Abstract
Over the past decade, multiple genetic and histological approaches have accelerated development of new breast cancer diagnostics and treatment paradigms. Multiple distinct genetic subtypes of breast cancers have been defined, and this has progressively led toward more personalized medicine in regard to treatment options. There still remains a deficiency in the development of molecular diagnostic assays that can be used for breast cancer detection and pretherapy clinical decisions. In particular, the type of cancer-specific biomarker typified by a serum or tissue-derived protein. Progress in this regard has been minimal, especially in comparison to the rapid advancements in genetic and histological assays for breast cancers. In this review, some potential reasons for this large gap in developing protein biomarkers will be discussed, as well as new strategies for improving these approaches. Improvements in the study design of protein biomarker discovery strategies in relation to the genetic subtypes and histology of breast cancers is also emphasized. The current successes in use of genetic and histological assays for breast cancer diagnostics are summarized, and in that context, the current limitations of the types of breast cancer-related clinical samples available for protein biomarker assay development are discussed. Based on these limitations, research strategies emphasizing identification of glycoprotein biomarkers in blood and MALDI mass spectrometry imaging of tissues are described.
Introduction
Breast cancer is a heterogeneous group of different tumor subtypes that vary in prognosis and response to therapy. Recent years have seen great success in defining the genetic and histological basis of this heterogeneity, leading to multiple molecular genetic assays that offers the promise of targeted personalized treatment strategies for those newly diagnosed. However, there remains a significant lack of molecular diagnostic assays that can be used for breast cancer detection or pretherapy clinical decision making. In particular, a serum protein biomarker analogous to prostate-specific antigen and prostate cancer diagnostics is lacking for breast cancer, and despite many attempts and studies, potential candidates are few. A proteomic biomarker has several inherent advantages over genomics in that proteins are more reflective of the tumor microenvironment and can undergo cancer specific posttranslational modifications. Additionally, measured mRNA levels do not necessarily correlate to corresponding protein levels. This article will summarize the current successes in molecular genetics and histology of breast cancers, and in that context, contrast this with the many challenges to developing protein-based biomarkers for use in clinical diagnostics. The goal of this report is not to exhaustively evaluate every potential proteomic approach or breast cancer-related system that could be evaluated for biomarkers, but concentrate on addressing the basis for the large gap in clinical proteomic biomarker assay development for breast cancers. The emphasis will therefore focus on the current limitations of the types of breast cancer-related clinical samples available for protein biomarker assay development. Based on these limitations, a research strategy emphasizing identification of glycoprotein biomarkers in blood and tissue using mass spectrometry approaches will be discussed.
Breast Cancer Subtypes and Genetic Diagnostics
Breast cancer diagnostics have significantly changed over the last decade, increasingly relying on gene expression analyses combined with immunohistochemistry of specific receptor proteins (Harris et al., 2007; Perou et al., 2000; van't Veer et al., 2005). Multiple breast cancer subtypes have been defined, with their names reflecting their area of origin within the breast, termed broadly as mesenchymal, basal, or luminal, and histologic classification based on immunohistochemistry staining of Her2 receptor, estrogen receptor (ER), and progesterone receptor (PR) (Fan et al., 2006; Parker et al., 2009). Two primary luminal subtypes are termed luminal A (ER+/Her2−) and luminal B (ER+/Her2+), with better prognosis associated with luminal A-type tumors compared to intermediate prognosis for luminal B tumors (Cheang et al., 2009). The basal-like tumors have the worst prognosis, and are commonly associated with a triple negative phenotype of ER−/PR−/Her2− (Irvin and Carey, 2008; Sorlie, et al., 2001). Treatment regimens targeting the estrogen receptor and Her2 expressing tumors have proven to be effective. However, there are as yet no targeted agents for the basal-like/triple-negative breast tumors. Understandably, these designations are broad classifiers, as there is emerging evidence to suggest that individual breast tumors can have molecular signature phenotypes that do not fit neatly into each category, representing a heterogeneous gradient of subtypes across the entire range of mesenchymal, basal, and luminal cell lineages (Lim et al., 2009). For example, a subpopulation of the basal-like triple-negative tumors have also been linked with the majority of BRCA1-associated breast cancers (Foulkes et al., 2003; Lakhani et al., 2005), with more aggressive clinicopathologic features including onset at a younger age, higher mean tumor size, and higher grade tumors (Carey et al., 2006; Dent et al., 2007). Several population-based studies have also indicated that triple-negative breast cancers are more likely to occur among premenopausal women of African-American descent than in other races (Bauer et al., 2007; Morris et al., 2007). Last, the triple-negative subtype in general is associated with reduced breast cancer-specific survival compared with luminal phenotypes (Carey et al., 2006; Dent et al., 2007). Comparatively, those with triple-negative breast cancer were also much more likely to develop a recurrence during the first 3 years following therapy, with rapid declines thereafter (Dent et al., 2007).
These subtype classifications have been determined in conjunction with many gene microarray expression studies for breast cancers, particularly as applied to improving prognostic and therapeutic prediction (Kim and Paik, 2010; Turaga et al., 2010). From these approaches, multiple prognostic assays to determine risk of recurrence or treatment responsiveness, typified by Oncotype DX (Paik et al., 2005) (Genomic Health, Inc., Redwood City, CA), Mammaprint (van de Vijver et al., 2002) (Agendia Inc., Irvine, CA), and other genetic assays (Turaga et al., 2010), have been evolving into wider clinical use. Larger clinical trials are ongoing to determine the power of these gene expression signatures in reclassifying patients after conventional risk classification (Turaga et al., 2010). The cumulative genetic-based research into the different breast cancer subtypes, evaluation of additional associated immunohistochemical targets (reviewed in Choo and Nielsen, 2010), continued commercial assay development, and larger clinical studies assessing risk classification and treatment responsiveness issues will continue to accelerate all aspects of related breast cancer research in these areas.
