Abstract
The view of biology as goal-directed engineering has deep historical roots in developmental biology, a field currently benefitting from an influx of ideas and methods from systems biology. Systems biology draws on non-biological paradigms to explain developmental mechanisms of control, the specific type of regulation that achieves or maintains a desired end. This review highlights some of the current efforts designed to elucidate basic design principles underlying the engineering objectives of robustness, precision, and scaling that are required during developmental control of growth and pattern formation. Examples from vertebrate and invertebrate development are used to illustrate general principles including the value of integral feedback in achieving set-point control; the usefulness of self-organizing behavior; the importance of recognizing and appropriately handling noise; and the No Free Lunch theory. Through the examination of such principles, systems biology offers a functional framework to make sense of the mechanistic complexity of organismal development.
Introduction
The practice of Developmental Biology is much like re-reading a good book. Even when the ending is well known, much can be learned from exploring the plot twists and character developments that lead to the dénouement. The habit of seeing developmental events as being inevitably directed towards fixed, predetermined ends is deeply ingrained in developmental biology, a view that is understandable, given the remarkable abilities of embryos to come out normally after drastic manipulations. Embryonic regulation has fascinated scientists since the 19th Century, when Driesch derived normally patterned sea urchin larvae from single embryo blastomeres. Extending this concept to genetic, as opposed to surgical, manipulation is Waddington’s notion of canalization, the idea that the normal phenotype is selected to be especially insensitive to genetic variation. In the modern developmental biology literature, terms like robustness and precision are finding increasing use. Robustness is the further generalization of canalization, including insensitivity to all kinds of perturbations, environmental and genetic. Precision—the magnitude of natural variation in developmental outcomes—is a measure of robustness with respect to natural perturbations (e.g., standing genetic variation, normal environmental fluctuations, and the randomness of biochemical processes).
The frequency and degree with which embryonic regulation, canalization, robustness, and precision are encountered in development raises many questions. Is there a common principle underlying all such phenomena? Are there conserved mechanisms? Can we explain how (and why) such processes evolved? These questions have, of course, been around for a very long time. What’s new these days is an influx of ideas and concepts from methodologies outside of traditional biology, including control theory, information theory, and network analysis. The application of these computational methods to biological systems provides a more comprehensive view of the design principles underlying the functions of complex biological systems. This review discusses, exclusively, the regulation of morphogenesis in selected vertebrate and invertebrate animal species (Figure 1). However, it should be noted that systems biology approaches to plant morphogenesis are currently bearing (if the pun may be forgiven) considerable fruit (e.g. Jiao and Meyerowitz, 2010; Sahlin et al., 2011).
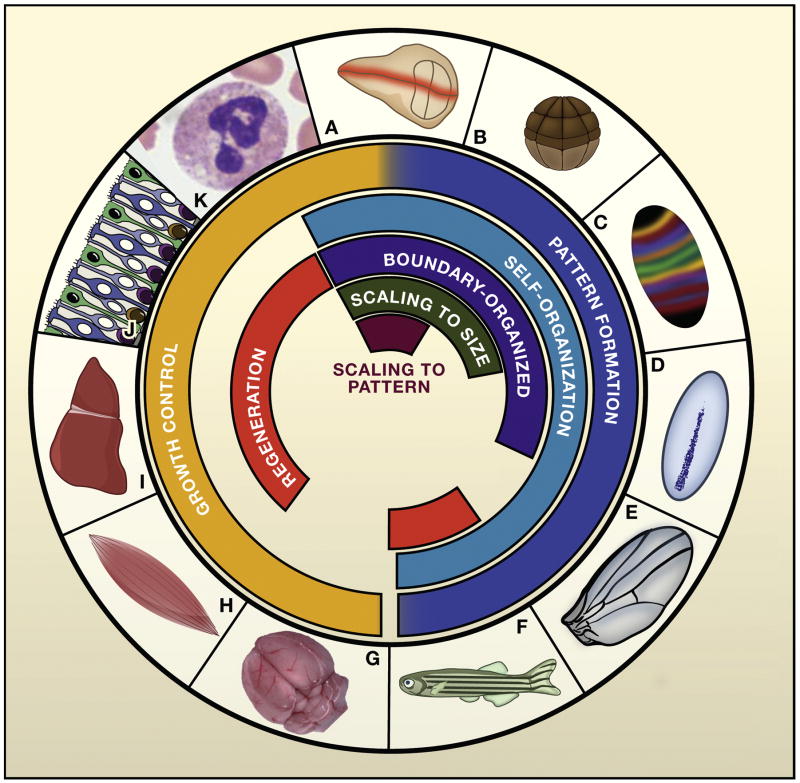
Figure 1.
