When Salmonella invade mammalian epithelial cells, some populations are surrounded by the autophagy protein LC3. We found that LC3 was recruited in proximity to Salmonella independently of both Atg9L1 and FIP200, which are required for formation of autophagosomes. The dynamics of the ULK1 complex and Atg9L1 were dependent on one another.
Abstract
Salmonella develops into resident bacteria in epithelial cells, and the autophagic machinery (Atg) is thought to play an important role in this process. In this paper, we show that an autophagosome-like double-membrane structure surrounds the Salmonella still residing within the Salmonella-containing vacuole (SCV). This double membrane is defective in Atg9L1- and FAK family-interacting protein of 200 kDa (FIP200)-deficient cells. Atg9L1 and FIP200 are important for autophagy-specific recruitment of the phosphatidylinositol 3-kinase (PI3K) complex. However, in the absence of Atg9L1, FIP200, and the PI3K complex, LC3 and its E3-like enzyme, the Atg16L complex, are still recruited to Salmonella. We propose that the LC3 system is recruited through a mechanism that is independent of isolation membrane generation.
INTRODUCTION
Salmonella enterica serovar Typhimurium (S. typhimurium) is a Gram-negative bacterial pathogen that causes food poisoning and gastroenteritis in humans (Finlay and Brumell, 2000; Haraga et al., 2008). Salmonella infects small intestinal epithelial cells and develops as a resident bacterium in this niche, which allows this bacterium to cause widespread infection. Therefore understanding the mechanisms that Salmonella uses to develop into a resident bacterium in host cells is important in controlling Salmonella infection. Salmonella can invade epithelial cells by forming a specialized single-membrane organelle, termed the Salmonella-containing vacuole (SCV), using a type III secretion system (TTSS), which is a needle-like structure that injects effector proteins into the host cell cytosol (Hueck, 1998; Finlay and Brumell, 2000). In the early phase following invasion, SCVs temporarily acquire early endosome markers, such as Rab5, transferrin receptor, and EEA1; late endosomal proteins, such as LAMP1, are then substituted for these proteins (Bakowski et al., 2008; Drecktrah et al., 2008; Huang and Brumell, 2009). A few hours postinfection, Salmonella rearranges the SCVs into long tubular structures called Salmonella-induced filaments (Sifs), where the bacteria stably propagate (Bakowski et al., 2008; Drecktrah et al., 2008; Huang and Brumell, 2009).
It was previously reported that a portion of invading S. typhimurium is associated with LC3, a typical marker of autophagy, which is an intracellular bulk protein degradation system (Kabeya et al., 2000; Birmingham et al., 2006). The intracellular replication of S. typhimurium is increased in autophagy-defective host cells compared with wild-type cells (Birmingham et al., 2006; Sun et al., 2008; Fujita et al., 2009), and ultimately leads to host cell death. Therefore autophagy protects host cells from pathogenic bacteria. On the other hand, it is possible that the autophagic machinery plays a role in establishing resident bacteria. These processes are collectively called xenophagy, and the molecular mechanisms that govern these processes are just now beginning to be analyzed (Levine, 2005; Deretic and Levine, 2009; Huang and Brumell, 2009; Noda and Yoshimori, 2009). Macroautophagy is a nonselective bulk degradation system; however, S. typhimurium appears to be selectively isolated in xenophagy. Therefore analyzing xenophagy will provide additional information on how the machinery of autophagy functions. The autophagy machinery is generally composed of Atg proteins that are highly conserved from yeast to mammals (Yoshimori and Noda, 2008; Nakatogawa et al., 2009). Therefore we decided to analyze how Atg proteins function in S. typhimurium xenophagy. In this paper, we show that Atg proteins are recruited near S. typhimurium, and we provide new insight into their functions during this process. Atg9L1 is absolutely required for the formation of an autophagosome-like double membrane around the SCV. Furthermore, LC3 systems are recruited to proximity to Salmonella, even in the absence of autophagosome-like structures in Atg9L1 knockout mouse embryonic fibroblasts (KO MEFs). These results indicate the existence of a recruitment mechanism of the LC3 system independent of isolation membrane generation.
RESULTS
LC3 associates with S. typhimurium through an LC3-lipidation reaction
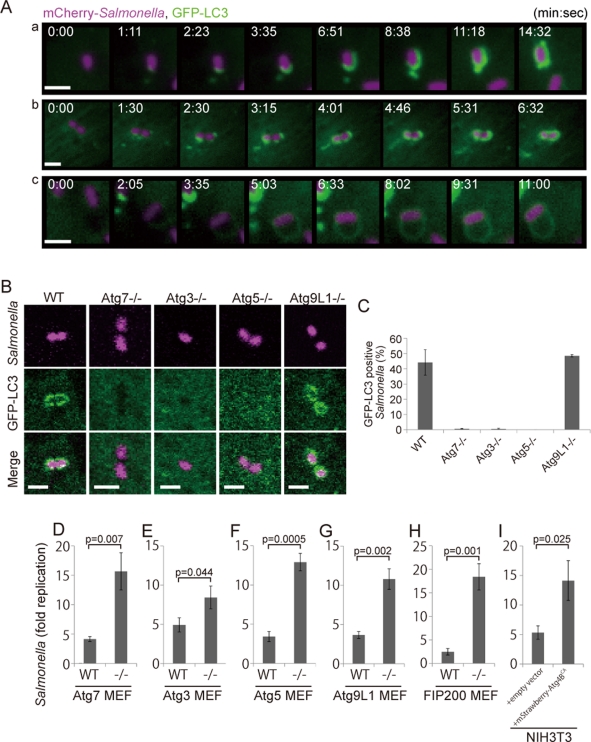
It was previously reported that a population of S. typhimurium in infected mammalian epithelial cells was surrounded by green fluorescent protein (GFP)-LC3 (Birmingham et al., 2006). To examine the detailed process by which GFP-LC3 associates with S. typhimurium, we performed time-lapse imaging of live cells. MEFs stably expressing GFP-LC3 were infected with S. typhimurium expressing the fluorescent protein mCherry. Single (Figure 1Aa and Supplemental Movie S1) or multiple (Figure 1Ab and Movie S2) GFP-LC3 puncta appeared in close contact with S. typhimurium, and the GFP-LC3 signal then elongated laterally, along with the bacteria, and finally completely surrounded the bacteria. We speculate that the multipuncta pattern represents a focus located on the posterior surface of S. typhimurium that elongated with several finger-like edges, which appear as distinct puncta. The other pattern was also observed in some instances (∼23%; Figure 1Ac and Movie S3). These observations clearly established that LC3 actively associates with S. typhimurium.
