Abstract
Multipotent stem/progenitor cells from bone marrow stroma (mesenchymal stromal cells or MSCs) were previously shown to enhance proliferation and differentiation of neural stem cells (NSCs) in vivo, but the molecular basis of the effect was not defined. Here coculturing human MSCs (hMSCs) with rat NSCs (rNSCs) was found to stimulate astrocyte and oligodendrocyte differentiation of the rNSCs. To survey the signaling pathways involved, RNA from the cocultures was analyzed by species-specific microarrays. In the hMSCs, there was an upregulation of transcripts for several secreted factors linked to differentiation: bone morphogenetic protein 1 (BMP1), hepatocyte growth factor (HGF), and transforming growth factor isoforms (TGFβ1 and TGFβ3). In both the hMSCs and the rNSCs, there was an upregulation of transcripts for Notch signaling. The role of TGFβ1 was verified by the demonstration that hMSCs in coculture increased secretion of TGFβ1, the rNSCs expressed the receptor, and an inhibitor of TGFβ signaling blocked differentiation. The role of Notch signaling was verified by the demonstration that in the cocultures hMSCs expressed a Notch ligand at sites of cell contact with rNSCs, and the rNSCs expressed the receptor, Notch 1. Increased Notch signaling in both cell types was then demonstrated by assays of transcript expression and by a reporter construct for downstream targets of Notch signaling. The results demonstrated that glial differentiation of the rNSCs in the cocultures was driven by increased secretion of soluble factors such as TGFβ1 by the hMSCs and probably through increased cell contact signaling between the hMSCs and rNSCs through the Notch pathway.
Introduction
Neural stem cells (NSCs) are currently generating interest for their potential therapeutic benefits as well as their intrinsic role in central nervous system (CNS) processes, including memory, aging, depression, and a host of neurodegenerative diseases [1]. Endogenous NSCs are known to reside in at least 2 germinal zones in the adult CNS and are an inherently proliferating population capable of undertaking migration in addition to differentiating down neuronal as well as glial lineages [2]. Because of their multipotentiality and capacity for self-renewal, NSCs were originally seen as an endogenous source for cellular replacement following CNS injury [3]. In addition to cellular replacement, recent evidence suggests that NSCs may be therapeutically beneficial through immunomodulation of the damaged environment, protective mechanisms limiting the degree of damage, and enhancement of endogenous repair mechanisms following neurologic insult [4]. These effects may be mediated by NSC secretion of trophic support or through direct cell–cell contact.
The nonhematopoietic population of adult stem/progenitor cells from bone marrow referred to as mesenchymal stem cells or multipotent mesenchymal stromal cells (MSCs) were shown to produce therapeutic effects in numerous animal models of neurodegenerative diseases, including cerebral ischemia, Parkinson's disease, multiple sclerosis, spinal cord injury, and traumatic brain injury [5–10]. Given their ability to home to multiple tissues, including the CNS, MSCs may be a viable means to stimulate the endogenous NSC population to assume a neuroprotective or neuroregenerative role. Indeed MSCs produce a large number of growth-stimulating factors, including neurotrophins in vitro and in vivo [11,12]. An intriguing recent observation was that following implantation into the hippocampi of immunodeficient mice, human MSCs (hMSCs) stimulated the proliferation and dorsal migration of endogenous BrdU-labeled NSCs [12–14]. Despite limited survival of the MSCs, BrdU-labeled NSCs persisted for up to 30 days and expressed markers for differentiated neurons, astrocytes, and oligodendrocytes. Subpopulations of BrdU-labeled NSCs also expressed potentially beneficial trophic factors, including ciliary neurotrophic factor, neurotrophin-4/5, nerve growth factor, and vascular endothelial growth factor.
Here we examined cocultures of hMSCs and rat NSCs (rNSCs) for potential mechanisms underlying the effects on NSCs seen in vivo. Our data suggest that hMSCs stimulated glial differentiation of rNSCs in part through increased secretion of soluble factors such as transforming growth factor β (TGFβ) and possibly in part by cell–cell contact-mediated effects, including increased Notch signaling in both the hMSCs and the rNSCs.
Materials and Methods
Preparation and culture of hMSCs
hMSCs from normal healthy donors were obtained from the Tulane Center for the Preparation and Distribution of Adult Stem Cells (www.som.tulane.edu/gene_therapy/distribute.shtml). The cells were prepared as previously described [15,16] with protocols approved by an Institutional Review Board. Frozen vials of passage-1 hMSCs (1 × 106) were thawed, plated in 25 mL complete hMSC medium—α-MEM (Gibco/BRL); 20% FBS (lot selected for rapid growth; Atlanta Biologicals); 100 units/mL penicillin (Gibco/BRL); 100 μg/mL streptomycin (Gibco/BRL); and 2 mM L-glutamine (Gibco/BRL)—and incubated at 37°C with 5% humidified CO2. After 24 h, the medium was removed and adherent, viable cells were washed twice with PBS, harvested with 0.25% trypsin/1 mM ethylenediaminetetraacetic acid (EDTA), replated at 100 cells/cm2 in hMSC medium, and incubated with a medium change every 3–4 days. The cells (passage-2) were incubated until they reached 70% confluence (approximately 7 days) at which time they were harvested with trypsin/EDTA for assays or further expansion under the same conditions. For some assays green fluorescent protein (GFP) transduced hMSCs were prepared as previously described [17]. Briefly, hMSCs were transduced by lentiviral GFP construct (WPT-CAG-hrGFP-WPRE). Transduded cells were then purified by fluorescence activated cell sorting (FACS, Vantage SE cell sorter; Becton-Dickinson) before expansion and subsequent freezing. Frozen vials of passage-3 GFP expressing hMSCs were thawed and cultured as outlined above. After thawing and culturing, over 95% of passage-4 hMSCs expressed GFP as determined by FACS analysis (MoFlo XDP Cell Sorter; Beckman Coulter).
Preparation and culture of rNSCs
Cryopreserved adult rat hippocampal neural stem cells (NSCs) were obtained from Chemicon/Millipore. For cell expansion, frozen vials of approximately 1 × 106 rNSCs were rapidly thawed and diluted dropwise with 9 mL optimal rNSC growth medium: 9 parts NeuroCult® NS-A Basal Medium (Stemcell Technologies) to 1 part NeuroCult Proliferation Supplements (Stemcell Technologies) with 0.0002% Heparin (Stemcell Technologies); 20 ng/mL recombinant human epidermal growth factor (EGF; Stemcell Technologies); and 10 ng/mL recombinant human basic fibroblast growth factor (FGF-b; Stemcell Technologies). Cells were pelleted, resuspended at 60,000 cells/mL in 20 mL optimal rNSC growth medium, and grown in a T-175 culture flask (Nunc) at 37°C with 5% humidified CO2. Half of the medium was replaced with fresh medium every other day in culture. The nonadherent cells gradually generated neurospheres and were passaged after 4–7 days by gentle pipetting of the neurospheres and medium to dissociate the spheres. Cells and medium were transferred to a collection tube, centrifuged, resuspended in optimal rNSC growth medium at 60,000 cells/mL, and returned to a flask. For the coculture experiments, the cultures were recovered after 2–4 days. To confirm the differentiation potential of the rNSCs, cells were plated at 5,000 cells/cm2 on glass coverslips precoated with Poly-D-lysine/laminin (BD Biosciences) and incubated in rNSC differentiation medium: 9 parts NeuroCult NS-A Differentiation Medium (Stemcell Technologies) to 1 part NeuroCult Differentiation Supplements (Stemcell Technologies). Cells were fixed for immunostaining after 4–7 days in culture.