Missing in this discussion are any corresponding proteomic-based analyses to identify clinical biomarkers that complement the genetic-based approaches. Despite the continued need for proteomic-based assays, the development of genetic-based assays linked to developmental breast histology has far outpaced current proteomic-based studies. One of the reasons for the lack of proteomic studies is due to limitations with available breast cancer clinical samples. Three main clinical sample types that are the most commonly used for proteomic studies will be discussed in different applied contexts: ductal fluids, serum/plasma and tissues. Cell lines, related xenograft animal models, and other animal models of breast cancer could, of course, be sources for tissue and fluids for biomarker discovery. However, in the context of the many heterogenous subtypes of breast cancers, these systems lack broad representation and are not ideal to develop applicable biomarker candidates for clinical diagnostics. Their use in developing proteomic biomarker assays will not be further discussed.
Detection of Breast Cancer Biomarkers in Nipple Aspirate/Ductal Lavage
The majority of breast cancers originate in the ductal system of the breast, and therefore, it is feasible that the composition of nipple fluids could reflect the local microenvironment, including benign and cancerous conditions. Eight to 10 milk ducts exit the nipple and nipple aspirates can be obtained using various noninvasive techniques in the majority of women (King and Love, 2006). Collection of nipple aspirate fluid (NAF) is noninvasive, inexpensive, and much easier to obtain than more invasive procedures like ductal lavage. However, it has serious limitations for routine diagnostic use as acquisition rates vary from 25–67% (Fabian et al., 2005; Higgins et al., 2005), and rates are even lower for older women and those of Asian descent (Wrensch et al., 1990). In general, for most collected NAF samples, only microliter volumes are obtained. However, this certainly does not preclude using NAF as a source for protein biomarker discovery efforts. A recent study employed multifractionation strategies prior to peptide analysis on an LTQ Orbitrap mass spectrometer and reported over 800 unique proteins present in NAF (Pavlou et al., 2010). Approximately 40% of these proteins are also found in serum/plasma, and over 50% of them were secreted or of plasma membrane origin. This fluid, when available, could certainly prove useful in identifying biomarker candidates for further assay development in breast cancer related tissue or blood samples, as described in the next sections.
A BIRADS 4 Serum Collection Model
The concept of collecting serum or plasma in the clinic is straightforward, and in general, standard operating procedures are well established. This ease of collection and general acceptance of individuals to provide blood specimens, makes this fluid an ideal source for developing diagnostic assays. However, variability exists for each patient sample depending on the facility or clinic setting where the blood is drawn with respect to processing and transporting of the specimens. Except for specialized centers, blood samples additionally could be coming from clinic visits to general practitioners, surgeons or oncologists, reflecting multiple points along a diagnostic and treatment course. Standardizing this collection to reflect the many potential phases of breast cancer diagnostics remains a challenge. However, the overall breadth of collection remains attractive for proteomic-based studies, as samples can easily be stratified to reflect breast cancer-related clinical parameters such as menopause/age, body mass index (BMI)/obesity, race, pathology stage, cell origin (ductal vs. lobular), ER/PR/Her2 status, and genetic subtypes.
One model serum collection cohort that could be used to design protein-based diagnostic assays is as follows. For the past 5 years, we have collected over 1,000 serum samples at two sites in a study design focused on women given a BIRADS 4 mammogram report. The Breast Imaging-Reporting and Data System (BIRADS) is used by radiologists to make treatment or follow-up recommendations by categorizing mammographic findings based on the likelihood of malignancy (Obenauer et al., 2005) across seven categories ranging from 0 (“incomplete assessment”) to 6 (“confirmed malignant”). A BIRADS 4 lesion is categorized as a “suspicious abnormality,” even though the majority (80%) will be benign conditions. Because a malignancy cannot be ruled out, a biopsy will be performed, even for those lesions further stratified into BIRADS 4a, 4b, and 4c (Lazarus et al., 2006), with a 4a lesion designation as a “low level of suspicion.” Thus, each year, a large number of women undergo a biopsy that will yield a benign diagnosis. Beyond pathological cytology, biomarkers of benign disease from any sample type, genetic, metabolite or protein, are lacking.
Our study was designed to collect prebiopsy serum from women with a BIRADS 4 mammogram who were about to have a breast biopsy procedure. The ultimate goal is to develop a proteomic-based blood test that would be used prior to biopsy that could better discriminate those BIRADS 4 designated women that are more likely to have benign disease, or confirm cancer suspicions, and therefore better justify a tissue biopsy decision. The specifics regarding sample collection have been reported (Shin et al., 2006), and are summarized as follows. Patients underwent either stereotactic or ultrasound-guided core biopsy, or needle localized excisional breast biopsy. After informed consent, which included a short questionnaire for assessment of each subject's risk factors for breast cancer, serum was collected immediately prior to the biopsy procedure. The questionnaire data included age, race, BMI, age at menarche, parity, age at first live birth, and age at menopause. Each serum specimen was assigned a diagnosis retrospectively after final pathology review. As would be expected, final pathologies for the benign cohort included fibroadenomas, fibrocystic disease, papillomas, atypical ductal hyperplasia without associated carcinoma or carcinoma in situ, lobular carcinoma in situ, and benign breast tissue (Shin et al., 2006). The cancer cohort included ductal carcinoma in situ and primarily early stage invasive ductal and lobular carcinomas (Shin et al., 2006). An initial cohort was used for extensive MALDI-TOF serum protein profiling following weak cation exchange affinity bead enrichment. Expression of a panel of 12 low molecular weight peptides could be used with a genetic algorithm to correctly classify 88% of the cancer samples and 86% of the benign disease samples (Shin et al., 2006). Despite evidence of protein differences between the two disease classifications, use of this single affinity method alone has proven to be not sensitive enough to routinely distinguish cancer from benign disease classifications.