FIGURE 1:
GFP-LC3 association with infected S. typhimurium. (A) MEFs stably expressing GFP-LC3 were infected with S. typhimurium expressing mCherry for 5 min and then washed to remove the inoculum. The cells were imaged at ∼15-s intervals using a fluorescence inverted microscope. Live-cell imaging is shown and the elapsed time after the association of GFP-LC3 is indicated. Scale bar: 1 μm. (B and C) MEFs with deletions in each of the noted Atg genes and their parental cells stably expressing GFP-LC3 were infected with S. typhimurium expressing mCherry for 1 h at an MOI of 10 and then washed. The samples were fixed, and the GFP-LC3 association rate was examined by fluorescence microscopy. The average ± SD is shown for three independent experiments where at least 100 bacteria were counted. Scale bar: 2 μm. (D−I) MEFs of the noted Atg KOs and their parental cells or NIH3T3 cells with or without Atg4BC74A expression were infected with S. typhimurium for 10 min in DMEM without antibiotics at an MOI of 100. After infection, the cells were washed to remove extracellular S. typhimurium and incubated in DMEM containing gentamicin for 6 h. The cell lysates were plated on LB plates and the number of colonies was counted after a 1-d incubation at 37°C. The average ± SD of three independent experiments is shown.
We next examined the molecular mechanism of LC3 association. LC3 is a ubiquitin-like protein that undergoes phosphatidylethanolamine modifications at its C-terminus through a reaction that is catalyzed by the E1 (Atg7), E2 (Atg3), and E3 (Atg16L1 complex consisting of Atg12, Atg5, and Atg16L1) enzymes (Ichimura et al., 2000; Kabeya et al., 2000; Mizushima et al., 2001; Fujita et al., 2008b). In macroautophagy, LC3 recruitment is defective in KO MEFs lacking these genes (ATG7, ATG3, and ATG5) (Mizushima et al., 2001; Kuma et al., 2004; Komatsu et al., 2006; Sou et al., 2008). Similarly, we found that GFP-LC3 recruitment during S. typhimurium infection was totally defective in Atg7, Atg3, and Atg5 KO cells (Figure 1, B and C). These results indicate that the dynamic recruitment of LC3 to S. typhimurium requires the LC3-lipidation reaction.
Previously, it was reported that S. typhimurium growth was highly accelerated in Atg5 KO cells compared to wild-type cells (Birmingham et al., 2006). We investigated the growth of intracellular bacteria in a series of Atg KO cells. The infection efficiency of S. typhimurium was similar in wild-type and Atg KO cells (Supplemental Figure S1). In addition, we used an inactive Atg4B mutant, Atg4BC74A, which is a protease that processes LC3s and its paralogues, such as GABARAP and GATE-16. Expression of the Atg4BC74A mutant resulted in the sequestration of LC3 in a stable complex with the Atg4BC74A mutant, and thus inhibited LC3 recruitment to the autophagosome (Fujita et al., 2008a). As shown in Figure 1, D−I, S. typhimurium grew more efficiently in Atg7, Atg3, and Atg5 KO cells and Atg4BC74A-expressing cells than in wild-type cells (Figure 1, D−F and I, and Figure S1, A–F). These results indicate that LC3s must be recruited to S. typhimurium to suppress the growth of this bacterium.
Atg9L1 is dispensable for LC3 recruitment but indispensable for S. typhimurium growth suppression
Atg9 is a membrane-spanning protein that is essential for autophagosome formation in macroautophagy (Noda et al., 2000; Yamada et al., 2005; Young et al., 2006; Saitoh et al., 2009). We next examined the role of Atg9L1 in Salmonella xenophagy by monitoring the localization of GFP-LC3. GFP-LC3 efficiently associated with S. typhimurium in Atg9L1 KO cells (Figure 1, B and C), indicating that LC3 can be recruited to S. typhimurium through an Atg9L1-independent mechanism. Therefore we asked whether Atg9L1 plays a role in suppressing S. typhimurium growth by performing a colony-forming assay. Interestingly, bacterial growth was not suppressed in Atg9L1 KO cells. These results suggest that LC3 recruitment to S. typhimurium is not sufficient to suppress intracellular bacterial growth (Figure 1G).
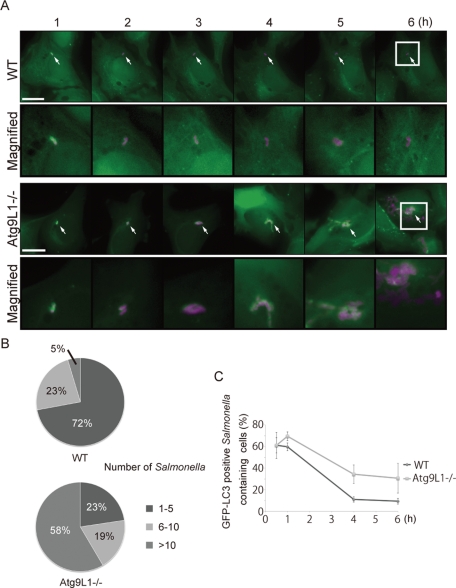
These results prompted us to investigate how LC3 recruitment is related to S. typhimurium growth in Atg9L1 KO cells. We examined the fate of a single S. typhimurium cell from 1 to 6 h postinfection by time-lapse imaging. Wild-type or Atg9L1 KO cells stably expressing GFP-LC3 were infected with S. typhimurium expressing mCherry and incubated for 1 h. Then a GFP-LC3–positive S. typhimurium was recorded for 5 h with time-lapse imaging at 4-min intervals. In wild-type cells, more than 70% of the GFP-LC3–positive S. typhimurium cells did not grow to more than 5 cells after 6 h, and only a small population grew to >10 cells (Figure 2, A and B, and Movie S4). On the other hand, in Atg9L1 KO cells, almost 60% of the GFP-LC3–positive S. typhimurium grew to >10 cells after 6 h (Figure 2, A and B, and Movie S5). The growth of GFP-LC3−negative S. typhimurium was similar between wild-type and Atg9L1 KO cells (Figure S2, A and B, and Movies S6 and S7). Significantly, the GFP-LC3 signal was diminished at a relatively early phase in wild-type cells (Figure 2C and Movie S4). However, the GFP-LC3 signal surrounding the bacteria was retained until late in infection in Atg9L1 KO cells (Figure 2, A [4−5 h postinfection] and C, and Movie S5). These results clearly indicate that GFP-LC3−positive membranes in Atg9L1 KO cells lack the ability to suppress S. typhimurium proliferation.
FIGURE 2:
Extended trace of GFP-LC3 association with Salmonella. (A) Wild-type or Atg9L1 KO MEFs stably expressing GFP-LC3 were infected with S. typhimurium expressing mCherry for 5 min at an MOI of 100, washed, and then incubated for 1 h. One GFP-LC3–positive S. typhimurium was followed by time-lapse imaging at 4-min intervals with the automated trace system of MetaMorph until 6 h postinfection. The elapsed time is indicated. Scale bar: 20 μm. (B) The final number of S. typhimurium in each cell was determined after 6 h of time-lapse imaging and grouped as indicated. (C) Cells stably expressing GFP-LC3 were infected with S. typhimurium expressing mCherry and incubated. The cells were fixed at the indicated time points and examined by fluorescence microscopy. At least 100 cells were counted for each experiment. The average ± SD is shown for three independent experiments.