Coculture experiments
Passage-2 hMSCs suspended in complete hMSC medium (20 mL) were plated at 900–2,000 cells/cm2 on 148-cm2 culture dishes or glass chamber slides (Sigma-Aldrich) coated with 10 μg/mL poly-L-ornithine (Sigma-Aldrich)/5 μg/mL laminin (Sigma-Aldrich) or on glass coverslips precoated with Poly-d-lysine/laminin (BD Biosciences). Following 1 day to allow for cell adherence, cultures were washed with PBS and overlaid with rNSCs (5,000 cells/cm2) in optimal rNSC growth medium (20 mL for 148-cm2 dishes or 1 mL per coverslip). Fresh growth factors were added to all of the cultures after 2 days including 20 ng/mL EGF and 10 ng/mL FGF-b. Cultures were grown for 4 days after the addition of the rNSCs. hMSC and rNSC cultures incubated alone were plated in identical conditions. To inhibit TGFβ signaling in cultures, optimal rNSC growth medium was supplemented with either 4 or 10 μM of a TGFβ receptor type 1 inhibitor (SB431542; Sigma-Aldrich) at the start of the coculture and again after 2 days. To inhibit Notch signaling in cultures, optimal rNSC growth medium was supplemented with 4 μM of a γ-secretase inhibitor (DAPT; Sigma-Aldrich). For conditioned medium experiments, rNSCs were plated at 5,000 cells/cm2 on coated coverslips in optmal rNSC growth medium. After 1 day medium was aspirated from cultures and replaced with hMSC conditioned medium. To prepare hMSC conditioned medium, hMSCs were cultured in optimal rNSC growth medium supplemented with EGF and FGF-b for 4 days. Medium was then collected and centrifuged, fresh EGF and FGF-b were added to the supernatant.
Immunocytochemistry
After 4 days of incubation, cells on coverslips or in chamber slides were fixed with 4% paraformaldehyde in PBS for 15 min and then washed twice with PBS. Cells were permeabilized with 0.4% Triton X-100 (Sigma-Aldrich) and nonspecific binding was blocked by a 1 h incubation at room temperature in PBS containing 5% normal serum from the species in which the secondary antibody was raised and 0.4% Triton X-100. Slides were subsequently incubated over night at 4°C with the following primary antibodies diluted in blocking buffer: Mouse antinestin (clone 401; BD Bioscience, cat# 556309; 1:1,000) and rabbit anti-Sox2 (Thermo Scientific, cat# PA1-16968; 1:250) were used to stain neural stem/progenitors; rabbit anti-Ki67 (Novocastra, cat# NSL-Ki67p; 1:8,000) to stain proliferating cells; rabbit anti-glial fibrillary acidic protein (GFAP; Chemicon, cat# AB5804; 1:1,000 or Sigma-Aldrich, cat# G9269; 1:1,000) or mouse anti-GFAP (clone GA-5; Cell Signaling, cat# 3670; 1:500) to stain astrocytes; mouse anti-oligodendrocytes (RIP, clone NSC-1; Chemicon, cat# MAB1580; 1:20,000) and mouse anti-2′,3′-cyclic nucleotide 3′-phosphodiesterase (2′,3′-cyclic nucleotide 3′-phospho-diesterase [CNPase], clone 11-5B; Sigma-Aldrich, cat# C5922; 1:100) to stain oligodendrocytes; mouse anti-β-III tubulin (TUJ1; Covance, Cat# MMS-435P; 1:1,000) or rabbit anti-β-III tubulin (Abcam, cat# AB18207; 1:2,000) to stain neurons; rabbit anti-Jagged 1 (Abcam, cat#Ab7771; 1:200) and goat anti-Notch 1 (R&D Systems, cat# AF1057; 1:100) to stain Notch signaling proteins. After PBS washes, coverslips were incubated with flourescent dye-conjugated secondary antibodies for 1 h at room temperature. Secondary antibodies used were Alexa-conjugated goat anti-mouse, goat anti-rabbit, donkey anti-goat, or donkey anti-rabbit antibodies (Invitrogen; 1:1,000). After final washes in PBS, coverslips were mounted onto glass slides (Superfrost Plus Microscope Slides; Fisher Scientific) with a mounting medium containing DAPI to label nuclei (Vectashield, Vector Laboratories). Cells were observed with a spinning disk confocal microscope (Olympus BX51-DSU) equipped with a CCD camera (Hamamatsu C9100 EM-CCD; MBF Bioscience) and fluorescent images were acquired using Slidebook 4.2 software (Intelligent Imaging Innovations). Cells were counted in a minimum of 3 frames at either 100 × or 200 × magnification to survey at least 200 total cells per condition. A minimum of at least 3 independent cultures were analyzed for each marker counted.
Western blotting
After 4 days in culture, cells were harvested with trypsin/EDTA, resuspended in PBS, and sorted into GFP+ or GFP− populations (Vantage SE cell sorter; Becton-Dickinson). Cells were lysed using NP40 cell lysis buffer (BioSourse/Invitrogen) with protease inhibitors (Roche) and phosphatase inhibitors (eBioscience, San Diego, CA) for 30 min at 4°C with frequent vortexing. The samples were centrifuged at 18,000 g for 10 min and the supernatants were realiquoted and assayed for total protein content (DC protein kit; Bio-Rad). One microgram of total protein per lane was electrophoresed on a 4%–12% BisTris NuPage Gel (Invitrogen). Proteins were electrophoretically transfered to a polyvinylidene fluoride (PVDF) membrane (Invitrogen) and then blocked with 5% milk (Bio-Rad) in Tris buffered saline containing 0.1% tween-20 (TBST). Membranes were probed overnight in 2.5% milk in TBST with primary antibodies against GFAP (clone GA-5; Sigma-Aldrich, cat# G3893; 1:500), β-III tubulin (Abcam; cat# AB18207 1:1000), TGFβ receptor I (Santa Cruz, Cat# SC9048; 1:200), TGFβ receptor II (Cell Signaling, Cat# 3713; 1:1,000), or β-actin (clone AC-15; Sigma-Aldrich, cat# A1978; 1:40,000). Membranes were washed and incubated for 1 h at room temperature with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG secondary antibodies (Pierce; 1:1,000) diluted in 1% milk TBST solution. Membranes were then washed and exposed to a chemiluminescent substrate (SuperSignal West Femto Substrate; Pierce). Blots were observed by exposure to X-ray film (Biomax XAR, Fisher). For semiquantitative analysis, the volume (mean intensity × mm2) of each band was measured using Quantity One software (Bio-Rad). Percent expression was calculated using the following equation: (volume of GFAP/volume of β-actin) × 100. Three independent cultures were analyzed per condition.
Microarrays
Seven micrograms of total RNA extracted from cultures of hMSCs alone, rNSCs alone, or cocultures was used for assays on human (HG-U133 Plus 2.0) or rat (RG-230 2.0) arrays (Affymetrix) according to manufacturer's instructions. The scanned signal intensities were transferred to the dChip program for analysis [18,19].