The advantages of using this type of serum cohort center on the study design, as several known biases related to sample collection are minimized. All of the samples are collected in the same way in a similar clinic environment, from patients with the same BIRADS 4 designation. Because it is done just prior to biopsy, there is no diagnosis at the time of collection, so psychological and stress issues for each subject are similar. All associated risk factor information for breast cancer is collected, and can readily be combined with subsequent pathology and genetic properties associated with each sample. The ease and breadth of collection also facilitate their use in any developed high throughput detection platform. The clinical decisions made that brought each subject to the point of biopsy are also very similar. Because the BIRADS 4 diagnosis is based on a radiological test, samples are collected from all possible types of malignant and benign breast diseases. The collection protocol itself is potentially transportable to any breast cancer-related surgical facility that routinely performs breast biopsy procedures. Overall, collection protocols can be standardized and readily integrated into current clinical decision-making workflow associated with a benign or malignant diagnosis.
The disadvantages to use of this cohort are multifaceted. The primary disadvantage is related to all of the challenges of developing any blood-based protein biomarker diagnostic for cancer. A core problem is the enormous dynamic protein concentration range of blood proteins, with 20 proteins accounting for over 98% of the total protein amounts. Detecting a cancer specific protein in blood derived from the tumor is thus hampered by the very low concentrations of these proteins that would be present in blood. Previous studies that have used proteomic profiling approaches for detecting low molecular weight serum proteins (reviewed in Laronga and Drake, 2007) including our own BIRADS 4 study (Shin et al., 2006), are weakened by lack of protein identification and the dynamic concentration range issues (Anderson and Anderson, 2002; Diamandis, 2004), as well as study design bias, overgeneralization of results, and sample processing issues (Hu et al., 2005; McLerran et al., 2008a, 2008b; Ransohoff, 2005). Although our BIRADS 4 collection strategy addresses the bias and processing issues, the dynamic concentration range and tumor specificity remains a large barrier to assay development. There is also a sample collection time line issue in our approach, in that we have to collect mostly benign disease samples (4 to 1) to obtain enough cancers to have statistically relevant numbers for even the most basic breast cancer subtype stratifications based on BMI, age/menopause status, tumor pathology, and tumor genetic subtypes. Addressing these cumulative weaknesses form the basis for the proposed glycoprotein targeting strategy described in the next section.
Glycoproteomic Strategies for Breast Cancer Serum Biomarkers
Despite major efforts to identify blood-based biomarkers, currently no standardized blood test exists for breast cancer screening or staging purposes. The MUC-1 mucin glycoproteins CA 15.3 and CA 27.29 are the best characterized serum markers related to breast cancer (Duffy, 2006; Mathelin et al., 2006), but they have not been recommended for diagnostic use due to low sensitivity (Harris et al., 2007; Molina et al., 2006). As described below, designing assays to detect specific glycan differences remains a largely wide-open research area. Glycoproteomics is a newly emerging branch of proteomic diagnostics that focuses on characterizing any differences in the glycosylation patterns of carbohydrate residues added posttranslationally to proteins. Changes in glycosylation patterns may be of great significance, as this is known to influence many cellular processes including cell adhesion, signaling, and stabilization of protein structure, protein trafficking, as well as oncogenesis (Dwek et al., 1998). Interestingly, aberrant glycosylation of glycoproteins and glycolipids has been observed in many malignancies, and previous research indicates that these changes influence disease progression (Couldrey and Green, 2000; Dwek et al., 2001). Handerson et al. (2005) found an increase of β1–6 branched oligosaccharides within metastatic lymph nodes of breast carcinomas. This increase in branched oligosaccharides was associated with poor prognosis. A recent breast cancer tissue-based study targeted these increased β1–6 branched oligosaccarides to identify 34 glycoproteins that were overexpressed in invasive ductal carcinoma tissues relative to nontumor tissues (Abbott et al., 2009). The role of sialylated oligosaccharides was evaluated within primary breast tumors, and it was found that an overall reduction in the diversity of sialylated and neutral oligosaccharides occurred with disease progression (Dwek et al., 2001). Although there is apparently a wealth of glycosylation changes associated with breast cancer development and progression, they remain to be translated into a clinical assay format.
A wealth of new strategies targeting the characterization of serum glycoproteins and their carbohydrate constituents are emerging (Drake et al., 2006; Miura et al., 2008; Wada et al., 2007; Wuhrer et al., 2006). Structural elucidation of glycoproteins has long relied upon the use of chemical derivation and cleavage reactions, and differential capture by lectins, a class of proteins known to bind specific oligosaccharide moieties (Drake et al., 2006; Hirabayashi, 2004). The most common approaches for characterizing the protein constituents of N-linked glycoproteins is to use some combination of the following: peptides are generated by digestion with trypsin, the glycopeptides are isolated with one or more lectins or hydrazine linked to a support resin, the bound peptides are treated with protein N-Glycanase F (PNGaseF) to release glycans, then sequencing of the peptides is done by tandem mass spectrometry (Liu et al., 2005; Wang et al., 2006; Zhang et al., 2003; Zielinska et al., 2010). New strategies that probe different lectins bound on multiple array platforms are also emerging (Chen and Haab, 2009; Kuno et al., 2008; Mao et al., 2004; Zhao et al., 2007).