Atg9L1-positive puncta surround S. typhimurium prior to the appearance of LC3
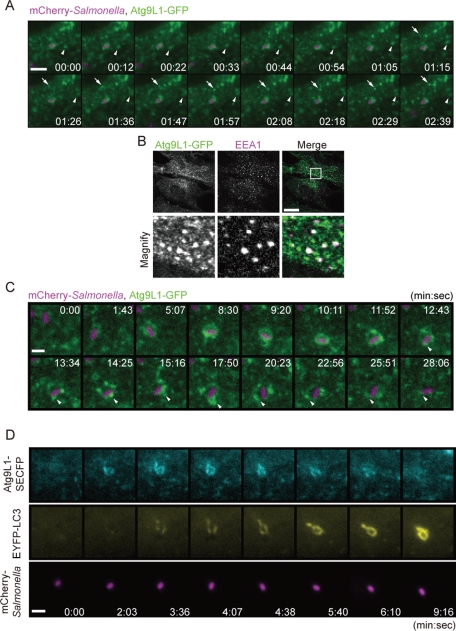
To gain further insight into how Atg9L1 is involved in these processes, we next performed time-lapse live-cell imaging of Atg9L1-GFP during bacterial infection. MEFs stably expressing Atg9L1-GFP were infected with S. typhimurium expressing mCherry. In addition to localizing to the Golgi, as previously reported (Young et al., 2006), numerous Atg9L1-GFP−positive puncta moved within the cytosol irrespective of S. typhimurium infection (Figure 3A [arrows and arrowheads], and Movie S8). Using immunofluorescence, we determined that at least some of these puncta colocalized with EEA1, an early endosomal marker (Figure 3B). There were several intense punctate signals that gradually associated with S. typhimurium, and these signals moved dynamically (Figure 3C [arrowheads] and Movie S9). Furthermore, over a longer time period, the Atg9L1-GFP signals gradually surrounded S. typhimurium, and after a certain period of time (10−60 min), the signal disappeared from the area containing S. typhimurium (Figure 3C and Movie S9). To relate these findings with the localization of LC3, we next generated cells stably expressing Atg9L1 tagged with superenhanced cyan fluorescent protein (SECFP)- and enhanced yellow fluorescent protein (EYFP)-LC3, and performed time-lapse live-cell imaging. Atg9L1-SECFP signals surrounding S. typhimurium become apparent before LC3 was recruited to the bacteria (Figure 3D [2 min, 3 s]). Next the EYFP-LC3 signals appeared and colocalized with Atg9L1-SECFP (Figure 3D [4 min, 38 s]). Finally, the Atg9L1-SECFP signals disappeared from S. typhimurium even when the EYFP-LC3 signals remained (Figure 3D [9 min, 16 s] and Movie S10). Swapping the SECFP and EYFP tags did not change the results (Figure S3 and Movie S11). These results raised the possibility that Atg9 plays a role during S. typhimurium infection that is independent of the function of LC3.
FIGURE 3:
Live-cell imaging of Atg9L1 localization. (A and C) MEFs stably expressing Atg9L1-GFP were infected with S. typhimurium expressing mCherry for 5 min. The cells were washed and images were then taken using a fluorescence inverted microscope at ∼3-s intervals for 4 min or at ∼50-s intervals for 28 min. The elapsed time is indicated. Scale bar: 5 μm (A) or 1 μm (C). (B) MEFs stably expressing Atg9L1-GFP were fixed and immunostained with anti-EEA1 antibodies. Scale bar: 20 μm. (D) MEFs stably expressing Atg9L1-SECFP and EYFP-LC3 were infected with S. typhimurium expressing mCherry for 5 min at an MOI of 100 and images were taken as in (A) and (C) at ∼30-s intervals for 10 min. Scale bar: 1 μm.
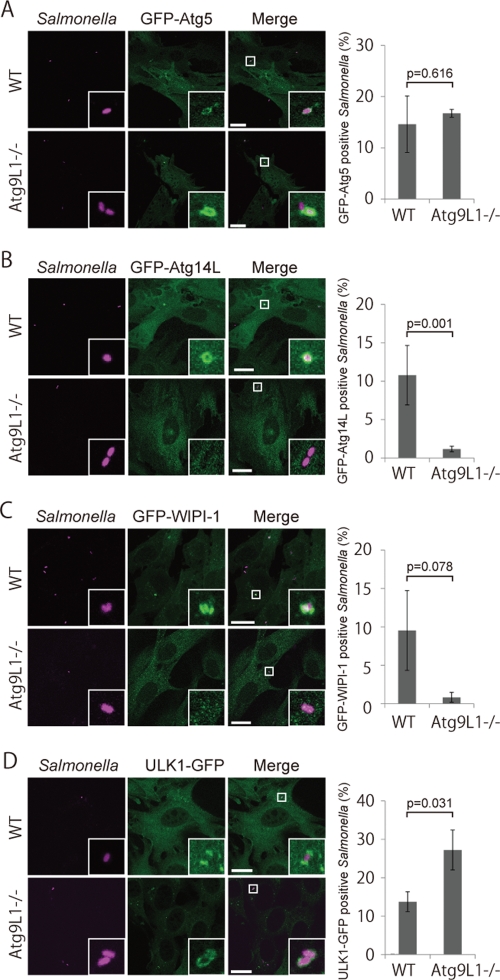
We examined the recruitment of other Atg proteins to S. typhimurium to determine if it is affected in Atg9L1 KO cells. We previously reported that the membrane localization of the Atg16L1 complex (E3) determines the site where LC3 lipidation occurs (Fujita et al., 2008b). Consistent with LC3 recruitment, GFP-Atg5, a subunit of the Atg16L1 complex, was also recruited near S. typhimurium in Atg9L1 KO cells as well as wild-type cells (Figure 4A). Therefore the Atg16L1 complex is specifically recruited to S. typhimurium independently of Atg9L1.
FIGURE 4:
Localization of Atg proteins in Atg9L1 KO cells. Atg9L1 KO MEFs and the parental wild-type cells stably expressing GFP-Atg5 (A), GFP-Atg14L (B), GFP-WIPI-1 (C), or ULK1-GFP (D) were infected with S. typhimurium expressing mCherry for 1 h. After fixation, images were taken. The number of Atg-positive bacteria for each condition was determined and the percentages per all bacteria are shown. The average ± SD is shown for three independent experiments where at least 100 bacteria were counted. Scale bar: 20 μm.
