Abstract
The role of microRNA-1 (miR-1) has been studied in cardiac and skeletal muscle differentiation. However, it remains unexplored in vascular smooth muscle cells (SMCs) differentiation. The aim of this study was to uncover novel targets of and shed light on the function of miR-1 in the context of embryonic stem cell (ESC) differentiation of SMCs in vitro. miR-1 expression is steadily increased during differentiation of mouse ESC to SMCs. Loss-of-function approaches using miR-1 inhibitors uncovered that miR-1 is required for SMC lineage differentiation in ESC-derived SMC cultures, as evidenced by downregulation of SMC-specific markers and decrease of derived SMC population. In addition, bioinformatics analysis unveiled a miR-1 binding site on the Kruppel-like factor 4 (KLF4) 3′ untranslated region (3′UTR), in a region that is highly conserved across species. Consistently, miR-1 mimic reduced KLF4 3′UTR luciferase activity, which can be rescued by mutating the miR-1 binding site on the KLF4 3′UTR in the reporter construct. Additionally, repression of the miR-1 expression by miR-1 inhibitor can reverse KLF4 downregulation during ESC-SMC differentiation, which subsequently inhibits SMC differentiation. We conclude that miR-1 plays a critical role in the determination of SMC fate during retinoid acid-induced ESC/SMC differentiation, which may indicate that miR-1 has a role to promote SMC differentiation.
Introduction
The recent discovery of microRNAs (miRs) introduces a novel type of regulatory control over gene expression during plant and animal development [1,2]. The finding that some of these miRs are expressed in specific mesoderm and muscle tissues has rekindled an interest in post-transcriptional regulation during muscle development and pathology and has raised the question of the role of miRs during this process. MiRs comprise a large family of ∼22-nucleotide single-stranded RNAs that decrease gene expression by binding to target mRNAs and causing translational repression. Subsequently, miRs can cause partial or full silencing of respective target genes. Further, it seems that miRs can form extensive regulatory networks with a complexity comparable to that of transcription factors [3,4]. Bioinformatics prediction tools for miR targets allow us to get a glimpse at the complexity of cross-regulation, multiplicity, and redundancy in targeting, and also the multilayered nature of potential miR regulatory networks. Several miRs have been shown to be involved in skeletal muscle cell, cardiomyocyte, and smooth muscle cell (SMC) biology [5–10]. Specifically, emerging data suggest that miR-1 is involved in skeletal muscle cell and cardiomyocyte differentiation, cardiac hypertrophy and the maintenance of normal cardiac function [9,11]. However, the role of miR-1 in SMC differentiation remains unexplored. Here, we analyzed miR-1 expression and function during mouse ESC/SMC differentiation and our results define miR-1 as a positive regulator of SMC differentiation through specific targeting of Kruppel-like factor 4 (KLF4), an antimyogenic factor [12–16]. Thus, our studies establish miR-1 as a factor to enhance SMC differentiation.
Methods
Mouse embryonic stem cell/SMC differentiation system
Dicer−/− embryonic stem cells (ESCs) (kindly provided by Dr. Gregory J. Hannon of Cold Spring Harbor Lab) [17] and wild-type (WT) ESCs (CMTI-1) were routinely expanded and induced to differentiate into SMCs in vitro treated with 10 μM all trans-retinoid acid (RA) following the protocol described in our previous report [18].
TaqMan miR assay
For measurement of miR expression, specific primers for miR-1 and U6 (Applied Biosystem) were used. miR-1 expression was quantified using TaqMan miR assays (Applied Biosystem) following the manufacturer's protocol. All TaqMan miR assays were performed in triplicate. Total RNA input was normalized based on the threshold cycle (Ct) values of the U6 assay as an endogenous control. The fold change was calculated based on delta Ct between endogenous U6 control and miR-1.