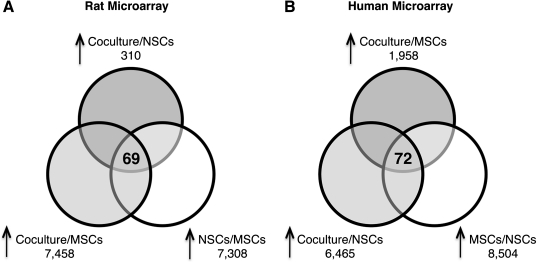
Rat and human microarray data were filtered to identify genes that were either up- or downregulated due to coculture. To correct for cross-hybridization of rat mRNA to the human microarray or human mRNA to the rat microarray, genes were considered upregulated if they were (1) scored P (present) in the coculture data and were at least 2-fold higher (with 90% confidence) than in data from cultures of either cell type alone and (2) scored P in the alone culture of interest data and were at least 2-fold higher than in the opposite alone culture data (Fig. 2). This filtering resulted in 72 rat and 95 human genes (69 rat and 72 human nonredundant genes). Genes were considered downregulated if they were (1) scored P in the alone culture of interest data and were at least 2-fold lower in the coculture data, and were not (2) scored P in the opposite alone culture data and were not at least 2-fold higher than in the alone culture of interest data. This filtering resulted in 75 rat and 7135 human genes (75 rat and 5635 human nonredundant genes).
FIG. 2.
Assays with species-specific microarrays. Total RNA was isolated from individual cultures of hMSCs and rNSCs as well as cocultures. All samples were hybridized to both rat and human microarray chips. (A) Venn diagram of 69 nonredundant rat genes upregulated with coculture. (B) Venn diagram of 72 nonredundant human genes upregulated with coculture. In both Venn diagrams, lower 2 circles are genes that met the first and second filtering criteria (see Materials and Methods).
Enzyme-linked immunosorbent assays
Medium from 4-day cultures was collected and treated with protease inhibitors (Roche). Human TGFβ1 and hepatocyte growth factor (HGF) were assayed using commercial ELISA kits (TGFβ1 cat# DB100B and HGF cat# DHG00 Quantikine, R&D Systems) with samples standardized by volume. All samples were assayed in triplicate.
Real-time reverse transcription-polymerase chain reaction
After 4 days of incubation, cells were sorted by FACS as previously described. Cells were lysed and total RNA was isolated according to manufacturer's instructions (Rneasy Mini Kit, Qiagen). All RNA samples were DNase-treated. Primer sequences for hairy and enhancer of split (HES1, NM_024360.3), hairy and enhancer of split related with YRPW motif 1 (HEY1, XM_342216.3), and β-actin (NM_031144.2) were designed and synthesized commercially (Superarray Bioscience). Assays with primers for β-actin mRNA were used to normalize samples. First-strand cDNA synthesis and real-time reverse transcription-polymerase chain reaction (RT-PCR) were performed (RT2 First Strand Synthesis Kit, RT2 qPCR Primer Assays, Superarray Bioscience). One microgram total RNA was used per sample, and amplification was performed using an automated instrument (Model 7900; Applied Biosystems). For all RT-PCR assays RNA was isolated from 2 independent experiments. Samples were assayed in triplicate.
Luciferase reporter assay
Notch signaling was monitored using a reporter construct for the recombination signal binding protein for immunoglobulin kappa J region (RBP-Jk) protein (Cignal RBP-Jk) Reporter Kit; Superarray Bioscience). rNSCs were transfected with a 40:1 mixture of an inducible reporter construct encoding the firefly luciferase gene under control of the RBP-Jk transcriptional response element (sequence: CGTGGGAA) and a constitutive reporter construct encoding the renilla luciferase gene under control of a minimal (m) cytomegalovirus (CMV) promoter. Approximately 5 × 105 rNSCs in a 24-well plate were transfected with 1000 ng of the nucleic acid mixture for 24 h using a transfection reagent (SureFECT; Cignal RBP-Jk Reporter Kit, Superarray Bioscience). At the same time, hMSCs were plated into 96-well cell culture plates at 1,000 cells/cm2 in hMSC medium for coculture experiments. Following 1 day to allow for cell adherence, the hMSC cultures were washed with PBS and overlaid with 2,000 transfected rNSCs/cm2 in complete rNSC medium. After 24 h, cells were lysed and assayed for luciferase activity using an automated instrument (FLUOstar OPTIMA; BMG Labtech) and commerical reagents (Dual-luciferase Reporter Assay System; Promega). Renilla luciferase activity was used to normalize RBP-Jk-responsive firefly luciferase activity. For all reporter assays, samples were assayed in triplicate.
Statistical analysis
Statistical significance was evaluated between 2 groups using Student's t-test. RT-PCR and luciferace reporter assay data were compared by 1-way ANOVA with post hoc parwise comparisons using Tukey's test. All error bars represent standard deviations. P values <0.05 were considered statistically significant.
Results
hMSCs drive the glial differentiation of rat NSCs
In initial experiments, the phenotype of rNSCs cultured in optimal growth medium as a monolayer was established by immunocytochemistry (ICC) (Supplementary Fig. S1, available online at www.liebertonline.com/scd). Most cells showed dispersed expression of the immature neural marker nestin and nuclear expression of the characteristic rNSC marker Sox2 (Supplemental Fig. S1C and D, respectively). The cells were actively dividing based on expression of the cell cycle marker Ki-67 and the presence of double nuclei in rare cells (Supplemental Fig. S1E). To confirm the differentiation potential of the rNSC, the cells were incubated in optimal rNSC differentiation medium. Subpopulations of the cells expressed markers for oligodendrocytes (RIP and CNPase), neurons (β-III tubulin), and astrocytes (GFAP) (Supplemental Fig. S1F–K). In parallel experiments, control cultures of hMSCs were incubated in optimal rNSC growth medium. The hMSCs survived in optimal rNSC growth medium and continued to proliferate although at a slower rate than in the hMSC culture medium (not shown). In addition, hMSCs assumed a more-elongated spindle-shaped morphology.
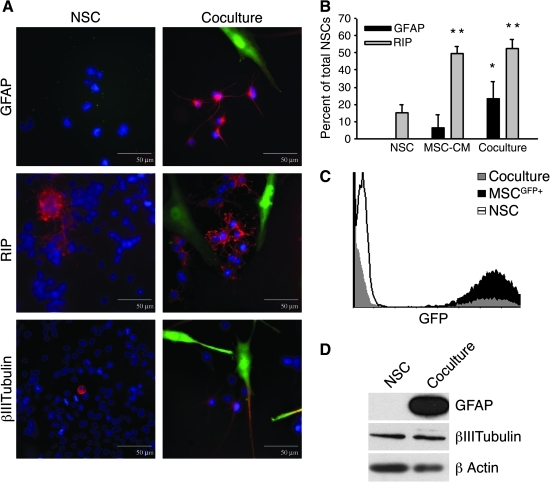
To investigate cellular signaling, rNSCs were cocultured with GFP+ hMSCs in the optimal rNSC growth medium. After 4 days of coculture, sub-populations of the rNSCs differentiated into astrocytes, oligodendrocytes, and neurons as indicated by a marked increase in GFP− cells expressing GFAP, RIP, and β-III tubulin (Fig. 1A). Astrocytes and a few neurons were widely dispersed in cultures, and positive antibody staining was typically peri-nuclear. Positively stained oligodendrocytes were observed more frequently than the other cell types and were typically seen as clusters of cells in culture. The undifferentiated GFP− rNSC typically had characteristic small nuclei and soma, whereas the GFP+ hMSCs typically demonstrated large, fibroblast-like morphology. The GFAP+ astrocytes demonstrated thin, multipolar processes extending from the central cell body, the RIP+ oligodendrocytes large cell bodies with numerous shorter processes, and the β-III tubulin+ neurons a single far-reaching extension.