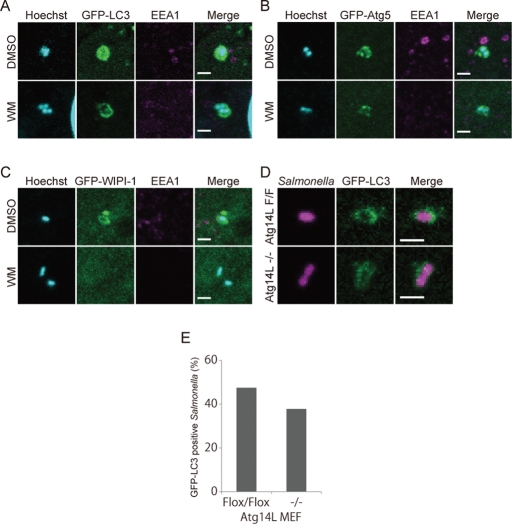
Phosphatidylinositol 3-kinase is not necessary for LC3 recruitment during S. typhimurium infection
Atg14L is a specific subunit of the class III phosphatidylinositol 3-kinase (PI3K) complex (Vps34) involved in macroautophagy (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009, 2010; Zhong et al., 2009). WD-repeat protein interacting with phosphoinositides (WIPI)-1 is a homologue of the essential yeast autophagy protein, Atg18, and binds to phosphatidylinositol 3-phosphate (PI3P) (Proikas-Cezanne et al., 2004). They localize to the autophagosomes as the autophagosomes form (Proikas-Cezanne et al., 2004; Itakura et al., 2008; Matsunaga et al., 2009). As shown in Figure 4, B and C, Atg14L and WIPI-1 were also recruited to S. typhimurium in wild-type cells. This result indicated the autophagy-specific PI3K complex might also be involved in Salmonella xenophagy. Interestingly, these localizations were defective in Atg9L1 KO cells (Figure 4, B and C).
Because Atg14L and WIPI-1 localization was not observed in Atg9L1 KO cells, we investigated whether PI3K helps recruit LC3 to S. typhimurium. During macroautophagy, LC3 recruitment is abolished by treating with wortmannin, a potent PI3K inhibitor (Kabeya et al., 2000). Cells stably expressing GFP-LC3 were treated with wortmannin 15 min prior to S. typhimurium infection, and then the number of S. typhimurium-infected cells was determined 60 min postinfection. As shown in Figure 5A, wortmannin treatment abolished EEA1 localization, which binds to endosomal membranes via PI3P (Patki et al., 1997). However, the percentage of GFP-LC3−positive S. typhimurium was not decreased in wortmannin-treated cells (36.3 ± 6.7%) compared to control cells (25.9 ± 5.4%). Likewise, Atg5 was efficiently recruited to S. typhimurium in cells treated with wortmannin during S. typhimurium infection (Figure 5B), although the localization of Atg5 during macroautophagy was also affected by wortmannin treatment (Mizushima et al., 2001). Furthermore, GFP-LC3 was efficiently and comparably recruited in Atg14L KO and Atg14L flox/flox MEFs (Figure 5, D and E). In sharp contrast to these findings, GFP-WIPI-1, the localization of which depends on the PI3P-binding capacity, was dispersed from S. typhimurium by wortmannin treatment (Figure 5C). Collectively, these data indicate that Atg5 and LC3 are recruited to S. typhimurium independent of the Atg9L1-PI3K axis.
FIGURE 5:
The effects of wortmannin on the association of each Atg protein with S. typhimurium. MEFs stably expressing GFP-LC3 (A), GFP-Atg5 (B), or GFP-WIPI-1 (C) were treated with 100 nM wortmannin for 15 min prior to infection. The cells were infected with S. typhimurium for 30 min at an MOI of 40, washed, fixed, and then immunostained with anti-EEA1 antibodies and stained with Hoechst 33342. Scale bar: 2 μm. (D, E) MEFs stably expressing GFP-LC3 were infected with S. typhimurium expressing mCherry for 1 h at an MOI of 10 and then washed. The samples were fixed, and the GFP-LC3 association rate was examined by fluorescence microscopy. At least 100 bacteria were counted. Scale bar: 2 μm.
Atg9L1 and FAK family-interacting protein of 200 kDa are required for the formation of the double-membrane structure surrounding S. typhimurium
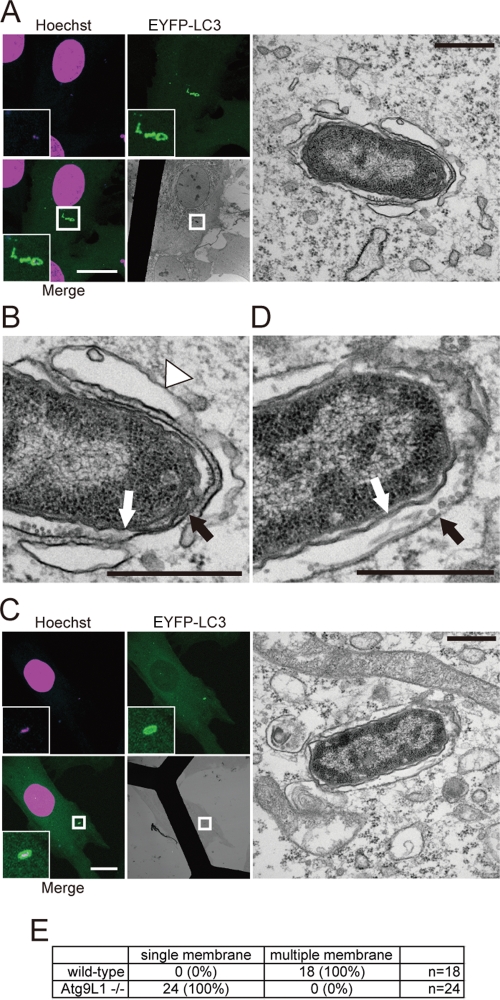
We further pursued the nature of the LC3-positive structures in Atg9L1 KO cells by using electron microscopy (EM) to examine the LC3-positive membrane structure during S. typhimurium infection. GFP-LC3−expressing MEFs were infected with S. typhimurium, and the cells were fixed 30 min postinfection. Correlative light microscopy–electron microscopy (CLEM) revealed that S. typhimurium was surrounded by GFP-LC3 signals. All of the obtained images showed that S. typhimurium was enclosed in a single membrane structure, and additionally, a double-membrane structure surrounded the single-membrane structure (Figure 6A). In some instances, a bent single-membrane sac, which is homologous to the isolation membrane in macroautophagy, was associated instead of the double membrane (Figure 6A). These findings indicate that an autophagosome-like double membrane captured S. typhimurium while it was enclosed within the SCV. By contrast, GFP-LC3−positive S. typhimurium in Atg9L1 KO cells was surrounded only by a single membrane and lacked the double membrane or isolation membrane-like structure (Figure 6B). Furthermore, we could not observe any obvious membrane and/or vesicular structures surrounding the single membrane containing Salmonella (Figures 6 and S4, A and B). The same results were obtained in FAK family-interacting protein of 200 kDa (FIP200) KO cells (Figure S5B). Therefore we suggest that formation of the autophagosome-like double-membrane structure is greatly abolished in Atg9L1 and FIP200 KO MEFs. These results indicate that Atg9L1 and FIP200 are absolutely required for the formation of the double membrane. Furthermore, GFP-LC3 is recruited to S. typhimurium-containing vacuoles even in the absence of the Atg9L1- and FIP200-dependent double-membrane structure. We contend that LC3s are less important for membrane elongation, because the autophagosome-like membrane surrounding Salmonella was observed in Atg7 KO MEFs (Figure S5A) and Atg5 KO MEFs (Zheng et al., 2009). During S. typhimurium infection, the cells were treated with Lysotracker to label the acidic compartment. After 2 h, we compared the labeling of GFP-LC3−positive S. typhimurium. The ratio of Lysotracker-positive S. typhimurium was similar between wild-type and Atg9L1 KO cells (Figure S6, A and B). These results indicated that the maturation of the GFP-LC3−positive acidic compartment is not affected by the absence of Atg9L1.