Quantitative real time-polymerase chain reaction
Total RNA was extracted by using the RNeasy mini kit (Qiagen) following previous report [18]. cDNA was synthesized and subjected to polymerase chain reaction (PCR) amplification with primers specific for mouse SMC genes and other control genes with 18S RNA as an internal standard. PCR primers and reaction conditions are described in Supplementary Table S1 (Supplementary Materials available online at www.liebertonline.com/scd) [19].
Transient transfection of miR inhibitor or mimic and infection of adenovirus expressing KLF4
For inhibiting the function of cellular miR-1, a commercially available miR-1 inhibitor or mimic and inhibitor negative control (NC) or mimic NC (Dharmacon) [20] were used for transfection. A complex of Lipofectamine 2000 (Invitrogen) and 50 nM miR-1 inhibitor/mimic or inhibitor/mimic NC were prepared according to the manufacturer's instructions and previous report [21]. Adenovirus expressing KLF4 (Ad-KLF4, kindly provided by Dr. GK Owens) [14] was used to infect differentiating ESCs, with adenovirus expressing green fluorescence protein (Ad-GFP) serving as control.
Immunofluorescence staining and fluorescence-activated cell sorting analysis
Immunofluorescence staining and fluorescence-activated cell sorting (FACS) analysis was performed following a previous report [22]. Briefly, cell pellets were fixed and permeabilized using a BD cytofix/cytoperm™ kit (BD Bioscience) and incubated overnight with antibodies (anti-α-smooth muscle actin [α-SMA] antibody from Millipore and anti-smooth muscle myosin heavy chain [SMMHC] from BTI). Mouse IgG2a served as isotypic control (Dakocytomation), respectively. Goat anti-mouse IgG Alexa fluor 488 (green) or 594 (Red) served as secondary antibody (Molecular Probes). Finally, fluorescence was imaged by fluorescence microscopy (Olympus) and analyzed using the FACSCalibur™ system (BD Biosciences) following the user's guide.
Construction of reporter plasmid and reporter assays
A construct in which a fragment of the 3′ untranslated region (3′-UTR) of KLF4 mRNA containing the putative or mutated (MU) miR-1 binding sequence was used. pMIR-REPORT™ vectors (Promega) harboring KLF4 (951 bp) sequences with WT (5′-ggauggaucuucuaucauuccaa-3′, from 286 to 292 bp of the 3′ UTR region) or MU (5′ ggauggaucuucuaucauCUUGa- 3′, with the MU positions indicated by capital letters and underscore) miR-1 binding sites were generated by cloning into the SacI and HindIII restriction sites of pMIR-REPORT, independently. Cloning primers are shown in Supplementary Table S2. Luciferase activity assays were performed in 293T cells co-transfected with (1) miR-1 mimic or mimic NC (Dharmacon; 50 nM) and (2) pMIR-REPORT vectors containing WT or MU miR-1 binding sites (200 ng), together with Renilla luciferase control reporter vector [pRL-thymidine kinase (TK)] using Lipofectamine 2000 (Invitrogen). Cells were further grown in Dulbecco's modified Eagle's medium/F12 supplemented with 10% fetal bovine serum, and relative luciferase measurements were performed 48 h post-transfection using the dual Luciferase Reporter Assay System (Promega) by a Luminescence Counter (PerkinElmer). Luciferase activity of each sample was normalized to the responding thymidine kinase (TK) promoter–Renilla-luciferase activity.
miR target search
Target genes for miR-1 were predicted using open-source software PicTar (http://pictar.mdc-berlin.de/) and TargetScan 4.2 (www.targetscan.org/).
Western blot analysis
Protein samples from cells and tissues were extracted using the mammalian protein extraction reagent M-Per (Promega) supplemented with a protease inhibitor cocktail (Roche). Antibodies against α-SMA (1:3,000; Millipore), SMMHC (1:2,000; BTI), β-tubulin (1:10,000; Millipore), Myocardin (MyoCD, 1:1,000; Abcam), and KLF4 (1:500; Abcam) were used for testing individual protein expression. Immunoactivity was observed by the enhanced chemiluminescence detection system (Amersham Biosciences) according to the manufacturer's instructions.