Control objectives in morphogenesis. The figure compares some of the experimental systems, discussed in this review, that are being used to study developmental regulation, canalization, robustness, and precision. The Drosophila wing imaginable disc (A) is an excellent model for both growth control and pattern formation (through the action of long-range morphogens, such as Hedgehog, Decaptentaplegic [Dpp], and Wingless). The wing disc demonstrates both scaling of pattern to size and scaling of size to pattern. The early Xenopus embryo (B) provides an excellent system for studying pattern formation in the absence of growth, as well as the scaling of pattern to size. Pattern formation has been extensively studied along the anteroposterior axis (C) and the dorsoventral axis (D) of the Drosophila embryo. Anteroposterior patterning is initiated by the transcription factor Bicoid, which acts as a long-range morphogen within the cytoplasm of the syncytial early embryo, controling a cascade of long-range and self-organizing events that segment the embryo into specific regions (stripes). Dorsoventral patterning utilizes the long-range morphogen Dpp to trigger, among other things, a self-organizing process at the dorsal midline. Self-organization also characterizes the mechanism by which narrow, straight veins are positioned on the Drosophila wing (E) during the pupal stage. The development of pigment stripes in teleost fish, such as the zebrafish (F), provides another opportunity to investigate self-organizing patterns, especially in the context of regeneration, which has shed new light on mechanism. Mammalian brain (G) and muscle (H) are good models of organ size control; in the case of muscle, genetic studies have revealed a critical role for feedback from chalones. Feedback regulation of growth has long been known about through studies on regeneration of the mammalian liver (I). More recently, studies in the mouse olfactory epithelium (J) have shed light on mechanisms underlying feedback control of both size and regenerative speed. Analogous mechanisms appear to be at work in mammalian hematopoiesis (K). Other excellent experimental systems, not shown here, include the early vertebrate spinal cord and hindbrain (pattern formation); vertebrate limb buds (pattern formation, growth control); vertebrate and invertebrate retinas (growth control); and plant shoot apical meristems (pattern formation). Figure 1J courtesy of Kim Gokoffski and Anne Calof.
Complexity, performance and control
A complex system can be defined as any system in which large enough numbers of elements interact in simple ways to produce non-obvious behavior. There are two types of such systems: those that are complex by chance, and those that are complex by necessity. The former are often studied by physicists, and typically involve situations in which orderly properties of matter at one level of description emerge out of collective chaos at lower ones. Such emergent properties are often summarized in terms of physical laws like the Universal Gas Law, or Fick’s Laws of Diffusion.
The second type of complex system is encountered by engineers, who design systems to meet specific performance objectives. When engineered systems are dynamic (changing in time), and the performance goals require control (steering behavior toward desired goals), the numbers and types of interacting components can quickly reach the point at which system behavior is sufficiently non-obvious and sophisticated mathematics or computer simulation is required to both understand and predict it.
There is clearly a strong affinity between this second type of complexity—deriving from dynamics and control—and the picture developmental biologists have of embryos: dynamic, yet reliably achieving pre-specified ends. Indeed, the idea that complex engineering performances of technology could be a pertinent model for the nature of morphogenesis was postuated more than 60 years ago (Weiss, 1950). Yet in those times, conditions were not right for exploiting the natural affinity between engineering and morphogenesis. As we shall see, there are essentially two types of engineering: forward engineering, which involves knowing a set of performance objectives and building a system that fulfills them, and reverse engineering, which involves knowing how a system is built, and inferring the performance objectives that necessitated it being built that way. Throughout most of the 20th century, developmental biologists were ready to do neither. Now they are increasingly doing both.