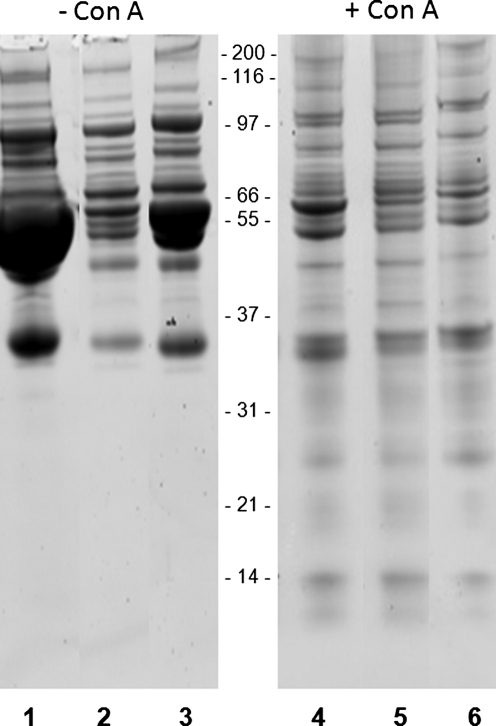
Because of the abundance of glycoproteins in serum, and the relative ease of collecting large serum sets associated with breast cancers, we propose that glycoproteomic targeted strategies could be the most effective approach to identify diagnostic biomarkers. As illustrated in the next two figures, use of lectins address some of the previously stated challenges in performing serum proteomic analyses. In Figure 1, a broad specificity lectin that binds most glycoproteins due to its affinity for mannose residues, Concanavalin A (Con A), was used with and without upfront albumin depletion strategies to isolate serum glycoproteins. Depletion of albumin by either an antibody column or affinity column prior is shown in the left panel of Figure 1, and these approaches are effective at reducing the levels of the abundant 65-kDa serum albumin. Use of just Con A alone with human serum resulted in significant enrichment of glycoproteins, and minimal amounts of bound albumin. Depletion of albumin prior to Con A binding yielded similar results. Thus, lectins like Con A, and both fucose targeting lectins used in Figure 2, enrich for serum glycoproteins while essentially minimizing the amount of albumin carried into the next phase of proteomic analyses. In our experience, many lectins have this low affinity for serum albumin binding, but not all of them have this property, which must be determined empirically.
FIG. 1.
Concavalin A (Con A) lectin capture of serum glycoproteins. A nondisease human pooled serum sample (20 μL) was processed under different conditions, and the resulting proteins separated on an 8–16% SDS-gel, visualized by Coomassie blue staining. Lane 1, unprocessed serum, 1 μL loaded; lane 2, serum depleted of albumin and immunoglobulins using a ProteoPrep AI column from Sigma Chemicals; lane 3, serum depleted of abundant proteins using a Montage column from Millipore; lane 4, serum incubated with agarose bound Con A lectin. Bound glycoproteins were eluted with 0.2 mM alpha-methyl mannoside; lane 5, ProteoPrep AI depleted serum proteins incubated with Con A lectin, followed by elution; lane 6, montage-depleted serum proteins incubated with Con A lectin, followed by elution.
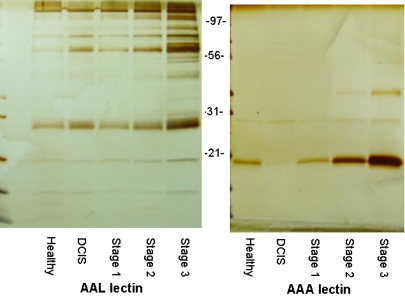
FIG. 2.
Staged breast cancer pooled sera with fucose lectin affinity capture (AAL) and (AAA). Pooled sera (20 μL) from each condition was incubated with the indicated agarose-bound lectin, aleuria aurantia (AAL), or anguilla anguilla (AAA), for 16 h. Bound glycoproteins were eluted with 0.2 mM L-Fucose, separated on an 8–16% SDS-gel and silver stained.
In Figure 2, pooled sera (n = 10 per groups) representative of healthy clinic control subjects, and sera from ductal carcinoma in situ, stage 1, stage 2, and stage 3 breast cancer diagnoses (Laronga et al., 2004), were incubated with two different fucose binding lectins. As seen in the gel separations of the bound and eluted proteins, there is a clear enrichment of different serum glycoproteins associated with different stages of disease, particularly for the more severe stage 3 samples. We have used this type of approach with multiple lectins like Con A, the two fucose lectins, and sialic acid binding lectins, followed by protease digestion and determination of protein identities by LC-electrospray mass spectrometry (Drake et al., 2006). A cumulative list of target serum glycoproteins that we have determined to be differentially expressed in breast cancer-related serum across many conditions is provided in Table 1. Not surprisingly, these proteins are all relatively abundant serum proteins and are generally associated with the stress or “acute-phase” response to disease. From what is known about the individual proteins, these are not necessarily breast cancer- or even cancer-specific proteins and may be more associated with the immune system. However, what has not been determined or most of these proteins is whether there are subpopulations of these abundant glycoproteins that carry cancer specific glycan signatures. This theme is slowly emerging, as for example cancer-specific glycoforms of serum haptoglobin and alpha-1-antitrypsin have been reported (Abbott et al., 2009; Comunale et al., 2010; Fujimari et al., 2008; Zeng et al., 2010). In particular, for our BIRADS4 serum cohort that contains many examples of benign breast disease and other patient demographics (race, BMI, age), targeting of individual serum glycoproteins listed in Table 1 for site-specific glycan analysis could yield distinct disease state biomarker candidates. Specific proteotypic peptides of most of these glycoproteins listed in Table 1 have also been reported in a multiple reaction monitoring (MRM) panel of 45 plasma proteins (Kuzyk et al., 2009). The relative abundance of these serum glycorproteins is also an advantage for mass spectrometry based assays, particularly those involving direct glycopeptide-based strategies using a triple quadropole mass spectrometer, which we have reported previously (White et al., 2009). Continued improvements in mass spectrometry instrumentation, and in software applications like SimGlycan (Premier Biosoft, Palo Alto, CA) for analysis of complex glycopeptide fragmentation patterns, will facilitate the characterization of these potential serum glycoprotein biomarker candidates. Using this type of mass spectrometry platform and software capability, we are currently exploring development of specific MRM assays targeting different glycopeptide species reflective of a given disease state.
Table 1.