Statistical analysis
Data were analyzed by analysis and tested for statistical significance by the Student-Newman-Keuls test using SYSTAT software (SYSTAT) with values of P < 0.05 considered to be significant. All experiments were independently repeated at least 3 times.
Results
miR-1 is induced during SMC differentiation from mouse ESCs
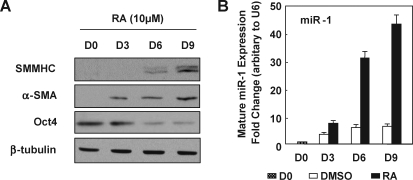
SMC differentiated from ESC differentiation with the treatment of RA, as we showed previously [21] and in Fig. 1A and Supplementary Fig. S1. miR-1 was quantified in undifferentiated and differentiated cells at different time points by TaqMan miR assay. As shown in the Fig. 1B, miR-1 was significantly upregulated ∼30-fold in RA-induced SMC differentiation (Fig. 1B) at day 6 and kept increasing with extended culture times. Expression of several reported SMC-related miRs analyzed (Supplementary Fig. S2A) showed that miR-21 and miR-145 increased along with SMC differentiation from ESCs, whereas miR-221 and miR-222 had no significant change, at least by 3-day induction.
FIG. 1.
miR-1 is upregulated during SMC differentiation. (A) Representative Western blot showed that the pluripotent gene octamer-binding protein 4 (OCT4) was downregulated and SMC-specific markers upregulated dramatically along with the RA treatment on mouse ESCs at the indicated days. (B) miR-1 expression during RA-induced SMCs from ESCs cultured for 0, 3, 6, or 9 days, at which time miR-1 were measured by TaqMan miR assay and normalized to U6. Fold changes are shown with respect to DMSO-treated cells, where miR-1 levels on day 0 were set to a value of 1. miR-1, microRNA-1; SMC, smooth muscle cell; RA, retinoic acid; SMMHC, smooth muscle myosin heavy chain; ESC, embryonic stem cell; α-SMA, α-smooth muscle actin; DMSO, dimethyl sulfoxide.
miR-1 is required for mouse ESC/SMC differentiation
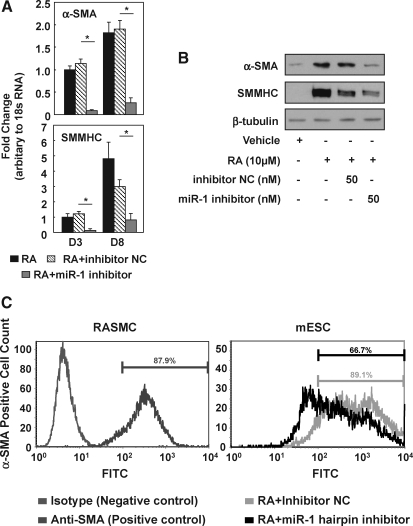
To test the hypothesis that miR-1 may have an active role as a positive regulator of SMC differentiation, we blocked miR-1 expression during ESC/SMC differentiation by transfecting miR-1 inhibitor into mouse ESCs. Results showed a dramatic repression of miR-1 upregulation, which was sustained up to day 8 (Supplementary Fig. S2B). Such inhibitory effect on miR-1 was specifically targeted on miR-1, with no effect on other miRs, as determined by TaqMan miR assay (data no shown). Transfection with miR-1 inhibitor resulted in major repression of SMC-specific markers, including α-SMA and SMMHC, as determined by quantitative real time PCR (qRT-PCR) and Western blotting (Fig. 2A, B). Further, FACS analysis showed that the efficiency of SMC differentiation from mouse ESCs was reduced after the introduction of miR-1 inhibitor (Fig. 1C, 90.09% ± 3.12% in the inhibitor NC group vs. 66.24% ± 5.73% in the miR-1 inhibitor group, n = 4, P < 0.05). Compared to the inhibitor NC, miR-1 inhibitor slightly increased octamer-binding protein 4 (OCT4) mRNA level; however, no effect was observed on other germline marker expression, including NeuroD1, alpha-fetoprotein (AFP), and GATA binding protein 2 (GATA2) (Supplementary Fig. S2C). In addition, miR-1 mimic introduction into Dicer−/− ESCs has significant beneficial effect in the context of SMC differentiation (Supplementary Fig. S2D). Taken together, the results indicate that miR-1 upregulation is required for efficient RA-mediated mouse ESC/SMC differentiation.