FIGURE 6:
CLEM analysis of EYFP-LC3−associated membranes. Wild-type (A and B) or Atg9L1 KO MEFs (C and D) stably expressing EYFP-LC3 were cultured on gridded, glass bottom dishes and then infected with S. typhimurium for 30 min. The cells were fixed and stained with Hoechst 33342. GFP-LC3−positive S. typhimurium was observed using a confocal laser scanning microscope. The same specimens were further examined by EM. The electron micrographs were taken in the same sample field as the transmission electron microscope. White scale bar: 20 μm; black scale bar: 500 nm. (E) Quantification of the number of single or multiple membranes surrounding S. typhimurium in wild-type or Atg9L1 KO MEFs.
FIP200 and Atg9L1 recycle interdependently from the proximity of S. typhimurium
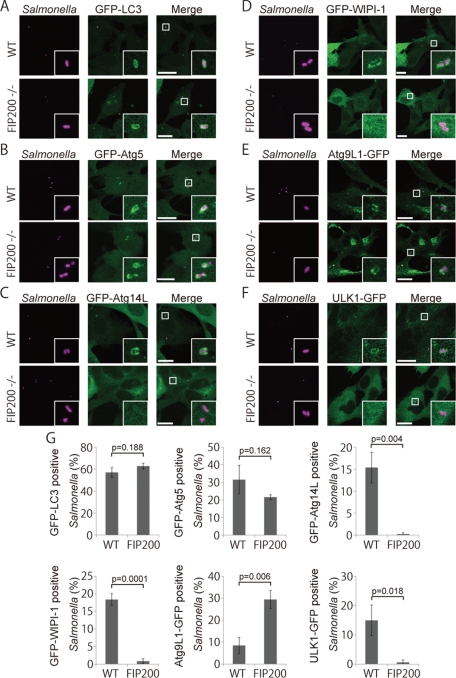
FIP200 is a subunit of an uncoordinated 51-like kinase (ULK1) protein kinase complex consisting of FIP200, Atg101, Atg13, and ULK1 (Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009). This complex is thought to be the mammalian counterpart to the yeast Atg1 protein kinase complex that is essential for autophagy (Mizushima, 2010). FIP200 KO cells have severe defects in autophagy (Hara et al., 2008). We investigated the growth of S. typhimurium in FIP200 KO cells, and as in other Atg cells, the growth of S. typhimurium was not suppressed (Figure 1H). As shown in Figure 7, A, B, and G, GFP-LC3 and GFP-Atg5 were efficiently recruited to S. typhimurium in FIP200 KO cells, as in Atg9L1 KO cells. Similar to Atg9L1 KO cells, GFP-Atg14L and GFP-WIPI-1 localization was severely affected in FIP200 KO cells (Figure 7, C, D, and G). One potential cause for these similar localization defects in Atg9L1 KO cells and FIP200 KO cells is that Atg9L1 is not properly recruited to S. typhimurium in FIP200 KO cells. However, Atg9L1 was recruited to S. typhimurium even in FIP200 KO cells (Figure 7E). Another possibility is that the ULK1 complex is not targeted to S. typhimurium in Atg9L1 KO cells. However, this possibility is also not the case, as ULK1 localization to S. typhimurium was not affected in Atg9L1 KO cells, although it was defective in FIP200 KO cells (Figure 7F). Thus, both Atg9L1 and FIP200 are required for Atg14L-WIPI-1 to localize to S. typhimurium, but are not required for each other's localization. Interestingly, high levels of Atg9L1 accumulated around S. typhimurium in FIP200 KO cells compared to wild-type cells (Figure 7G). These findings indicate that the recycling of Atg9L1 from the proximity of S. typhimurium is defective in FIP200 KO cells. Likewise, ULK1 accumulated to high levels in Atg9L1 KO cells compared to wild-type cells (Figure 4D). These results suggest that Atg9L1 and the ULK1 complex are important for each other's recycling from the area containing S. typhimurium.
FIGURE 7:
Association of Atg proteins with S. typhimurium in FIP200 KO MEFs. Wild-type or FIP200 KO MEFs stably expressing GFP-LC3 (A), GFP-Atg5 (B), GFP-Atg14L (C), GFP-WIPI-1 (D), Atg9L1-GFP (E), or ULK1-GFP (F) were infected with S. typhimurium expressing mCherry for 1 h. The cells were fixed and observed. (G) The percentage of bacteria associated with the Atg proteins was determined by fluorescence microscopy. The average ± SD is shown for at least three independent experiments where at least 100 bacteria were examined. Scale bar: 20 μm.
DISCUSSION
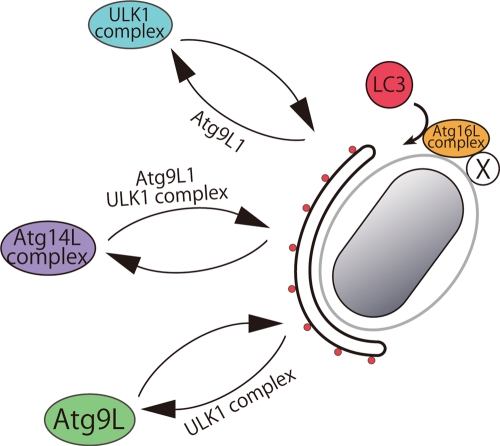
In this study, we have identified three key points regarding Salmonella xenophagy: 1) An autophagosome-like double-membrane structure captures S. typhimurium while it is still within the SCV. 2) Atg9L1 is required to form the isolation membrane-like double-membrane structure. 3) The recruitment of LC3 to S. typhimurium is independent of both the Atg9L1-PI3K axis and ULK1 complex.