Cumulative List of Lectin Captured, High Abundance, and Differentially Expressed Glycoproteins Identified as Biomarker Candidates from Breast Cancer Serum
| Alpha-2-HS-glycoprotein (Fetuin) |
| Alpha-1-acid glycoprotein 1 |
| Alpha-1-acid glycoprotein 2 |
| Haptoglobin |
| Haptoglobin-related protein |
| Alpha-1-antitrypsin |
| Alpha-2-macroglobulin |
| Alpha-1-antichymotrypsin |
| Beta-2-glycoprotein 1 |
| Ceruloplasmin |
| Galectin-3-binding protein |
| Clusterin |
| Histidine-Rich Glycoprotein |
| Transthyretin |
| Zinc-alpha-2-glycoprotein |
| Lactotransferrin |
| Hemopectin |
| Immunoglobulins |
MALDI-MS Imaging and Detection of Breast Cancer Biomarkers in Tissue
Using tumor tissues as a primary source of protein for biomarker discovery is self-evident, but there are many issues to address associated with obtaining quality breast cancer-related tissue samples (Sherman et al., 2010). One issue is ironically related to the overwhelming emphasis and success of the genetic and histology approaches. Tissue samples obtained via biopsy or mastectomy are most commonly held back for pathology use, whether for histological analysis or for isolation of genetic material. The tumors themselves are highly heterogenous, and also the quality of the sample can be greatly affected by the amount of fat in the breast. Obtaining enough cancer versus nontumor/uninvolved epithelial cell tissues can thus be challenging, and this requires much additional input from pathologists to select the most appropriate regions of tissue with disease. An additional confounder is that the types of tumor tissue obtained at biopsy in outpatient clinics are much more likely to be from smaller, less developed tumors compared to tissues obtained at major medical centers, which are more likely to be from larger, more aggressive tumors. Also, fresh-frozen tissues are expensive to obtain and store, and require established standard operating procedures for informed consent and collection at the time of surgery. Compared with collection of serum, tissues require many more clinical resources and more complex follow-up evaluations.
Although some of these limitations can be overcome, the amounts of breast tissue available for in-depth and large-scale proteomic analyses is frequently limiting. A new proteomic approach that is relatively conservative for sample sparing is MALDI-based mass spectrometry imaging approaches (Cornett et al., 2006; Schwamborn and Caprioli, 2010). In this approach, breast cancer tissues mounted on slides are directly analyzed by MALDI-TOF mass spectrometry. Two recent papers highlight its application in relation to genetic and histologic approaches for breast cancers. In one study (Rauser et al., 2010), the MALDI tissue profiles of Her2+ versus Her2− breast cancers were compared. A definitive marker of 8,404 m/z was detected in most Her2+ samples, but not in the most Her2− tissues. It was identified as CRIP1 (cysteine-rich intestinal protein 1), and mRNA levels of CRIP1 correlated with Her2 mRNA levels (Rauser et al., 2010). In another study (Bauer et al., 2010), MALDI imaging was applied to pretreatment biopsy samples obtained from a cohort of women treated by taxane/radiation and surgery of high risk breast cancers. The pretreatment biopsies were classified based on complete pathologic response following treatments versus those with residual disease. It was found that expression of alpha-defensins in the prebiopsy tissues as detected by MALDI imaging correlated with taxane responsiveness (Bauer et al., 2010).
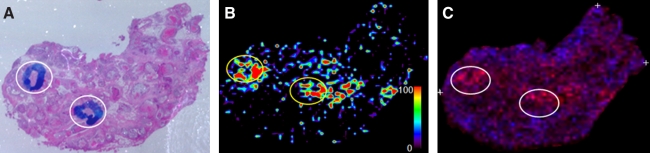
An example MALDI imaging workflow as applied to a breast tissue containing regions of invasive ductal carcinoma cells is described as follows, and a representative image is shown in Figure 3. An automated matrix sprayer was used to deposit an even layer of micron-sized droplets of matrix directly across the tissue containing normal epithelial cells, stromal areas, and invasive cancer cells. The spectra generated from each defined laser shot area were compared to each other, as well as the underlying pathology of the cell type present at each spot. Instead of being represented as peak heights, an individual m/z value is assigned a pixel intensity and color. Therefore, the more abundant a protein detected at a given m/z is in a given region, the brighter the pixel color will be in the image at that location. An example of the expression of a tumor associated peptide mass at 4,588 m/z in the invasive ductal carcinoma sample regions is shown in Figure 3B. In contrast, the expression of an abundant peptide not associated with the invasive ductal region is shown in blue pixel with the invasive ductal associated m/z peptide ion in red pixel (Fig. 3C).
FIG. 3.
MALDI-TOF mass spectrometric analysis of human breast tissue with areas of invasive ductal carcinoma. A 10-μm slice of Sakura/UMFix preserved breast tissue was processed for MALDI imaging and coated with SPA matrix followed by spectra acqusition in linear mode on a Bruker UltraFlexIII MALDI-TOF/TOF mass spectrometer. Spectra were normalized and individual m/z values visualized using FlexAnalysis and FlexImaging software, as previously described (Cazares et al., 2009). (A) Representative histology image of H&E stained breast tissue showing pathologist determined areas of invasive ductal carcinoma in the highlighted ovals. (B) Colorimetric intensity of expression levels of a peptide at m/z 4,588. Areas of most intense expression are in red (see color bar for scale). (C) Differential color intensity comparison of two peaks at 4,588 m/z (red) in the invasive ductal tissue sections relative to a noncancer associated peak at 4,990 m/z (blue).
In principle, MALDI imaging approaches can facilitate simultaneous profiling of all of the many different cell types present in a heterogeneous breast tumor tissue for each individual patient, and all of this data is linked integrally to any pathology, cytology, and genetic determinations. For breast cancers, this MALDI-MS imaging approach has the potential to improve biopsy diagnoses, better define tumor margins, and aid in multiple prognostic and treatment decision-making processes. It is also consistent with current pathology and genetic testing workflows, in that the tissues can be read by pathologists prior to analysis to ensure that regions of interest are targeted, and no more tissue is required beyond that normally done for standard H&E staining. The disadvantage and challenge with the approach is determining the sequence and identification of the differentially expressed species. Proteins and peptides of masses in the 4,000–10,000-Da range are notoriously difficult to sequence by mass spectrometry methods. This is achievable for proteins in this mass range, but requires additional sample fractionation and purification strategies to enrich for the target peptide or protein specie. Besides bulk tissue homogenization approaches, which can be limiting as described for breast cancer tissues, laser capture microscopy-based strategies combined with mass spectrometry analysis can be used to achieve the requisite enrichment (Umar et al., 2009).