FIG. 2.
Inhibition of miR-1 represses SMC differentiation. (A, B) Regulation of SMC differentiation by miR-1 inhibitor. About 50 nM miR-1 inhibitor repressed expression of SMC-specific markers, including α-SMA and SMMHC, as evidenced by Western-blotting (C). *P < 0.05. (C) miR-1 inhibitor reduced the SMC differentiation efficiency determined by fluorescence-activated cell sorting. Left panel: Green histogram represents IgG2a isotype control and red represents anti-α-SMA of rat aorta SMCs (RASMCs) as positive control. Right panel: the proportion of the α-SMA-positive cells with green and blue representing inhibitor NC and miR-1 inhibitor, respectively. n = 4. PCR, polymerase chain reaction; NC, negative control; FITC, fluorescein isothiocyanate.
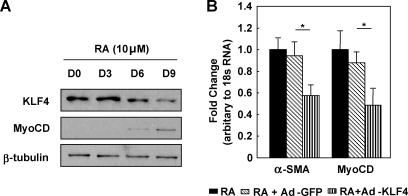
miR-1 inhibits KLF4 expression
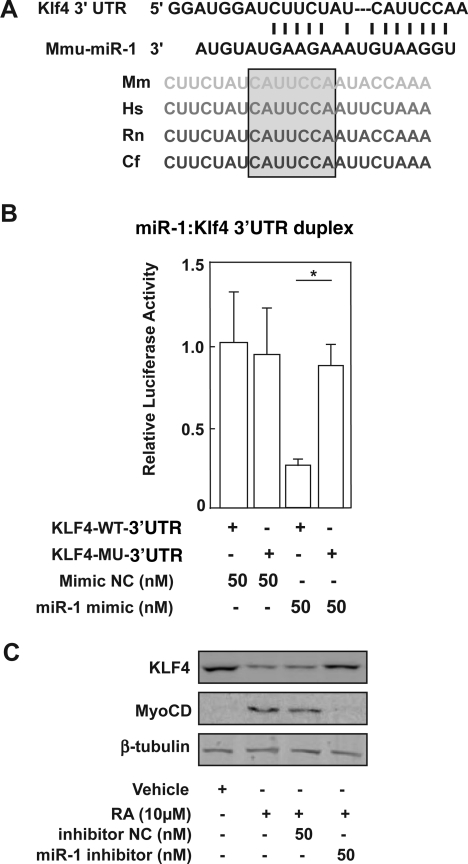
An inverse relationship between miR-1 expression (Fig. 1B) and KLF4 protein (Fig. 3A) during mouse ESC/SMC differentiation was unveiled. Consistently, with overexpression of KLF4 in mouse ESCs infected with an Ad-KLF4 vector (Supplementary Fig. S3), expression of SMC-specific markers, including α-SMA and MyoCD, was attenuated (Fig. 3B). Bioinformatics search for putative miR-1 binding sites within the KLF4 mRNA revealed that miR-1 is predicted to hybridize to a seed sequence in the KLF4 3′-UTR, which is evolutionarily conserved among vertebrate species (Fig. 4A). Secondary structure analysis also showed a favorable minimum free energy (−18.8 kcal/mol) in the formation of the miR-1: KLF4 3′UTR duplex stem-loop (Supplementary Fig. S4A). Further, a predicted binding exists between the seed region of miR-1 and the 3′UTR of KLF4, suggesting that miR-1 is involved in translational repression of KLF4. To determine whether KLF4 is a direct target of miR-1 we cloned a 951-bp fragment of the KLF4 3′ UTR containing the predicted miR-1 target sequence (5′-ggauggaucuucuaucauuccaa-3′, from 286 to 292 bp of the 3′ UTR region) into downstream of the firefly luciferase gene in the pMIR-Report vector (Promega) (Supplementary Fig. S4B). Co-transfection of miR-1 with the KLF4 3′ UTR reporter resulted in a dose-dependent repression of luciferase activity (P < 0.05) (Fig. 4B). This was specific to miR-1 binding since the effect was prevented by mutation of the putative miR-1 complementary seed sequence (Fig. 4B). In addition, miR-1 did not show binding to KLF2, KLF5, and KLF13 3′UTR, evidenced by individual luciferase activity (Supplementary Fig. S4C). Further, KLF4 expression was restored by miR-1 inhibitor (Fig. 4C), consistent with the functional prediction that miR-1 binding to the KLF4 3′UTR would lead to KLF4 translational repression.
