Our study involved two different sample collections, striatum preparations, and sets of mass spectrometry runs separated by several months, which can be typical in experiments involving multiple biological replicates. Using cluster analyses (see Figure 1) and principal component analyses (Supporting Information Part 2), we found that significant batch effects (additional sources of nonbiological variability) that altered protein ranks were still present even after quantile normalization. Our study design, where two pairs of samples were run at each time point, allowed for correction of batch effects using empirical Bayesian methods.(24) Removal of nonbiological variation resulted in lower p-values from statistical tests and thresholds had to be adjusted accordingly. Batch corrections can be aggressive and clear evidence that they are necessary should be demonstrated. Quantitative proteomic study designs must also be compatible with batch correction assumptions.
Genome-Sequence Informed Databases
We identified 0.44% more peptides when we used a protein database that took into account the strain’s genome sequence. As these two strains of mice are roughly as similar to each other as two humans are, we expect similar results would be obtained in human data. Although the increase in spectral counts is low, most of the observed differences are concentrated in only a handful of families and may alter their differential expression status. When we used the Ensembl reference database rather than the strain-specific database in the analysis for differential expression, we observed a false positive rate of 0.64% and a false negative rate of 9.9%. These values show that protein-based expression techniques are more robust to underlying genomic sequence variation than mRNA hybridization techniques.(2) This was not too surprising as there are many more genomic polymorphisms than amino acid substitutions due to codon redundancy in the genetic code. We conclude that the vast majority of proteins do not have quantitative estimates that are influenced by underlying sequence differences, but in the few that do, the influence can be significant.
Annotation for known amino acid substitutions is growing in databases such as Swiss-Prot. This will continue to improve as the availability of genome sequence data increases exponentially. Search algorithms that incorporate this annotation for known amino acid substitutions will increase their spectrum-to-peptide assignments and will avoid some false positive and negative conclusions.
Differential Expression in Mouse Strains
The number of protein families found to be differentially expressed in striatum between strains B6 and D2 was only about one-hundred. Of those that were, a large proportion (83%) contained proteins that lie within previously identified genomic regions of interest for alcohol-related behavioral traits. These proteins will serve as good candidates for proteins that may explain the vast behavioral differences between these strains.
Acknowledgments
We thank OHSU’s Proteomics Shared Resource for the expertise, time, and resources needed to complete this project. Materials and mass spectrometer time was paid for by a pilot grant from the Portland Alcohol Research Center (PARC). The PARC is funded by the National Institutes of Health (NIH) National Institute on Alcohol Abuse and Alcoholism (NIH/NIAAA-5-P60-AA010760-13). S.F. was supported by an NIH National Library of Medicine Biomedical Informatics Training Grant (NIH/NLM-5-T15-LM07088-15). We thank the Hitzemann lab for providing the striatal tissue, analysis advice, mRNA data, and additional funding (R01-AA011034 and U01-AA013484). The Proteomics Shared Resource is funded by NIH grants R01-EY007755, P30-EY10572, and P30-CA069533, as well as support from the Oregon Opportunity Fund. DBA/2J sequencing data and variant calling was provided by the Sanger Mouse Genomes Project: http://www.sanger.ac.uk/resources/mouse/genomes/.