FIG. 1.
hMSCs enhanced glial differentiation of rNSCs. rNSCs were cultured alone (NSC), with hMSC-conditioned medium (MSC-CM), or in cocultures with GFP+ hMSCs (coculture). (A) Coculture increased expression in rNSCs of the astrocyte marker GFAP (green in upper left panel; red in upper right) and the oligodendrocyte marker RIP (red) but not the neuron marker β-III tubulin (red). Nuclei were stained with DAPI (blue). The GFAP+, RIP+, and β-III tubulin+ cells had the typical morphologies of astrocytes, oligodendrocytes, and neurons, respectively. The GFP+ cells had the typical morphology of hMSCs. Scale bar = 50 μm. (B) MSC-CM increased number of rNSCs expressing RIP, and coculture increased the number expressing either GFAP or RIP (t-test: *P < 0.05; **P < 0.01 vs. NSC). (C) Histogram of GFP expression used for FACS isolation of GFP+ hMSCs and GFP− rNSCs from cocultures. (D) Western blot assay demonstrating that coculture increased expression of GFAP. Expression of β-III tubulin was unchanged. β-actin expression was used as a loading control. hMSC, human mesenchymal stromal cell; rNSC, rat neural stem cell; GFAP, glial fibrillary acidic protein. GFP, green fluorescent protein; RIP, anti-oligodendrocytes antibody.
To obtain quantitative data, systematic random sampling was performed on cultures. rNSCs incubated alone were 15.0% ± 4.8% (mean ± SD) positive for RIP and none were positive for GFAP (Fig. 1B). After coculture with GFP+ hMSCs, rNSCs were 52.4% ± 5.2% positive for RIP and 23.6% ± 9.9% positive for GFAP. By immunolabeling about 70% of the rNSCs expressed differentiation markers. The results were confirmed in a separate coculture experiment utilizing nontransfected hMSCs harvested from a different bone marrow donor (not shown). Conditioned medium from hMSCs had a different effect: rNSCs incubated with conditioned medium from hMSCs were 49.2% ± 4.8% positive for RIP, but only 4.8% ± 5.5% were positive for GFAP labeling. Therefore, the conditioned medium drove rNSC differentiation toward oligodendrocytes but not toward astrocytes. GFP+ hMSCs did not express GFAP or RIP under any of the conditions.
We confirmed the differentiation of the rNSCs by Western blot analysis following cell sorting for discrete GFP+ and GFP− populations after 4 days in coculture (Fig. 1C). The results confirmed marked increases in expression of GFAP in GFP− cells isolated from the cocultures (Fig. 1D). No significant change in expression of β-III tubulin was detected.
Our results indicated that hMSCs promoted differentiation of rNSCs specifically along a glial lineage and did not affect neuronal differentiation.
Effects of hMSCs on the rNSC transcriptome
To survey for signaling pathways involved in rNSC differentiation, RNA extracted from rNSCs incubated alone or from cocultures was assayed with rat microarrays. To correct for human mRNAs that cross-hybridized to the rat microarray, RNA from hMSCs alone was assayed on a rat microarray and used as a control in filtering the data (Fig. 2A).
After filtering, comparison of the signal intensities on the rat microarrays of rNSCs alone and cocultures indicated that 69 nonredundant genes were upregulated in the rNSCs at least 2-fold by coculture with the hMSCs (Supplementary Table S1, available online at www.liebertonline.com/scd). A selective screen of the individual 69 upregulated genes revealed numerous genes involved in glial differentiation and development, and Notch signaling pathway (Table 1). Genes implicated in astrocyte and oligodendrocyte development included bone morphogenetic protein 4 (Bmp4), meteorin (Metrn), reelin (Reln), netrin1 (Ntn1), chondroitin sulfate proteoglycan 2 (Cspg2), and the mammalian Notch ligand Delta-like 1 (Dll1). The characteristic astrocyte phenotype gene Gfap was also found to be highly upregulated; however, upregulation was not found to be statistically significant due to high variance between individual perfect match and miss match probes.
Table 1.
Genes of Interest Upregulated in Cocultures
| Symbol | ID | Expression ratio | |
|---|---|---|---|
| Rat neural stem cell genes | |||
| Differentiation | |||
| Glial fibrillary acidic protein | Gfap | 24387 | 44.8a |
| Bone morphogenetic protein 4 | Bmp4 | 25296 | 2.7 |
| Meteorin | Metrn | 287151 | 2.2 |
| Notch signaling | |||
| Delta-like 1 | Dll1 | 84010 | 2.5b |
| Intracellular signaling/transcription factors | |||
| Lymphoid enhancer binding factor 1 | Lef1 | 161452 | 4.1 |
| Tribbles homolog 1 | Trib1 | 78969 | 4.0b |
| Protein tyrosine phosphatase N | Ptprn | 116660 | 3.4 |
| Protein tyrosine phosphatase 5 | Ptpn5 | 29644 | 2.7b |
| Protein kinase C eta | Prkch | 81749 | 2.5 |
| Early growth response 1 | Egr1 | 24330 | 2.3 |
| Signan transducer and activator of transcription 3 | Stat3 | 25125 | 2.1 |
| Myelin related proteins | |||
| Discoidin domain receptor family 1 | Ddr1 | 25678 | 2.3 |
| Plasma membrane proteolipid | Pllp | 64364 | 2.2 |
| Gelsolin | Gsn | 296654 | 2.2 |
| Regulators in development | |||
| Reelin | Reln | 24718 | 7.5 |
| Netrin 1 | Ntn1 | 114523 | 2.6 |
| Chondroitin sulfate proteoglycan 2 | Cspg2 | 114122 | 2.1 |
| Human mesenchymal stromal cell genes | |||
| Notch signaling | |||
| Hairy and enhancer of split 1 | HES1 | 3280 | 5.4 |
| Notch homolog 3 | NOTCH3 | 4854 | 5.0 |
| Jagged 1 | JAG1 | 182 | 4.2 |
| SMAD family member 3 | SMAD3 | 4088 | 2.7 |
| Secreted factors | |||
| Bone morphogenetic protein 1 | BMP1 | 649 | 3.3 |
| Hepatocyte growth factor | HGF | 3082 | 3.2 |
| Transforming growth factor beta 3 | TGFB3 | 7043 | 3 |
| Transforming growth factor beta 1 | TGFB1 | 7040 | 2.4 |
| Cell surface receptors | |||
| Thy-1 cell surface antigen | THY1 | 7070 | 2.3 |
| KIT ligand | KITLG | 4254 | 2.3 |
| Glypican 1 | GPC1 | 2817 | 2.3 |
Using a 90% confidence interval, change in transcript expression was not found to be significant due to high variance between individual perfect match and mismatch probes. Signal absent in NSCs alone.
Signal absent in NSCs alone.
NSC, neural stem cell.
Further comparison of the filtered data indicated that 75 nonredundant genes were downregulated in the rNSCs at least 2-fold by coculture with the hMSCs (Supplementary Table S2, available online at www.liebertonline.com/scd). The results indicated that hMSCs produce major cellular and developmental changes in the rNSC transcriptome.