Our time-lapse imaging data indicate that S. typhimurium is already enclosed within an Atg9L1-positive membrane structure before LC3 recruitment. Significantly, both Atg9L1 and LC3 surround S. typhimurium for a certain time period. This finding strongly suggests that the LC3-positive double membrane surrounds an Atg9L1-positive membranous structure. Our EM images also support this model. Therefore, as suggested in a recent review (Huang and Brumell, 2009), S. typhimurium is enclosed in a single membrane structure, the SCV, which is surrounded by a double-membrane structure. This membrane morphology, which is surrounded by a double membrane when the bacterium escapes into the cytosol, is not similar to the xenophagy of other bacteria, such as the group A streptococcus (Yamaguchi et al., 2009). In the case of Shigella, bacteria-induced rupture of the endosomal membrane provokes xenophagy (Paz et al., 2010). S. typhimurium that is associated with LC3 also has been shown to associate with ubiquitin, which suggests that there is a pore that is accessible from the cytosol (Birmingham et al., 2006). Thus, it is possible that SCV membrane rupture is also important for Salmonella xenophagy.
We showed that Atg9L1 is required to generate the isolation membrane during xenophagy. This finding is consistent with a previous observation that macroautophagosome formation is defective during macroautophagy in mammalian and yeast Atg9 mutants (Noda et al., 2000; Saitoh et al., 2009). Significantly, an autophagy-specific PI3K (Atg14L) and its downstream effector, WIPI-1, failed to be recruited to S. typhimurium in Atg9L1 KO cells (Figure 8). Atg14L/Barkor was also previously shown to be important in suppressing S. typhimurium growth (Sun et al., 2008). Thus defects in isolation membrane generation in Atg9L1 KO cells are at least partially due to defects in the autophagy-specific PI3K. It should be noted that Atg9L1 ultimately disappeared from S. typhimurium, although the LC3 signals remained. This suggests that Atg9L1 cycles between the SCV and a peripheral pool that includes the Golgi and early endosomes. We showed that FIP200 is important for this recycling (Figure 7, E and G, and Figure 8). A similar recycling defect has been observed during macroautophagy in ULK1-defective cells (Young et al., 2006; Hara et al., 2008), and also in a yeast Atg1 mutant (Reggiori et al., 2004). Furthermore, we noted that ULK1 accumulated near S. typhimurium in Atg9L1 KO cells, indicating that ULK1 recycling is abolished (Figures 4D and 8). Therefore Atg9L1 and the ULK1 complex are important for each other's recycling from proximity to S. typhimurium. We suggest that the recycling of these molecules is tightly coupled with the generation of the isolation membrane.
FIGURE 8:
Model of Atg protein dynamics in Salmonella xenophagy. Atg14L, ULK1, and Atg9L1 cycle between the membrane formation site near S. typhimurium and another cellular pool. The cycling to and from the membrane formation site requires the indicated Atg proteins. S. typhimurium provides an unknown recruitment factor (X), and the Atg16L1 complex is potentially recruited near the SCV. The isolation membrane, formed independently of LC3, may be the preferable target of LC3 lipidation. In Atg9L1 KO MEFs, the isolation membrane does not form, and the SCV is the target of LC3 lipidation. Even without the LC3 system, an incomplete isolation membrane is formed.
To our surprise, LC3 is recruited to S. typhimurium through a mechanism that is independent of Atg9L1, PI3K, and FIP200. Significantly, the Atg16L complex is also recruited near S. typhimurium. We previously showed that the localization of the Atg16L complex to any membrane is sufficient to recruit and convert LC3-I to LC3-II, and the Atg16L complex plays a key role in this process (Fujita et al., 2008b). Therefore we propose that the Atg16L-E3 complex is recruited in close proximity to Salmonella even in the absence of Atg9L1.
Among the examined Atg proteins, only the ubiquitination-like reaction system, including Atg7, Atg3, and Atg5, is required for LC3 recruitment. These results are not consistent with a previous report that LC3 recruitment to the autophagosome formation site is abolished by wortmannin treatment and in FIP200 KO cells (Hara et al., 2008; Itakura and Mizushima, 2010). However, LC3 lipidation still occurred in FIP200, Atg14L, Atg9L1, and Vps34 KO cells, and LC3 was targeted to an unidentified membrane other than the autophagosome (Hara et al., 2008; Matsunaga et al., 2009; Saitoh et al., 2009; Zhou et al., 2010). Thus we would like to point out that the defect in LC3 recruitment to the autophagosome formation site in macroautophagy may be due to the lack of a native isolation membrane as the target. During Salmonella xenophagy in Atg9L1 KO cells, the SCV acts as an alternative target membrane and therefore can recruit LC3 (Figure 8). In wild-type cells, the isolation membrane would be the preferential target of LC3 lipidation over the SCV (Figure 8). Wortmannin is a potent inhibitor of LC3 membrane targeting and lipidation in macroautophagy. We reasoned that the unnatural target membrane of LC3 observed in Vps34 KO and Atg14L KO cells during macroautophagy may be affected because wortmannin inhibits all classes of PI3K. We propose that LC3 targeting depends on the presence of three factors: a target membrane, a lipidation system, and some unknown targeting mechanism, but that this targeting is independent of Atg9L1, PI3K, and ULK1. What is the targeting mechanism of the LC3-lipidation machinery? It is possible that ubiquitin and its adaptor proteins (p62/SQSTM1, NDP52) directly recruit LC3, and/or formin-binding protein 1-like (FNBP1L) directly recruits Atg3 (Huett et al., 2009; Thurston et al., 2009; Zheng et al., 2009). However, we are certain that additional unknown mechanisms also recruit the E3 Atg16L1 complex (depicted as “X” in Figure 8).
Several studies have reported that a double-membrane autophagosome-like structure accumulates in cells with a defective LC3 system (Fujita et al., 2008a; Sou et al., 2008; Nishida et al., 2009). Even in Salmonella xenophagy, a double-membrane lamellar structure surrounds S. typhimurium in Atg5 KO and Atg9L1 KO cells as in wild-type cells, although this structure occurs less frequently (Figure 8; Zheng et al., 2009). Atg9L1 is absolutely required for the formation of the double membrane, while LC3s are less important for membrane elongation, because the autophagosome-like membrane surrounding Salmonella was observed in Atg7 KO MEFs (Figure S5A) and Atg5 KO MEFs (Zheng et al., 2009). Importantly, these autophagosomes are potentially not completely closed, and therefore LC3 is thought to be involved in the final closure of the isolation membrane (Fujita et al., 2008a; Noda et al., 2009). In this paper, we propose a novel xenophagy model as follows (Figure 8). First, the isolation membrane, which ultimately becomes the xenophagosome, is formed through an Atg9L1-dependent mechanism, and potentially requires PI3K and the ULK1 complex (Figure 8). The LC3-lipidation machinery is recruited independently of these proteins, but requires an unknown targeting mechanism (Figure 8). Finally, LC3 on the isolation membrane closes the isolation membrane (Figure 8). The most important point is that the LC3 recruitment system is independent of the isolation membrane formation system. We believe this is a decent working model of autophagic processes that may also be applicable to macroautophagy and should be further examined.