Conclusions and Summary
Genetic-based assays typified by Oncotype DX and Mammaprint will continue to be extended and refined for clinical use. With Oncotype DX, for example, its utility is already being evaluated for extended use in lymph node positive (one to three nodes) ER+ disease as well as in the neoadjuvant setting to predict complete pathologic response to chemotherapy. This is in addition to the two ongoing trials for reclassifying patients after conventional risk classification (Turaga et al., 2010). Standard of practice today is to treat all women with adjuvant chemotherapy if their invasive tumor is greater than 1 cm. However, all women will not have the same outcome. Per the Oncotype DX assay, those at low risk for the development of distant recurrence have no benefit of adjuvant chemotherapy, and therefore would be treated with hormone therapy alone. Determining those individuals at high risk for distant disease can receive a tremendous benefit from chemotherapy and would thus receive both hormone therapy and chemotherapy. As stated, there is little correlation of current proteomic study designs in developing new or complementary assays that address current clinical needs for breast cancers. Thus, in this overview, we have attempted to rationalize some of the reasons for this large disparity in lack of development of protein biomarkers for use in breast cancer diagnostics, and provide potential study design and research workflow solutions to better address these needs.
As presented, we believe that targeting of serum (or plasma) glycoproteins and use of tissue for MALDI mass spectrometry imaging studies. Knowing the limitations of both samples, study design issues should be addressed at the beginning of the process. The many lessons learned from the development of the genetic assays currently being used clinically are also applicable to the proteomic study design strategy. The goal of the proteomic assays should be to develop biomarkers that complement the existing assays. For these reasons, tissue and serum specimens are most likely to provide the best sources for developing proteomic assays, as these can be collected to best reflect the many variables associated with breast cancers. For both approaches, there are multiple studies already published that provide optimism for pursuit of this strategy. However, there remains an inherent tumor biology limitation for both genetic and protein biomarker-centric approaches. Just because mRNA or protein can be detected, this does not necessarily inform on the cellular and physiologic function of the biomarker, nor whether it could be a potential therapeutic target. This type of functional analysis for putative biomarkers will continue to need to be assessed in cell line or animal model systems that are amenable to direct manipulation and experimentation.
The serum and tissue systems are also not mutually exclusive to each approach, as clearly tissue-derived glycoproteins can be isolated for similar types of characterization. Identification of changes in glycan constituents for either serum or tissue glycoproteins also provides structural biology data that can be further evaluated for gene expression changes of glycosyltransferases or glycosidases involved in the biosynthesis of the glycoconjugate. The glycoproteins listed in Table 1 are all relatively high abundance serum constituents, which makes them attractive targets for lectin, tissue, or antibody array platforms to provide clinical throughput capabilities. Analogous to the protein MRM assays described for plasma proteins (Kuzyk et al., 2009), the abundance of the serum glycoproteins makes them candidates for developing glycopeptide specific MRM panels, which is an ongoing project in our laboratories. These same strategies could also be applied be applied to tissue glycoproteins, or those identified in ductal fluids like NAF. Conversely, we are pursuing the design of antibodies or small molecules targeting the many known carbohydrate antigens associated with cancers (i.e., CA 15.3, CA 19.9, sialyl Lewis X) linked with a MALDI-reactive reporter group (Thiery et al., 2008) to identify glycoprotein targets by MALDI imaging analysis. MALDI imaging approaches can also target differential expression of lipids and, distribution of cellular or drug metabolites (Cornett et al., 2008).
In conclusion, we describe two main strategies to assist in the development of proteomic-based breast cancer diagnostics that is consistent with current clinical practice and need. Each sample type, serum or tissue, can be directly linked with all clinical data, pathology, and histology properties and gene expression data generated for a given subject with breast cancer. The hope will be to apply these proteomic technologies to the continually emerging and complex factors that affect development of improved biomarkers for breast cancer screening, diagnosis, prognosis, and treatment monitoring.