Effects of rNSCs on the hMSC transcriptome
To survey changes in the hMSC transcriptome in response to coculture with rNSCs, RNA extracted from hMSCs incubated alone or from cocultures was assayed with human microarrays. To correct for rat mRNAs that cross-hybridized with the human microarray, RNA from rNSCs alone was assayed on a human microarray and used as a control in filtering the data (Fig. 2B).
After filtering, the data indicated that 72 nonredundant genes were upregulated in the hMSCs at least 2-fold by coculture with the rNSCs (Supplementary Table S3, available online at www.liebertonline.com/scd). A selective screen of the individual 72 upregulated genes confirmed numerous genes known to participate in cell signaling via direct cell–cell contact or secreted molecules (Table 1). Upregulated secreted factors included transforming growth factor beta 1 and 3 (TGFB1 and TGFB3) and HGF, all mitogens known to modulate NSC proliferation, differentiation, and migration. In addition, several genes implicated in the TGFβ signaling pathway were upregulated, including the TGFβ receptor-regulated Smad family member 3 (SMAD3) and the TGFβ regulatory gene BMP1. Several genes associated with the Notch signaling pathway were also upregulated. Notch signaling pathway inducers included the mammalian Notch ligand jagged 1 (JAG1) and Notch homolog 3 (NOTCH3). The Notch downstream target transcription factor hairy and enhancer of split (HES1) was also increased.
Further analysis of the filtered data indicated that 5635 genes were downregulated in the hMSCs at least 2-fold by coculture with the rNSCs. This finding may be explained by a high NSC to MSC ratio in the coculture condition diluting the hMSC signal. Therefore, we were unable to make any conclusions regarding rNSC inhibition of the hMSC transcriptome.
Soluble factors secreted by hMSCs in cocultures
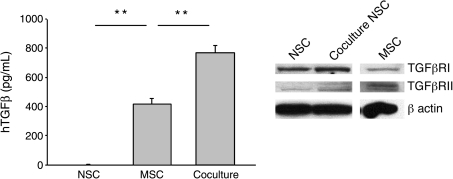
The microarray data allowed us to search for candidate genes that might explain the increased glial differentiation in the cocultures. Among the upregulated genes for secreted factors in the hMSCs were TGFB1 and TGFB3 (Table 1). Therefore, the medium from the cocultures was assayed with an ELISA specific for hTGFβ. As expected, coculture of the hMSCs with the rNSCs increased the secretion of hTGFβ (Fig. 3, left panel). A role for TGFβ was further supported by Western blot analysis demonstrating that rNSCs alone as well as those FACS isolated from cocultures expressed the appropriate receptors, TGFβR1 and TGFβR2 (Fig. 3, right panel). The gene for HGF was also upregulated in the hMSCs, and ELISA demonstrated increased secretion of HGF in the cocultures (Supplementary Fig. S2, available online at www.liebertonline.com/scd).
FIG. 3.
hMSCs increased secretion of TGFβ in cocultures. (Left panel) Human-specific ELISA of medium from cultures (t-test: **P < 0.01 vs. MSC). (Right panel) Western blot analysis of rNSCs cultured alone (NSC) or with hMSCs (coculture NSC) and hMSCs cultured alone (MSC) confirmed that hMSCs and rNSCs express both TGFβ receptors I and II. β-actin expression was used as a loading control. TGFβ, transforming growth factor β.
The TGFβ secreted by hMSCs in cocultures promotes the differentiation the rNSCs
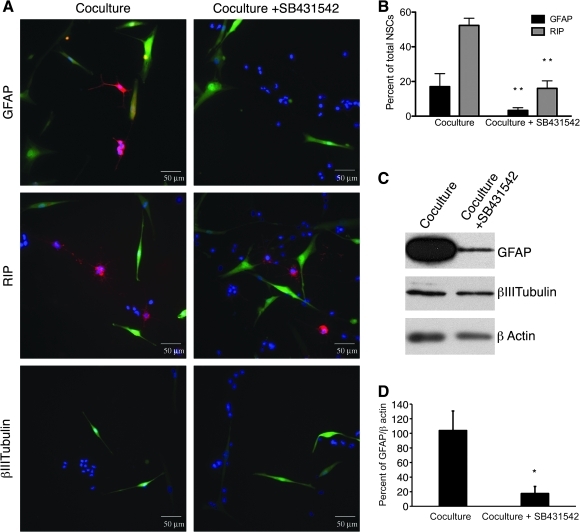
To confirm that secretion of TGFβ by hMSCs promoted glial differentiation, cocultures were incubated in the presence of SB431542, a selective inhibitor of TGFβ signaling. The inhibitor decreased the number of GFAP+ astrocytes, and clusters of RIP+ oligodendrocytes (Fig. 4A). A few β-III tubulin+ neurons were detected, but a majority of rNSCs did not stain positive for phenotypic markers of differentiated cells. GFP+ hMSCs assumed a more-elongated spindle-shaped morphology. rNSCs cocultured with hMSCs were 52.3% ± 4.2% positive for RIP and 17% ± 7.6% positive for GFAP. The TGFβ inhibitor decreased glial differentiation of the rNSC in cocultures, 16% ± 4.4% were positive for RIP and 3.3% ± 1.5% were positive for GFAP (Fig. 4B).
FIG. 4.
An inhibitor of TGFβ signaling (SB431542) decreased differentiation of the rNSCs in cocultures. (A) Immunocytochemistry for GFAP, RIP, or β-III tubulin (red) expression in rNSCs cocultured with GFP+ hMSCs. Nuclei were stained with DAPI (blue). In cocultures with the inhibitor more rNSCs had an immature morphology. Scale bar = 50 μm. (B) Treatment of cocultures with SB431542 decreased rNSC expression of GFAP and RIP (t-test: **P < 0.01 vs. coculture). (C) Western blots demonstrating that the inhibitor decreased expression of GFAP in rNSCs isolated from cocultures by FACS. (D) Semiquantitative analysis of rNSC expression of GFAP by Western blots (t-test: *P < 0.05 vs. coculture).
The effects of the inhibitor were confirmed by Western blot assays demonstrating decreased expression of GFAP in rNSCs isolated from cocultures (Fig. 4B). Recombinant human TGFβ added to rNSCs cultured alone did not increase the expression of GFAP as detected by Western blots (not shown). There was no change in rNSC expression of β-III tubulin between conditions.
Our results indicated that coculture with rNSCs activates hMSCs to secrete increased amounts of TGFβ and that TGFβ signaling is essential for hMSC-stimulated glial differentiation of rNSCs. However, we found that hTGFβ alone was not sufficient to stimulate glial differentiation of the rNSCs and that additional hMSC-derived factors might be playing a role.