MATERIALS AND METHODS
Plasmid construction and virus production
To generate pMRX-EGFP-C1-Atg5, pMRX-EGFP-C2-LC3, or pMRX-EGFP-C2-Atg14L, Atg5, LC3, or Atg14L were amplified by PCR and digested with KpnI-NotI, EcoRI-NotI, and EcoRI-NotI, respectively. pEGFP-C1 and pEGFP-C2 (Clontech, Mountain View, CA) were digested with NheI, treated with Klenow fragment (Takara Bio, Shigo, Japan), and then digested with KpnI and EcoRI, respectively. The pMRX vector was digested with EcoRI, treated with Klenow fragment, and then digested with NotI. Atg5, LC3, or Atg14L and the EGFP fragment were subcloned into the pMRX vector. To generate pMRX-ULK1-EGFP-N3 or pMRX-Atg9L1-EGFP-N1, ULK1 and Atg9L1 were PCR amplified and digested with EcoRI-BamHI and EcoRI-SacII, respectively. pEGFP-N3 and pEGFP-N1 were digested with BamHI-NotI and SacII-NotI, respectively. The PCR products and EGFP fragment were subcloned into the EcoRI-NotI sites of the pMRX vector. To generate pMRX- EYFP-C2-LC3, the EYFP sequence was PCR amplified from pEYFP-C1 (Clontech) and NheI-BamHI sites were added to the N-terminus of EYFP. The products were digested with NheI-BglII and subcloned into the pEGFP-C2 vector that was digested with NheI-BglII (pEYFP-C2B). The pEYFP-C2B vector was digested with BamHI-EcoRI. LC3 and the EYFP-C2B fragment were subcloned into the BamHI-NotI sites of the pMRX vector. The pMRX-EGFP-WIPI-1 plasmid was a generous gift from K. Matsunaga (University of Gunma, Gunma, Japan). pMRX-IRES-puro and pMRX-IRES-bsr were kindly provided by S. Yamaoka (Tokyo Medical and Dental University, Tokyo, Japan) (Saitoh et al., 2003). The pMRX-IRES-puro vector was kindly provided by T. Kitamura (University of Tokyo, Tokyo, Japan) (Morita et al., 2000). Recombinant retroviruses were prepared as previously described (Saitoh et al., 2003). Plat-E cells were generously provided by T. Kitamura (University of Tokyo, Tokyo, Japan) (Morita et al., 2000). To generate pMRX-Atg9L1-SECFP, Atg9L1 was subcloned into the pDONR221 plasmid (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Atg9L1-SECFP was constructed using the Multisite Gateway system (Invitrogen) (Sasaki et al., 2004; Sone et al., 2008). The DNA fragment containing attR1, ccdB, CmR, and attR2 was PCR amplified from pAd/CMV/V5-DEST (Invitrogen) and digested with BglII-NotI. The fragment was subcloned into the BamHI-NotI sites of pMRX (pMRX-DEST). The pENTR-Atg9L1-SECFP sequence was transferred into the pMRX-DEST vector using an LR reaction.
Host cell culture
MEFs, NIH3T3, and Plat-E cells were cultured in DMEM D6429 (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum (FBS), 4 mM l-glutamic acid, 5 U/ml penicillin, and 50 U/ml streptomycin at 37°C with 5% CO2. Stable transformants were selected in medium containing 1 μg/ml puromycin or 10 μg/ml blasticidin. For wortmannin treatment, 1 mg/ml wortmannin (Sigma) was dissolved in dimethyl sulfoxide (DMSO) and added at a final concentration of 100 nM.
Bacterial infection
Salmonella enterica serovar Typhimurium SR-11 x3181 was used as the wild-type strain and routinely cultured in supplemented LB. Salmonella harboring the pBR-mCherry or pBR-mRFP plasmid, which expresses mCherry or mRFP under a constitutive promoter in pBR322 (Birmingham et al., 2006), respectively, was routinely cultured in lysogeny broth (LB) supplemented with ampicillin (100 μg/ml). Bacteria were grown overnight at 37°C, and then subcultured at 1:33 for 3 h in LB without antibiotics. The bacterial inocula were prepared by pelleting at 10,000 × g for 2 min, and were then added to host cells at a multiplicity of infection (MOI) of 10–100 at 37°C with 5% CO2.
Indirect immunofluorescence analysis
Samples were fixed with 4% paraformaldehyde for 10 min, quenched with 50 mM NH4Cl/phosphate-buffered saline (PBS), permeabilized with 50 μg/ml digitonin/PBS, blocked with 0.5% bovine serum albumin (BSA)/PBS and then incubated with the indicated primary antibodies (anti-EEA1 antibodies were diluted 1:100). After washing with PBS, the samples were incubated with secondary antibodies and Hoechst 33342 (Sigma) and mounted with Slow Fade Gold (Invitrogen). Hoechst 33342/PBS was used according to the manufacturer's instructions. The samples were observed under an Olympus FV1000 confocal microscope with a 100×, 1.35 numerical aperture (NA) oil immersion objective (Olympus, Tokyo, Japan). The following antibodies were used: rabbit polyclonal anti–human EEA1 antibodies (1:100 dilution) (#2411; Cell Signaling Technology, Danvers, MA). All secondary antibodies were Alexa Fluor conjugates from Invitrogen and diluted 1:1000 in 0.5% BSA/PBS. Confocal images were imported into Adobe Photoshop and assembled in Adobe Illustrator (San Jose, CA).
Live-cell imaging
Host cells were grown in DMEM D6434 (Sigma) supplemented with 10% FBS without antibiotics on glass bottom dishes (D310300; Matsunami Glass, Osaka, Japan). After bacterial infection, the glass bottom dish was mounted onto the microscope stage, which was equipped with a humidified environment chamber (MI-IBC, Olympus) that maintained the dish at 37°C with 5% CO2. Images were acquired using an inverted microscope Olympus IX81 equipped with a 100×, 1.40 NA oil immersion objective (Olympus), a mercury or xenon lamp, a cooled charge-coupled device camera (Roper CoolSNAP HQ, Ottobrunn, Germany), and a ZDC system under the control of MetaMorph (Molecular Devices, MDS Analytical Technologies, Tokyo, Japan).
Colony formation assay
Colony formation assay was performed as previously described (Steele-Mortimer, 2008). Briefly, cells were cultured in DMEM without antibiotics and infected with S. typhimurium for 10 min at an MOI of 100. The cells were washed to remove extracellular S. typhimurium and incubated in DMEM without antibiotics for 20 min. The medium was replaced with DMEM containing 50 μg/ml gentamicin for 40 min and then replaced with medium containing 5 μg/ml gentamicin. The host cells were lysed with 1% Triton X-100 and 0.1% SDS/PBS solution, serially diluted with PBS, and spread onto LB plates.