Acknowledgments
This work was supported in part by funds from the National Institutes of Health (U01 CA 085067 to OJS) and the Susan G. Komen Breast Cancer Foundation (to R.R.D.).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Abbott K.L. Aoki K. Lim J.M. Porterfield M. Johnson R. O'Regan R.M., et al. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J. Proteome Res. 2009;7:1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N.L. Anderson N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- Bauer K. Brown M. Cress R. Parise C.A. Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triplenegative phenotype: a population-based study from the california cancer registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- Bauer J.A. Chakravarthy A.B. Rosenbluth J.M. Mi D. Seeley E.H. De Matos Granja-Ingram N., et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin. Cancer Res. 2010;16:681–690. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L.A. Perou C.M. Livasy C.A. Dressler L.G. Cowan D. Conway K., et al. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- Cazares L.H. Troyer D. Mendrinos S. Lance R.S. Nyalwidhe J.O. Beydoun H.A., et al. Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin. Cancer Res. 2009;15:5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- Cheang M.C. Chia S.K. Voduc D. Gao D. Leung S. Snider J., et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Haab B.B. Analysis of glycans on serum proteins using antibody microarrays. Methods Mol. Biol. 2009;520:39–58. doi: 10.1007/978-1-60327-811-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo J.R. Nielsen T.O. Biomarkers for basal-like breast cancer. Cancers. 2010;2:1040–1065. doi: 10.3390/cancers2021040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale M.A. Rodemich-Betesh L. Hafner J. Wang M. Norton P. Di Bisceglie A.M., et al. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornett D.S. Mobley J.A. Dias E.C. Andersson M. Arteaga C.L. Sanders M.E., et al. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol. Cell Proteomics. 2006;5:1975–1983. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- Cornett D.S. Frappier S.L. Caprioli R.M. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 2008;80:5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C. Green J.E. Metastases: the glycan connection. Breast Cancer Res. 2000;2:321–323. doi: 10.1186/bcr75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R. Trudeau M. Pritchard K.I. Hanna W.M. Kahn H.K. Sawka C.A., et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Diamandis E.P. Analysis of serum proteomic patterns for early cancer diagnosis: Drawing attention to potential problems. J. Natl. Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- Drake R.R. Schwegler E.E. Malik G. Diaz J. Block T.M. Mehta A., et al. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol. Cell Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- Duffy M.J. Serum tumor markers in breast cancer: are they of clinical value? Clin. Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- Dwek M.V. Lacey H.A. Leathem A.J. Breast cancer progression is associated with a reduction in the diversity of sialylated and neutral oligosaccharides. Clin. Chim. Acta. 1998;271:191–202. doi: 10.1016/s0009-8981(97)00258-1. [DOI] [PubMed] [Google Scholar]
- Dwek M.V. Ross H.A. Leathem A.J. Proteome and glycosylation mapping identifies post-translational modifications associated with aggressive breast cancer. Proteomics. 2001;1:756–762. doi: 10.1002/1615-9861(200106)1:6<756::AID-PROT756>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Fabian C.J. Kimler B.F. Mayo M.S. Khan S.A. Breast-tissue sampling for risk assessment and prevention. Endocr. Relat. Cancer. 2005;12:185–213. doi: 10.1677/erc.1.01000. [DOI] [PubMed] [Google Scholar]
- Fan C. Oh D.S. Wessels L. Weigelt B. Nuyten D.S. Nobel A.B., et al. Concordance among gene-expression-based predictors for breast cancer. N. Engl. J. Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- Foulkes W. Stefansson I. Chappuis P. Bégin L.R. Goffin J.R. Wong N., et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- Fujimura T. Shinohara Y. Tissot B. Pang P.C. Kurogochi M. Saito S., et al. Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int. J. Cancer. 2008;122:39–49. doi: 10.1002/ijc.22958. [DOI] [PubMed] [Google Scholar]
- Handerson T. Camp R. Harigopal M. Rimm D. Pawelek J. b1,6-Branched oligosaccharides are increased in lymph node metastases and predict poor outcome in breast carcinoma. Clin. Cancer Res. 2005;11:2969–2973. doi: 10.1158/1078-0432.CCR-04-2211. [DOI] [PubMed] [Google Scholar]
- Harris L. Fritsche H. Mennel R. Norton L. Ravdin P. Taube S., et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- Higgins S.A. Matloff E.T. Rimm D.L. Dziura J. Haffty B.G. King B.L. Patterns of reduced nipple aspirate fluid production and ductal lavage cellularity in women at high risk for breast cancer. Breast Cancer Res. 2005;7:R1017–R1022. doi: 10.1186/bcr1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi J. Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconj. J. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Hu J. Coombes K.R. Morris J.S. Baggerly K.A. The importance of experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief Funct. Genomic Proteomic. 2005;3:322–331. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- Irvin W.J. Carey L.A. What is triple-negative breast cancer? Eur. J. Cancer. 2008;44:2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Kim C. Paik S. Gene-expression-based prognostic assays for breast cancer. Nat. Rev. Clin. Oncol. 2010;7:340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
- King B.L. Love S.M. The intraductal approach to the breast: raison d'être. Breast Cancer Res. 2006;8:206. doi: 10.1186/bcr1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno A. Kato Y. Matsuda A. Kaneko M.K. Ito H. Amano K., et al. Focused differential glycan analysis with the platform antibody-assisted lectin profiling for glycan-related biomarker verification. Mol. Cell Proteomics. 2008;8:99–108. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]
- Kuzyk M.A. Smith D. Yang J. Cross T.J. Jackson A.M. Hardie D.B., et al. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani S. Reis-Filho J. Fulford L. Penault-Llorca F. van der Vijver M. Parry S., et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin. Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- Laronga C. Drake R.R. Proteomic approach to breast cancer. Cancer Control. 2007;14:360–368. doi: 10.1177/107327480701400406. [DOI] [PubMed] [Google Scholar]
- Laronga C. Becker S. Watson P. Gregory B. Cazares L. Lynch H., et al. SELDI-TOF serum profiling for prognostic and diagnostic classification of breast cancers. Dis. Markers. 2004;19:229–238. doi: 10.1155/2004/759530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus E. Mainiero M.B. Schepps B. Koelliker S.L. Livingston L.S. BI-RADS lexicon for US and mammography: interobserver variability and positive predictive value. Radiology. 2006;239:385–391. doi: 10.1148/radiol.2392042127. [DOI] [PubMed] [Google Scholar]