FIG. 3.
KLF4 attenuates ESC/SMC differentiation. (A) Representative Western blots probing for KLF4 and MyoCD in ESCs differentiating to SMCs at the indicated days. (B) Mouse ESCs were infected with adenovirus expressing KLF4 (Ad-KLF4) and subsequently induced to differentiate into SMCs with RA. The derived cells were subject to qRT-PCR analysis and the expression of SMC markers was detected in extracts from cells differentiated for 6 days. 18sRNA served as internal control for qRT-PCR. KLF4, Kruppel-like factor 4; MyoCD, myocardin. *P < 0.05.
FIG. 4.
KLF4 is a target of miR-1 during ESC/SMC differentiation. (A) Predicted target site of miR-1 in the 3′UTR of mouse KLF4, with the seed region, and alignment of the homologous regions of the 3′UTR of KLF4 from human, mouse, rat, and dog. Predicted miR-1 binding sites are highlighted in gray. (B) WT and MU reporter constructs were co-transfected into HEK 293 cells with miR mimic or mimic NC as indicated. Individual luciferase activity was normalized to the responding thymidine kinase (TK) promoter–Renilla-luciferase activity. Relative luciferase activities were expressed as mean ± standard deviation. Data shown are representative samples from at least 3 independent experiments, each done in triplicates. *P < 0.05. (C) Mouse ESCs were transfected with miR-1 inhibitor or inhibitor NC. KLF4 and MyoCD protein was detected in extracts from cells differentiated for 72 h. β-tubulin served as internal control. 3′UTR, 3′ untranslated region; WT, wild type; MU, mutant.
Taken together, these results strongly support that miR-1 regulates expression of KLF4 at the post-transcriptional level during mouse ESC/SMC differentiation.
Discussion
Compared to cardiac and skeletal muscle, only a few miRs, including miR-21 [5,23,24], miR-221 [8,25], miR-145, and miR-143 [7,10], have been investigated in SMCs, partially due to the limitations in availability of efficient and reproducible SMC differentiation models. We have recently developed a simple and highly efficient in vitro model for RA-induced SMC differentiation from mouse ESC [18]. This system allows us to explore the potential roles of individual miRs and miR:target pairs during SMC differentiation in vitro, and when combined with experimental angioplasty and transgenic mice models, it provides a powerful tool to study how they translate to vascular development and dysfunction in vivo. In the study presented here we have identified the miR-1:KLF4 pair for its ability to positively regulate mouse ESC/SMC differentiation in in vitro model.