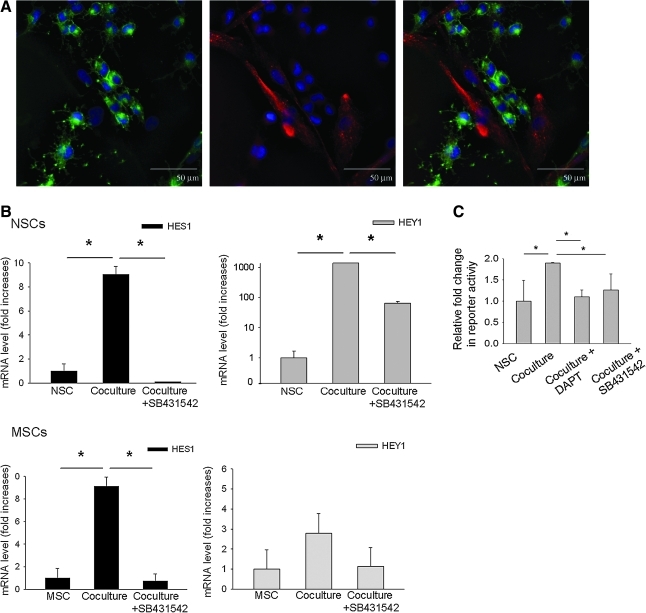
Increased Notch signaling in both the hMSCs and rNSCs
The microarray data also suggested that coculturing the cells increased Notch signaling in both the rNSCs and in the hMSCs (Table 1). The rNSCs in coculture robustly expressed the receptor Notch 1 (Fig. 5A). hMSCs at sites of contact with rNSCs also expressed the Notch ligand Jagged 1. The results therefore suggest direct cell–cell signaling increased activation of the Notch pathway in both cell types. The Notch signaling pathway has been extensively studied in the developing nervous system, and shown to drive glial differentiation of NSCs by enhancing expression of the transcriptional regulator Hes1 [20,21]. RT-PCR assays of rNSCs isolated from the cocultures (Fig. 5B) demonstrated that expression of the downstream Notch signaling targets Hes1 and Hey1 in rNSCs was increased (P < 0.01). In the hMSCs (Fig. 5B), there was increased expression of HES1 but not the coregulatory factor HEY1 (P < 0.01). Expression of HES1 in the hMSCs was confirmed by Western blot analysis; however, there was no detectable change in protein level between hMSCs cultured alone and those from coculture (not shown).
FIG. 5.
Notch signaling was increased in the cocultures and decreased by an inhibitor of TGFβ signaling. (A) rNSCs in coculture expressed Notch 1 (green). Also, hMSCs contacting rNSCs expressed the Notch ligand Jagged 1 (red). Nuclei were stained with DAPI (blue). Scale bar = 50 μm. (B) Real-time reverse transcription-polymerase chain reaction assays for the Notch downstream targets HES1 and HEY1. In coculture, expression of both HES1 and HEY1 was increased in rNSCs and that of HES1 was increased in hMSCs. An inhibitor of TGFβ signaling (SB431542) decreased expression of HES1 and HEY1 in the rNSCs and HES1 in the hMSCs (t-test: *P < 0.01 vs. coculture). (C) Notch intracellular signaling in the rNSCs was assayed using an inducible reporter construct for the recombination signal binding protein for immunoglobulin kappa J region (RBP-Jk) protein, a downstream modulator of Notch signaling. Coculture significantly increased intracellular Notch signaling. Both the TGF(inhibitor (SB431542) and the γ-secretase inhibitor (DAPT) decreased Notch signaling [1-way ANOVA, F(3,8) = 8.41, *P < 0.05 vs. coculture].
Because of reported synergy between the Notch and TGFβ pathways [22], we repeated the experiments in the presence of the inhibitor of the TGFβ pathway. As expected, incubation with the inhibitor decreased expression of both Hes1 and Hey1 in the rNSCs (Fig. 5B). It also decreased expression of HES1 in the hMSCs.
Notch signaling in the rNSCs assayed with reporter genes
To further confirm the Notch signaling, we transfected the rNSCs with 2 reporter constructs: 1 to assay for the Notch pathway and 1 to control for the efficiency of the transfections. The reporter construct for the pathway contained the luciferase gene under the control of an RBP-Jk response element. Upon activation, the intracellular domain of the Notch receptor (NICD) is cleaved and translocates to the nucleus. The NICD binds to RBP-Jk and thereby converts it from a repressor of transcription to an activator. Coculture with hMSCs significantly increased intracellular Notch signaling in the rNSCs (Fig. 5C). The Notch signaling was reduced by addition of DAPT, a gamma secretase inhibitor that inhibits the cleavage of NICD from the Notch extacellular domain. Also Notch signaling was reduced by addition of the TGFβ inhibitor.
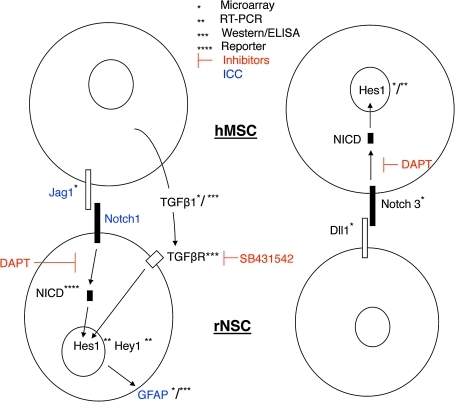
The results indicated that glial differentiation in the cocultures was driven by activation of the TGFβ signaling pathway, possibly through synergistic activity with the Notch signaling pathway (Fig. 6).
FIG. 6.
Schematic of cell signaling between rNSCs and hMSCs in coculture. Data obtained from multiple assays indicate activation of the classical Notch signaling pathway in both the hMSCs and rNSCs. Notch signaling is thought to be unidirectional; therefore, the data suggest that Notch signaling is directed from hMSCs to rNSCs in some sub-populations of the cocultures and from rNSCs to hMSCs in other sub-populations.
Discussion
Therapeutic benefits from administration of MSCs have been reported in a series of models of diseases of the CNS, but the mode of action of the cells has not been defined [8,23,24]. Since the cells differentiated into neural-like cells under some conditions, it was initially assumed that the therapeutic benefits were produced primarily by engraftment and differentiation of the MSCs to neural cells [25,26]. Subsequent observations suggested that therapeutic benefits were frequently obtained with only transitory engraftment and little evidence of differentiation of the cells [8,27–29]. Therefore, the cells apparently exerted their effects indirectly by secretion of soluble factors or cell-to-cell contact that modulated inflammatory and immune reactions [30,31], or by suppressing apoptosis [32,33].
Coculture experiments were used here to explore the recent observations [12,13] that hMSCs infused into the hippocampus-stimulated proliferation, migration, and differentiation of endogenous NSCs. To study the effects of hMSCs on a proliferative population of NSCs we chose to coculture the cells in growth medium as apposed to differentiation induction medium. The presence of hMSCs in the cocultures markedly stimulated differentiation of rNSCs to astrocytes and oligodendrocytes in the optimal NSC growth medium that did not in itself induce differentiation. The use of species-specific microarrays to assay RNA extracted from the cocultures made it possible to survey changes in the transcriptomes of both cell types. To validate the microarray findings a variety of techniques were used to assay both the rNSCs and hMSCs (see summary in Fig. 6).
The role of TGFβ1 was verified by the demonstration that hMSCs in coculture increased secretion of TGFβ1, the rNSCs expressed the receptors, and an inhibitor of the pathway decreased differentiation (Fig. 6). A role for HGF was in part verified by the observation that hMSCs in coculture increased secretion of HGF (Supplemental Fig. S1). The role of Notch signaling was verified by the demonstration that in the cocultures hMSCs expressed a Notch ligand at sites of cell contact with rNSCs, and the rNSCs expressed the Notch receptor. Increased Notch signaling in both cell types was then demonstrated by assays of transcripts for a downstream target for Notch (Hes1) in both cells and a reporter construct for downstream targets of Notch signaling in rNSCs.