Correlative light microscopy–electron microscopy (CLEM)
Host cells stably expressing EYFP-LC3 were cultured on glass bottom dishes with a grid pattern (P35G-2–14-C-GRID; MatTek, Ashland, MA) and infected with S. typhimurium for 30 min. The cells were fixed with 4% (wt/vol) formaldehyde in 30 mM HEPES buffer (pH 7.4) containing 100 mM NaCl, 2 mM CaCl2, and 1 μg/ml Hoechst 33342 for 30 min at room temperature; washed in 30 mM HEPES buffer (pH 7.4) containing 100 mM NaCl and 2 mM CaCl2; and examined using a confocal laser scanning microscope (FV1000; Olympus). The same specimens were further incubated with 2% (wt/vol) formaldehyde and 2.5% (wt/vol) glutaraldehyde in 30 mM HEPES buffer (pH 7.4) containing 100 mM NaCl and 2 mM CaCl2 at 4°C overnight. After three washes in 30 mM HEPES buffer (pH 7.4) containing 100 mM NaCl and 2 mM CaCl2, the samples were postfixed with 1% (wt/vol) osmium tetroxide in 30 mM HEPES buffer (pH 7.4) containing 0.5% (wt/vol) potassium ferrocyanide, 100 mM NaCl and 2 mM CaCl2 for 1 h, washed three times in distilled water, dehydrated in ethanol, and embedded in Epon812 (TAAB Laboratories Equipment, Aldermaston, UK). Ultrathin sections (70 nm thick) were stained with saturated uranyl acetate and Reynolds lead citrate solution. The electron micrographs were taken with a JEOL JEM-1011 transmission electron microscope (JEOL, Tokyo, Japan).
Statistical analysis
Statistical analyses were performed using a two-tailed unpaired t test; p values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The authors thank K. Matsunaga (Yoshimori lab) for the gift of pMX-GFP-WIPI-1 construct; Roger Y. Tsien (University of California, San Diego, CA) for kindly providing the mCherry cDNA; A. Miyawaki (Brain Science Institute, RIKEN, Japan) for kindly providing the SECFP cDNA; S. Yamaoka for the gifts of pMRX-IRES-puro vector and pMRX-IRES-bsr vector; T. Kitamura (University of Tokyo, Japan) for the gift of Plat-E cells and pMX-IRES-puro vector; N. Mizushima for the gift of Atg5−/− MEFs; M. Komatsu for the gift of Atg3−/− and Atg7−/− MEFs; the Kitasato Institute for Life Science (Tokyo, Japan), which provided Salmonella enterica serovar Typhimurium (SR-11 x3181); and all of the members of the Yoshimori laboratory for helpful discussions, especially N. Fujita (Yoshimori lab) for sharing the project idea. The work described in this report was supported in part by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, and by the Takeda Science Foundation and the Kato Memorial Bioscience Foundation.
Abbreviations used:
- BSA
bovine serum albumin
- CLEM
correlative light microscopy–electron microscopy
- DMSO
dimethyl sulfoxide
- EM
electron microscopy
- EYFP
enhanced yellow fluorescent protein
- FBS
fetal bovine serum
- FIP200
FAK family-interacting protein of 200 kDa
- FNBP1L
formin-binding protein 1-like
- GFP
green fluorescent protein
- KO MEF
knockout mouse embryonic fibroblast
- LB
lysogeny broth
- MOI
multiplicity of infection
- NA
numerical aperture
- PBS
phosphate-buffered saline
- PI3K
phosphatidylinositol 3-kinase
- PI3P
phosphatidylinositol 3-phosphate
- S. typhimurium
Salmonella enterica serovar Typhimurium
- SCV
Salmonella-containing vacuole
- SECFP
superenhanced cyan fluorescent protein
- Sifs
Salmonella-induced filaments
- TTSS
type III secretion system
- ULK
uncoordinated 51-like kinase
- WIPI
WD-repeat protein interacting with phosphoinositides
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0893) on April 27, 2011.
REFERENCES
- Bakowski MA, Braun V, Brumell JH. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic. 2008;9:2117–2129. doi: 10.1111/j.1600-0854.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, Brumell JH. Salmonella interactions with host cells: in vitro to in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355:623–631. doi: 10.1098/rstb.2000.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008a;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008b;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T. Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem. 2009;284:32602–32609. doi: 10.1074/jbc.M109.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. U.K. ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a U.K.interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, et al. Nutrient-dependent mTORC1 association with the U.K.-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Autophagy in immunity against intracellular bacteria. Curr Top Microbiol Immunol. 2009;335:189–215. doi: 10.1007/978-3-642-00302-8_9. [DOI] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huett A, Ng A, Cao Z, Kuballa P, Komatsu M, Daly MJ, Podolsky DK, Xavier RJ. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J Immunol. 2009;182:4917–4930. doi: 10.4049/jimmunol.0803050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. U.K.Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/U.K. complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ. 2009;16:984–990. doi: 10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Yoshimori T. Molecular basis of canonical and bactericidal autophagy. Int Immunol. 2009;21:1199–1204. doi: 10.1093/intimm/dxp088. [DOI] [PubMed] [Google Scholar]
- Patki V, Virbasius J, Lane WS, Toh BH, Shpetner HS, Corvera S. Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 1997;94:7326–7330. doi: 10.1073/pnas.94.14.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Nakayama M, Nakano H, Yagita H, Yamamoto N, Yamaoka S. TWEAK induces NF-kappaB2 p100 processing and long lasting NF-kappaB activation. J Biol Chem. 2003;278:36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Sone T, Yoshida S, Yahata K, Hotta J, Chesnut JD, Honda T, Imamoto F. Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J Biotechnol. 2004;107:233–243. doi: 10.1016/j.jbiotec.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Sone T, Yahata K, Sasaki Y, Hotta J, Kishine H, Chesnut JD, Imamoto F. Multi-gene gateway clone design for expression of multiple heterologous genes in living cells: modular construction of multiple cDNA expression elements using recombinant cloning. J Biotechnol. 2008;136:113–121. doi: 10.1016/j.jbiotec.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O. Infection of epithelial cells with Salmonella enterica. Methods Mol Biol. 2008;431:201–211. doi: 10.1007/978-1-60327-032-8_16. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–18290. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Nakagawa I, Yamamoto A, Amano A, Noda T, Yoshimori T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Noda T. Toward unraveling membrane biogenesis in mammalian autophagy. Curr Opin Cell Biol. 2008;20:401–407. doi: 10.1016/j.ceb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and U.K.-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang L, Hasegawa H, Amin P, Han BX, Kaneko S, He Y, Wang F. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proc Natl Acad Sci USA. 2010;107:9424–9429. doi: 10.1073/pnas.0914725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.