The observation that miR-1 expression was steadily upregulated during mouse ESC/SMC differentiation, whereas the miR-1 inhibitor partially repressed SMC differentiation, indicated that miR-1 expression, induced by RA treatment via a still unknown mechanism, can modulate the differentiation of SMC in this model. The inhibitor NC was used as the control for miR-1 inhibitor in parallel. Difference in expression of α-SMA and SMMHC shown here in the control group maybe due to (1) the vulnerability of stem cell differentiation to exogenous transfection; (2) SMMHC is the late-stage and more selective-SMC marker, while α-SMA early stage marker in the category of SMC differentiation. Although dimethyl sulfoxide-treated cells also showed moderate upregulation of miR-1 (5-fold by day 6), this upregulation was not as remarkable as the one observed in the RA-treated cells, and may result from a low percentage of spontaneous differentiation of ESCs into SMCs. Interestingly, miR-1 mimic per se cannot drive SMC differentiation in the absence of RA treatment, which implies that miR-1 upregulation is elegantly integrated in harmony with other regulatory mechanisms operating during RA-induced SMC differentiation.
We subsequently investigated the possible mechanism (miR:target pair/s) responsible for miR-1-positive effects on SMC differentiation in this system. Remarkably, KLF4 was consistently predicted by 2 algorithms, TargetScan [26] and PicTar [27]. KLFs are a subclass of evolutionarily conserved transcription factors [28] and KLF4 expression is associated with growth arrest and the inhibitory effect of SMC marker expression [29]. Previous studies have demonstrated that KLF4 potently represses expression of multiple SMC genes by repressing MyoCD levels during phenotypic switching of SMC in response to vascular injury, transforming growth factor-β, or platelet-derived growth factor-BB [14–16,30–34].
We observed an inverse relationship in expression between miR-1 and KLF4 during SMC differentiation from ESCs and luciferase assays demonstrated that there is a putative functional binding between mature miR-1 and the KLF4 3′UTR. Further, inhibition of miR-1 expression resulted in upregulation of KLF4 protein levels. Taken together, these results indicate that KLF4 is an miR-1 target, mediating its positive effects in modulation of SMC differentiation.
In summary, the results presented here derived from a systematic experimental approach that allowed us to identify novel “miR:target” gene pairs involved in SMC differentiation indicate that miR-1 plays a functional role in positively modulating SMC differentiation from ESCs. Moreover, miR-1-mediated inhibition of KLF4 may, in part, account for the observed effects on SMC differentiation in this study. Further investigation into the function of miR-1 during SMC differentiation, development, and dysfunction will provide additional insight on the role of small noncoding RNAs in these processes and establish miRs as potential therapeutic target for vascular disease.
Supplementary Material
Acknowledgments
This work was partially funded by National Institutes of Health (HL092421, HL068878, and HL89544 to Y.E.C.). J.Z. and C.X. were supported by American Heart Association (AHA) National Career Development Grants (0835237N to J.Z. and 09SDG2260023 to C.X.), respectively. Y.E.C. is an AHA established investigator (0840025N).
Author Disclosure Statement
The authors have no conflict of interest to disclose.