The observation that Notch signaling was increased in both the rNSCs and the hMSCs was surprising since Notch is generally recognized as a unidirectional signaling system [34–37]. Therefore, the results suggest that the Notch signaling was directed in opposite directions in different sub-populations of the cocultures (Fig. 6).
A role for TGFβ1 in glial differentiation is consistent with its being a member of a superfamily of growth factors known to produce a diverse range of biological effects in the developing CNS, including NSC expansion and fate determination [38–40]. Its role is also consistent with the observation that conditioned medium from hMSCs increased oligodendrogenesis by rNSCs incubated in medium containing 10% FBS [41,42]. hMSCs also increase both oligodendrogenesis and neuronogenesis by mouse neurospheres incubated in serum-free medium [43]. In the optimal NSC growth medium used here, conditioned medium from hMSCs increased differentiation to oligodendrocytes but not astrocytes. The different fates of the rNSCs are probably due to differences in the hMSC and rNSC preparations, and differences in the culture conditions.
The role of Notch signaling in glial differentiation is consistent with its extensive role in controlling cell fate in many different systems [34,37] and its role in the development of the CNS [44]. Notch signaling provides a mechanism to limit specific cell fates to single cells within a cluster of cells, but it can also restrict cells to an uncommitted fate [34–37]. The Notch signaling pathway has extensive interconnectedness with other signaling pathways [37]. For example, TGFβ can directly induce Hes1 or it can activate the Smad2/3 complex which interacts with NICD to recruit RBP-Jk and thereby induce Hes1 [22]. The results here suggest the possibility that Notch and TGFβ signaling were acting synergically in the rNSCs. This is supported by the observation that an inhibitor of TGFβ signaling inhibited both glial differentiation and increased expression of down stream targets of Notch signaling (HES1 and HEY1) in the rNSCs. It is likely that HGF and other factors that hMSCs were activated to secrete in the cocultures had roles in enhancing differentiation.
As we previously showed that hMSCs survive for short periods of time in the CNS [12,31], stimulation of the endogenous cycling cell population rather than cell replacement may be a more viable means to approach long-term cell therapy for treatment of neurologic insults. Glial cells are gaining appreciation for their ability to support the local environment structurally and functionally. Although long associated with the formation of a detrimental glial scar following neural injury, astrocytes are now being recognized as providing beneficial activities such as restricting inflammation and protecting neurons and oligodendrocytes [45,46]. Growth factor support supplied by astrocytes has been shown to increase neuronal survival, maturation, and function [47,48]. Stimulation of oligodendrocyte survival and function has long been a primary aim for effective treatment of demyelinating diseases and insults. The hMSC-stimulated increase in oligodendrocyte numbers and their maturation has been suggested to underlie remyelination and functional recovery following a demyelinating insult [49]. In addition, oligodendrocytes are now known to produce a number of trophic factors that can support neuronal survival and function [50,51]. Because MSCs are readily accessible and expanded in vitro, they may serve as an effective means to stimulate glial differentiation of the endogenous NSC population and thus enhance endogenous repair mechanisms following neurologic insult.
Supplementary Material
Acknowledgments
The work was supported in part by an NIH grant (P40 RR 17447), the Louisiana Gene Therapy Research Foundation, and a fellowship from the Louisiana Board of Regents Support Fund.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Zhao C. Deng W. Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 3.Arvidsson A. Collin T. Kirik D. Kokaia Z. Lindvall O. Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med. 2002;8:963–970. doi: 10.1038/nm747. [DOI] [PubMed] [Google Scholar]
- 4.Einstein O. Ben Hur T. The changing face of neural stem cell therapy in neurologic diseases. Arch Neurol. 2008;65:452–456. doi: 10.1001/archneur.65.4.452. [DOI] [PubMed] [Google Scholar]
- 5.Qu C. Mahmood A. Lu D. Goussev A. Xiong Y. Chopp M. Treatment of traumatic brain injury in mice with marrow stromal cells. Brain Res. 2008;1208:234–239. doi: 10.1016/j.brainres.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofstetter CP. Schwarz EJ. Hess D. Widenfalk J. El Manira A. Prockop DJ. Olson L. Marrow stromal cells form guiding strands in the injured spinal cord and promote recovery. Proc Natl Acad Sci USA. 2002;99:2199–2204. doi: 10.1073/pnas.042678299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y. Chen J. Wang L. Zhang L. Lu M. Chopp M. Intracerebral transplantation of bone marrow stromal cells in a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine mouse model of Parkinson's disease. Neurosci Lett. 2001;316:67–70. doi: 10.1016/s0304-3940(01)02384-9. [DOI] [PubMed] [Google Scholar]
- 8.Parr AM. Tator CH. Keating A. Bone marrow-derived mesenchymal stromal cells for the repair of central nervous system injury. Bone Marrow Transplant. 2007;40:609–619. doi: 10.1038/sj.bmt.1705757. [DOI] [PubMed] [Google Scholar]
- 9.Ukai R. Honmou O. Harada K. Houkin K. Hamada H. Kocsis JD. Mesenchymal stem cells derived from peripheral blood protects against ischemia. J Neurotrauma. 2007;24:508–520. doi: 10.1089/neu.2006.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zietlow R. Lane EL. Dunnett SB. Rosser AE. Human stem cells for CNS repair. Cell Tissue Res. 2008;331:301–322. doi: 10.1007/s00441-007-0488-1. [DOI] [PubMed] [Google Scholar]
- 11.Haynesworth SE. Baber MA. Caplan AI. Cytokine expression by human marrow-derived mesenchymal progenitor cells in vitro: effects of dexamethasone and IL-1 alpha. J Cell Physiol. 1996;166:585–592. doi: 10.1002/(SICI)1097-4652(199603)166:3<585::AID-JCP13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Munoz JR. Stoutenger BR. Robinson AP. Spees JL. Prockop DJ. Human stem/progenitor cells from bone marrow promote neurogenesis of endogenous neural stem cells in the hippocampus of mice. Proc Natl Acad Sci USA. 2005;102:18171–18176. doi: 10.1073/pnas.0508945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo SW. Kim SS. Lee SY. Lee HS. Kim HS. Lee YD. Suh-Kim H. Mesenchymal stem cells promote proliferation of endogenous neural stem cells and survival of newborn cells in a rat stroke model. Exp Mol Med. 2008;40:387–397. doi: 10.3858/emm.2008.40.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esneault E. Pacary E. Eddi D. Freret T. Tixier E. Toutain J. Touzani O. Schumann-Bard P. Petit E. Roussel S. Bernaudin M. Combined therapeutic strategy using erythropoietin and mesenchymal stem cells potentiates neurogenesis after transient focal cerebral ischemia in rats. J Cereb Blood Flow Metab. 2008;28:1552–1563. doi: 10.1038/jcbfm.2008.40. [DOI] [PubMed] [Google Scholar]