- Lim E. Vaillant F. Wu D. Forrest N.C. Pal B. Hart A.H., et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Liu T. Qian W.J. Gritsenko M.A. Camp D.G. Monroe M.E. Moore R.J., et al. Human Plasma N-Glycoproteome Analysis by Immunoaffinity Subtraction, Hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X. Luo Y. Dai Z. Wang K. Du Y. Lin B. Integrated lectin affinity microfluidic chip for glycoform separation. Anal. Chem. 2004;76:6941–6947. doi: 10.1021/ac049270g. [DOI] [PubMed] [Google Scholar]
- Mathelin C. Cromer A. Wendling C. Tomasetto C. Rio M.C. Serum biomarkers for detection of breast cancers: A prospective study. Breast Cancer Res. Treat. 2006;96:83–90. doi: 10.1007/s10549-005-9046-2. [DOI] [PubMed] [Google Scholar]
- McLerran D. Grizzle W.E. Feng Z. Thompson I.M. Bigbee W.L. Cazares L.H., et al. SELDI-TOF MS whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin. Chem. 2008a;54:53–60. doi: 10.1373/clinchem.2007.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLerran D. Grizzle W.E. Feng Z. Bigbee W.L. Banez L.L. Cazares L.H., et al. Analytical validation of serum proteomic profiling for diagnosis of prostate cancer: sources of sample bias. Clin. Chem. 2008b;54:44–52. doi: 10.1373/clinchem.2007.091470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y. Hato M. Shinohara Y. Kuramoto H. Furukawa J. Kurogochi M., et al. BlotGlycoABCTM, an integrated glycoblotting technique for rapid and large scale clinical glycomics. Mol. Cell Proteomics. 2008;7:370–377. doi: 10.1074/mcp.M700377-MCP200. [DOI] [PubMed] [Google Scholar]
- Molina R. Barak V. van Dalen A. Duffy M.J. Einarsson R. Gion M., et al. Tumor markers in breast cancer—European Group on Tumor Markers recommendations. Tumour Biol. 2006;26:281–293. doi: 10.1159/000089260. [DOI] [PubMed] [Google Scholar]
- Morris G. Naidu S. Topham A. Guiles F. Xu Y. McCue P., et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007;110:876–884. doi: 10.1002/cncr.22836. [DOI] [PubMed] [Google Scholar]
- Obenauer S. Hermann K.P. Grabbe E. Applications and literature review of the BI-RADS classification. Eur. Radiol. 2005;15:1027–1036. doi: 10.1007/s00330-004-2593-9. [DOI] [PubMed] [Google Scholar]
- Paik S. Shak S. Tang G. Kim C. Baker J. Cronin M., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Parker J.S. Mullins M. Cheang M.C. Leung S. Voduc D. Vickery T., et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou M.P. Kulasingam V. Sauter E.R. Kliethermes B. Diamandis E.P. Nipple aspirate fluid proteome of healthy females and patients with breast cancer. Clin. Chem. 2010;56:848–855. doi: 10.1373/clinchem.2009.136283. [DOI] [PubMed] [Google Scholar]
- Perou C.M. Sorlie T. Eisen M.B. van de Rijn M. Jeffrey S.S. Rees C.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Ransohoff D.F. Bias as a threat to the validity of cancer molecular-marker research. Nat. Rev. Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- Rauser S. Marquardt C. Balluff B. Deininger S.O. Albers C. Belau E., et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2010;9:1854–1863. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- Schwamborn K. Caprioli R.M. Molecular imaging by mass spectrometry–looking beyond classical histology. Nat. Rev. Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- Sherman M.E. Howatt W. Blows F.M. Pharoah P. Hewitt S.M. Garcia-Closas M. Molecular pathology in epidemiologic studies: a primer on key considerations. Cancer Epidemiol. Biomarkers Prev. 2010;19:966–972. doi: 10.1158/1055-9965.EPI-10-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. Cazares L. Schneider H. Mitchell S. Laronga C. Semmes O.J., et al. Serum biomarkers to differentiate benign and malignant mammographic lesions. J. Am. Coll. Surg. 2006;204:1065–1071. doi: 10.1016/j.jamcollsurg.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Sorlie T. Perou C. Tibshirani R. Aas T. Geisler S. Johnsen H., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery G. Anselmi E. Audebourg A. Darii E. Abarbri M. Terris B., et al. Improvements of TArgeted multiplex mass spectrometry IMaging. Proteomics. 2008;8:3725–3734. doi: 10.1002/pmic.200701150. [DOI] [PubMed] [Google Scholar]
- Turaga K. Acs G. Laronga C. Gene expression profiling in breast cancer. Cancer Control. 2010;17:177–182. doi: 10.1177/107327481001700306. [DOI] [PubMed] [Google Scholar]
- Umar A. Kang H. Timmermans A.M. Look M.P. Meijer-van Gelder M.E. den Bakker M.A., et al. Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol. Cell Proteomics. 2009;8:1278–1294. doi: 10.1074/mcp.M800493-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver M.J. He Y.D. van't Veer L.J. Dai H. Hart A.A.M. Voskul D.W., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- van't Veer L.J. Paik Cancer S. Hayes D.F. Gene expression profiling of breast cancer: a new tumor marker. J. Clin. Oncol. 2005;23:1631–1635. doi: 10.1200/JCO.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Wada Y. Azadi P. Costello C.E. Dell A. Dwek R.A. Geyer H., et al. Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17:411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- Wang Y. Wu S.L. Hancock W.S. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap—Fourier transform mass spectrometry. Glycobiology. 2006;16:514–523. doi: 10.1093/glycob/cwj091. [DOI] [PubMed] [Google Scholar]
- White K.Y. Rodemich L. Nyalwidhe J.O. Comunale M.A. Clements M.A. Lance R.S., et al. Glycomic characterization of prostate-specific antigen and prostatic acid phosphatase in prostate cancer and benign disease seminal plasma fluids. J. Proteome Res. 2009;8:620–630. doi: 10.1021/pr8007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch M.R. Petrakis N.L. Gruenke L.D. Ernster V.L. Miike R. King E.B., et al. Factors associated with obtaining nipple aspirate fluid: analysis of 1428 women and literature review. Breast Cancer Res. Treat. 1990;15:39–51. doi: 10.1007/BF01811888. [DOI] [PubMed] [Google Scholar]
- Wuhrer M. Catalina M.I. Deelder A.M. Hokke C.H. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2006;849:115–128. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Zeng Z. Hincapie M. Haab B.B. Hanash S. Pitteri S.J. Kluck S., et al. The development of an integrated platform to identify breast cancer glycoproteome changes in human serum. J. Chromatogr. A. 2010;1217:3307–3315. doi: 10.1016/j.chroma.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Patwa T.H. Qiu W. Shedden K. Hinderer R. Misek D.E., et al. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J. Proteome Res. 2007;6:1864–1874. doi: 10.1021/pr070062p. [DOI] [PubMed] [Google Scholar]
- Zhang H. Li X.J. Martin D.B. Aebersold R. Identification and quantification of N- linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- Zielinska D.F. Gnad F. Wiśniewski J.R. Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]