References
- 1.Lagos-Quintana M. Rauhut R. Lendeckel W. Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M. Rauhut R. Yalcin A. Meyer J. Lendeckel W. Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 3.Ke XS. Liu CM. Liu DP. Liang CC. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- 4.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Ji R. Cheng Y. Yue J. Yang J. Liu X. Chen H. Dean DB. Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y. Liu X. Yang J. Lin Y. Xu DZ. Lu Q. Deitch EA. Huo Y. Delphin ES. Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cordes KR. Sheehy NT. White MP. Berry EC. Morton SU. Muth AN. Lee TH. Miano JM. Ivey KN. Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X. Cheng Y. Zhang S. Lin Y. Yang J. Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams AH. Liu N. van Rooij E. Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin M. Small EM. Sutherland LB. Qi X. McAnally J. Plato CF. Richardson JA. Bassel-Duby R. Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rooij E. Marshall WS. Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deaton RA. Gan Q. Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am J Physiol Heart Circ Physiol. 2009;296:H1027–H1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai-Kowase K. Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y. Sinha S. McDonald OG. Shang Y. Hoofnagle MH. Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 15.Pidkovka NA. Cherepanova OA. Yoshida T. Alexander MR. Deaton RA. Thomas JA. Leitinger N. Owens GK. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circ Res. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida T. Kaestner KH. Owens GK. Conditional deletion of Kruppel-like factor 4 delays downregulation of smooth muscle cell differentiation markers but accelerates neointimal formation following vascular injury. Circ Res. 2008;102:1548–1557. doi: 10.1161/CIRCRESAHA.108.176974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein E. Kim SY. Carmell MA. Murchison EP. Alcorn H. Li MZ. Mills AA. Elledge SJ. Anderson KV. Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 18.Xie CQ. Huang H. Wei S. Song LS. Zhang J. Ritchie RP. Chen L. Zhang M. Chen YE. A comparison of murine smooth muscle cells generated from embryonic versus induced pluripotent stem cells. Stem Cells Dev. 2009;18:741–748. doi: 10.1089/scd.2008.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie CQ. Jeong Y. Fu M. Bookout AL. Garcia-Barrio MT. Sun T. Kim BH. Xie Y. Root S. Zhang J. Xu RH. Chen YE. Mangelsdorf DJ. Expression profiling of nuclear receptors in human and mouse embryonic stem cells. Mol Endocrinol. 2009;23:724–733. doi: 10.1210/me.2008-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruby JG. Jan C. Player C. Axtell MJ. Lee W. Nusbaum C. Ge H. Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 21.Huang H. Xie C. Sun X. Ritchie RP. Zhang J. Chen YE. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem. 285:9383–9389. doi: 10.1074/jbc.M109.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie CQ. Zhang J. Villacorta L. Cui T. Huang H. Chen YE. A highly efficient method to differentiate smooth muscle cells from human embryonic stem cells. Arterioscler Thromb Vasc Biol. 2007;27:e311–e312. doi: 10.1161/ATVBAHA.107.154260. [DOI] [PubMed] [Google Scholar]
- 23.Davis BN. Hilyard AC. Lagna G. Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y. Liu X. Cheng Y. Yang J. Huo Y. Zhang C. Involvement of microRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem. 2009;284:7903–7913. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis BN. Hilyard AC. Nguyen PH. Lagna G. Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP. Shih IH. Jones-Rhoades MW. Bartel DP. Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 27.Krek A. Grun D. Poy MN. Wolf R. Rosenberg L. Epstein EJ. MacMenamin P. da Piedade I. Gunsalus KC. Stoffel M. Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T. Aizawa K. Matsumura T. Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- 29.Chen X. Johns DC. Geiman DE. Marban E. Dang DT. Hamlin G. Sun R. Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y. Zhuang S. Gloddek J. Tseng CC. Boss GR. Pilz RB. Regulation of cGMP-dependent protein kinase expression by Rho and Kruppel-like transcription factor-4. J Biol Chem. 2006;281:16951–16961. doi: 10.1074/jbc.M602099200. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida T. Gan Q. Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. Am J Physiol Cell Physiol. 2008;295:C1175–C1182. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y. Sinha S. Owens G. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem. 2003;278:48004–48011. doi: 10.1074/jbc.M301902200. [DOI] [PubMed] [Google Scholar]
- 33.Kawai-Kowase K. Ohshima T. Matsui H. Tanaka T. Shimizu T. Iso T. Arai M. Owens GK. Kurabayashi M. PIAS1 mediates TGF{beta}-induced SM {alpha}-actin gene expression through inhibition of KLF4 function-expression by protein sumoylation. Arterioscler Thromb Vasc Biol. 2009;29:99–106. doi: 10.1161/ATVBAHA.108.172700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King KE. Iyemere VP. Weissberg PL. Shanahan CM. Kruppel-like factor 4 (KLF4/GKLF) is a target of bone morphogenetic proteins and transforming growth factor beta 1 in the regulation of vascular smooth muscle cell phenotype. J Biol Chem. 2003;278:11661–11669. doi: 10.1074/jbc.M211337200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.