- 15.Sekiya I. Larson BL. Smith JR. Pochampally R. Cui JG. Prockop DJ. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 16.Colter DC. Class R. DiGirolamo CM. Prockop DJ. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci USA. 2000;97:3213–3218. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spees JL. Olson SD. Whitney MJ. Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proc Natl Acad Sci USA. 2006;103:1283–1288. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong S. Li C. Wong WH. ChipInfo: Software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res. 2003;31:3483–3486. doi: 10.1093/nar/gkg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C. Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge W. Martinowich K. Wu X. He F. Miyamoto A. Fan G. Weinmaster G. Sun YE. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J Neurosci Res. 2002;69:848–860. doi: 10.1002/jnr.10364. [DOI] [PubMed] [Google Scholar]
- 21.Gaiano N. Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- 22.Blokzijl A. Dahlqvist C. Reissmann E. Falk A. Moliner A. Lendahl U. Ibanez CF. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003;163:723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dezawa M. Systematic neuronal and muscle induction systems in bone marrow stromal cells: the potential for tissue reconstruction in neurodegenerative and muscle degenerative diseases. Med Mol Morphol. 2008;41:14–19. doi: 10.1007/s00795-007-0389-0. [DOI] [PubMed] [Google Scholar]
- 24.Chopp M. Li Y. Zhang J. Plasticity and remodeling of brain. J Neurol Sci. 2008;265:97–101. doi: 10.1016/j.jns.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Kopen GC. Prockop DJ. Phinney DG. Marrow stromal cells migrate throughout forebrain and cerebellum, and they differentiate into astrocytes after injection into neonatal mouse brains. Proc Natl Acad Sci USA. 1999;96:10711–10716. doi: 10.1073/pnas.96.19.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermann A. Maisel M. Storch A. Epigenetic conversion of human adult bone mesodermal stromal cells into neuroectodermal cell types for replacement therapy of neurodegenerative disorders. Expert Opin Biol Ther. 2006;6:653–670. doi: 10.1517/14712598.6.7.653. [DOI] [PubMed] [Google Scholar]
- 27.Prockop DJ. “Stemness” does not explain the repair of many tissues by mesenchymal stem/multipotent stromal cells (MSCs) Clin Pharmacol Ther. 2007;82:241–243. doi: 10.1038/sj.clpt.6100313. [DOI] [PubMed] [Google Scholar]
- 28.Phinney DG. Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair—current views. Stem Cells. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 29.Caplan AI. Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem. 2006;98:1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 30.Uccelli A. Moretta L. Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 31.Ohtaki H. Ylostalo JH. Foraker JE. Robinson AP. Reger RL. Shioda S. Prockop DJ. Stem/progenitor cells from bone marrow decrease neuronal death in global ischemia by modulation of inflammatory/immune responses. Proc Natl Acad Sci USA. 2008;105:14638–14643. doi: 10.1073/pnas.0803670105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z. Deb A. Pachori A. He W. Guo J. Pratt R. Dzau VJ. Secreted frizzled related protein 2 protects cells from apoptosis by blocking the effect of canonical Wnt3a. J Mol Cell Cardiol. 2008;46:370–377. doi: 10.1016/j.yjmcc.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Block GJ. Ohkouchi S. Fung F. Frenkel J. Gregory C. Pochampally R. Dimattia G. Sullivan DE. Prockop DJ. Multipotent stromal cells (MSCs) are activated to reduce apoptosis in part by upregulation and secretion of stanniocalcin-1 (STC-1) Stem Cells. 2008 doi: 10.1634/stemcells.2008-0742. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanpain C. Horsley V. Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Artavanis-Tsakonas S. Rand MD. Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 36.Amsen D. Antov A. Flavell RA. The different faces of Notch in T-helper-cell differentiation. Nat Rev Immunol. 2009;9:116–124. doi: 10.1038/nri2488. [DOI] [PubMed] [Google Scholar]
- 37.Hurlbut GD. Kankel MW. Artavanis-Tsakonas S. Nodal points and complexity of Notch-Ras signal integration. Proc Natl Acad Sci USA. 2009;106:2218–2223. doi: 10.1073/pnas.0812024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitisin K. Saha T. Blake T. Golestaneh N. Deng M. Kim C. Tang Y. Shetty K. Mishra B. Mishra L. Tgf-Beta signaling in development. Sci STKE. 2007;2007:cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- 39.Golestaneh N. Mishra B. TGF-beta, neuronal stem cells and glioblastoma. Oncogene. 2005;24:5722–5730. doi: 10.1038/sj.onc.1208925. [DOI] [PubMed] [Google Scholar]
- 40.Aigner L. Bogdahn U. TGF-beta in neural stem cells and in tumors of the central nervous system. Cell Tissue Res. 2008;331:225–241. doi: 10.1007/s00441-007-0466-7. [DOI] [PubMed] [Google Scholar]
- 41.Rivera FJ. Kandasamy M. Couillard-Despres S. Caioni M. Sanchez R. Huber C. Weidner N. Bogdahn U. Aigner L. Oligodendrogenesis of adult neural progenitors: differential effects of ciliary neurotrophic factor and mesenchymal stem cell derived factors. J Neurochem. 2008;107:832–843. doi: 10.1111/j.1471-4159.2008.05674.x. [DOI] [PubMed] [Google Scholar]
- 42.Rivera FJ. Couillard-Despres S. Pedre X. Ploetz S. Caioni M. Lois C. Bogdahn U. Aigner L. Mesenchymal stem cells instruct oligodendrogenic fate decision on adult neural stem cells. Stem Cells. 2006;24:2209–2219. doi: 10.1634/stemcells.2005-0614. [DOI] [PubMed] [Google Scholar]
- 43.Bai L. Caplan A. Lennon D. Miller RH. Human mesenchymal stem cells signals regulate neural stem cell fate. Neurochem Res. 2007;32:353–362. doi: 10.1007/s11064-006-9212-x. [DOI] [PubMed] [Google Scholar]
- 44.Yoon K. Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 45.Faulkner JR. Herrmann JE. Woo MJ. Tansey KE. Doan NB. Sofroniew MV. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bush TG. Puvanachandra N. Horner CH. Polito A. Ostenfeld T. Svendsen CN. Mucke L. Johnson MH. Sofroniew MV. Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar-forming, reactive astrocytes in adult transgenic mice. Neuron. 1999;23:297–308. doi: 10.1016/s0896-6273(00)80781-3. [DOI] [PubMed] [Google Scholar]
- 47.O'Malley EK. Black IB. Dreyfus CF. Local support cells promote survival of substantia nigra dopaminergic neurons in culture. ExpNeurol. 1991;112:40–48. doi: 10.1016/0014-4886(91)90112-p. [DOI] [PubMed] [Google Scholar]
- 48.Muller HW. Seifert W. A neurotrophic factor (NTF) released from primary glial cultures supports survival and fiber outgrowth of cultured hippocampal neurons. J Neurosci Res. 1982;8:195–204. doi: 10.1002/jnr.490080209. [DOI] [PubMed] [Google Scholar]
- 49.Akiyama Y. Radtke C. Honmou O. Kocsis JD. Remyelination of the spinal cord following intravenous delivery of bone marrow cells. Glia. 2002;39:229–236. doi: 10.1002/glia.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkins A. Chandran S. Compston A. A role for oligodendrocyte-derived IGF-1 in trophic support of cortical neurons. Glia. 2001;36:48–57. doi: 10.1002/glia.1094. [DOI] [PubMed] [Google Scholar]
- 51.Dai X. Lercher LD. Clinton PM. Du Y. Livingston DL. Vieira C. Yang L. Shen MM. Dreyfus CF. The trophic role of oligodendrocytes in the basal forebrain. J Neurosci. 2003;23:5846–5853. doi: 10.1523/JNEUROSCI.23-13-05846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.