Supporting Information Available
Supporting Information Part 1. Additional details on the comparison of results using a nonredundant (Swiss-Prot) vs a complete (Ensembl) database. Supporting Information Part 2. A comparison of differential expression analysis workflows and a justification for choosing the methodology used in this work. Supporting Information Part 3. Additional details on the analysis of strain-specific databases, including spectra for the peptides in Table 3. Supplemental Table 1. The parsimonious lists of peptides and proteins identified by SEQUEST searches of the 3 databases. Data set summary statistics, false discovery analyses, and Tranche hash keys for downloading raw data files and databases are also included. Supplemental Table 2. Detailed lists of identified peptides for each sample searched against the Ensembl Reference B6 database. Supplemental Table 3. Detailed lists of identified peptides for each sample searched against the generated Ensembl D2 database. Supplemental Table 4. Detailed lists of identified peptides for each sample searched against the canonical Swiss-Prot database. Supplemental Table 5. A list of quantified protein families including raw and processed spectral counts, quantitative results, peptides identified, relevant annotation, and overlapping Portland Alcohol Research Center quantitative trait loci. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Crabbe J. C.; Phillips T. J.; Belknap J. K. The Complexity of Alcohol Drinking: Studies in Rodent Genetic Models. Behav. Genet. 2010, Nov;, 40 (6), 737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N. A.; McWeeney S. K.; Peters S. T.; Belknap J. K.; Hitzemann R.; Buck K. J. SNPs matter: impact on detection of differential expression. Nat. Methods 2007, 4 (9), 679–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovoy D.; Kwan T.; Majewski J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008, 36 (13), 4417–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin T. J.; Gygi S. P.; Ideker T.; Rist B.; Eng J.; Hood L.; Aebersold R. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 2002, 1 (4), 323–33. [DOI] [PubMed] [Google Scholar]
- Gygi S. P.; Rochon Y.; Franza B. R.; Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999, 19 (3), 1720–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRedmond J. P.; Park S. D.; Reilly D. F.; Coppinger J. A.; Maguire P. B.; Shields D. C.; Fitzgerald D. J. Integration of proteomics and genomics in platelets: a profile of platelet proteins and platelet-specific genes. Mol. Cell. Proteomics 2004, 3 (2), 133–44. [DOI] [PubMed] [Google Scholar]
- Mijalski T.; Harder A.; Halder T.; Kersten M.; Horsch M.; Strom T. M.; Liebscher H. V.; Lottspeich F.; de Angelis M. H.; Beckers J. Identification of coexpressed gene clusters in a comparative analysis of transcriptome and proteome in mouse tissues. Proc. Natl. Acad. Sci. U.S.A. 2005, 102 (24), 8621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y.; Choi P. J.; Li G. W.; Chen H.; Babu M.; Hearn J.; Emili A.; Xie X. S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329 (5991), 533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P.; Koller A.; Oshiro G.; Ulaszek R. R.; Plouffe D.; Deciu C.; Winzeler E.; Yates J. R. 3rd Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2003, 100 (6), 3107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.; Fu N.; Guo S.; Yan Z.; Xu Y.; Hu H.; Menzel C.; Chen W.; Li Y.; Zeng R.; Khaitovich P. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 2009, 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. W.; Miller S.; Klychnikov O.; Loos M.; Stahl-Zeng J.; Spijker S.; Mayford M.; Smit A. B. Quantitative Proteomics and Protein Network Analysis of Hippocampal Synapses of CaMKIIalpha Mutant Mice. J. Proteome Res. 2007, 6 (8), 3127–3133. [DOI] [PubMed] [Google Scholar]
- Li K. W.; Smit A. B. Subcellular proteomics in neuroscience. Front. Biosci. 2008, 13, 4416–25. [DOI] [PubMed] [Google Scholar]
- Wilmarth P. A.; Tanner S.; Dasari S.; Nagalla S. R.; Riviere M. A.; Bafna V.; Pevzner P. A.; David L. L. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to Crystallin insolubility?. J. Proteome Res. 2006, 5 (10), 2554–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S.; Wilmarth P. A.; David L. L. The membrane proteome of the mouse lens fiber cell. Mol. Vis. 2009, 15, 2448–63. [PMC free article] [PubMed] [Google Scholar]
- Wilmarth P. A.; Riviere M. A.; David L. L. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2009, 2 (4), 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Tan G.; Levenkova N.; Li T.; Pugh E. N. Jr.; Rux J. J.; Speicher D. W.; Pierce E. A. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 2007, 6 (8), 1299–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Wen Z.; Washburn M. P.; Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 2010, 82 (6), 2272–81. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 2005, 4 (10), 1419–40. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Chambers M. C.; Tabb D. L. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 2007, 6 (9), 3549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B. M.; Irizarry R. A.; Astrand M.; Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19 (2), 185–93. [DOI] [PubMed] [Google Scholar]
- Tusher V. G.; Tibshirani R.; Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (9), 5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Gray W.; Zhang H.; Chung C. H.; Billheimer D.; Yarbrough W. G.; Liebler D. C.; Shyr Y.; Slebos R. J. Comparative Shotgun Proteomics Using Spectral Count Data and Quasi-Likelihood Modeling. J. Proteome Res. 2010, 9 (8), 4295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D.; McCarthy D. J.; Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26 (1), 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E.; Li C.; Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8 (1), 118–27. [DOI] [PubMed] [Google Scholar]
- Leek J. T.; Scharpf R. B.; Bravo H. C.; Simcha D.; Langmead B.; Johnson W. E.; Geman D.; Baggerly K.; Irizarry R. A. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010, 11 (10), 733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L.; McDonald W. H.; Yates J. R. 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002, 1 (1), 21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeley P.; Siepen J. A.; Lawless C.; Hubbard S. J. Investigating protein isoforms via proteomics: a feasibility study. Proteomics 2010, 10 (6), 1127–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.