Forward engineering pattern
If we logically understand “performance objectives” in biology—as corresponding to whatever natural selection selects for, i.e. what evolutionary biologists call “fitness”—then we see that one problem with forward engineering is that biologists rarely have a thorough understanding of what contributes to fitness (except perhaps for unicellular organisms in simple environments). Moreover, even if the performance objectives that drove the evolution of an individual biological system were known, there is no guarantee that a forward engineering approach would come up with the same solution as nature. Ask an engineer to build a bridge, and it may not look like any other bridge. This point is illustrated by the history of Turing patterns in developmental biology.
The name, Turing pattern, derives from Alan Turing’s seminal paper, which also introduced the word morphogen (Turing, 1952). It describes a solution to the general problem of creating repeating patterns in space. Through further elaboration of Turing’s work (Gierer and Meinhardt, 1972; Meinhardt and Gierer, 2000), we know that such patterns tend to arise in systems of spatially arrayed, equivalent components (e.g. cells), when they produce both “activating” and “inhibiting” signals that spread at different rates. Depending upon the details, steady states may be reached in which peaks and troughs of signal production occur in repeated patterns of spots or stripes (Figure 2a). Turing patterns exemplify a class of mechanisms termed “self-organizing”, because the location and spacing of elements emerge out of local interactions, and not through instructions that come from elsewhere.
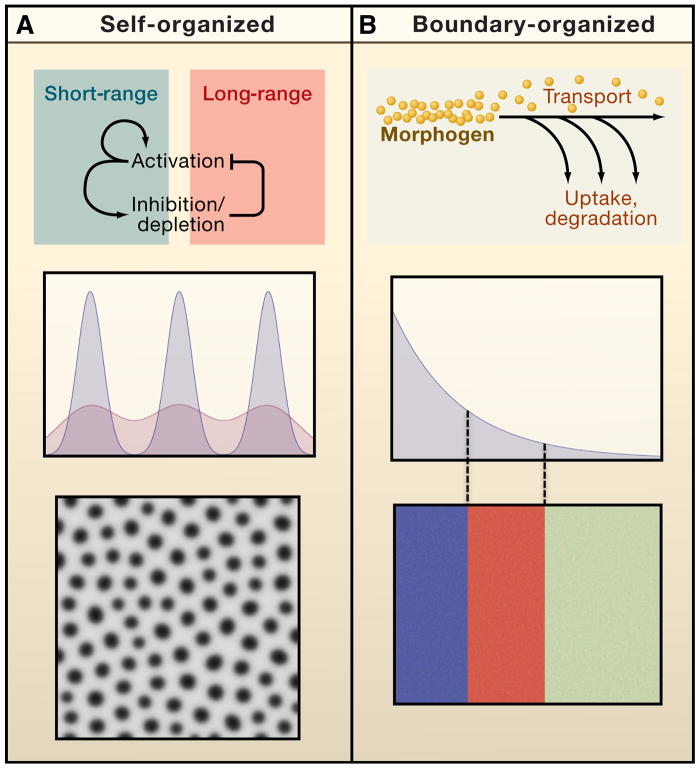
Figure 2.
Two modes of organization in the control of pattern. The performance objectives of patterning systems include both controlling the locations of events relative to each other, and controlling them relative to pre-specified landmarks. Turing patterns are one example of self-organizing pattern (A). Repeated patterns form spontaneously, and exhibit spacings that depend primarily upon the details of local signal activation, inhibition and spread, with relatively little influence from events outside the system. In contrast, long-range morphogen gradients typify boundary-driven organization (B). They inform cells of their location relative to fixed landmarks. In both cases, morphogens establish a characteristic “length scale” or “wavelength”. In the first case (A), pattern is a direct reflection of that scale, such that elements (rows of spots) occur once per length scale. In the second case (B), the length scale simply determines how gradually “positional information” decays over space; where pattern elements occur (blue, red, green blocks) depends upon how cells interpret the positional information they receive.
Initial hopes that Turing processes would provide a simple explanation for all the periodic patterns of development—from skin markings to seashell patterns to embryonic segmentation—have not been realized. Particularly with respect to early, high-precision events, such as the specification of embryonic segments, 30 years of intensive experimental genetics has failed to produce simple, diffusible activator/inhibitor pairs for such cases. Instead, such work has tended to support the view that pattern is organized by morphogens that form long-range gradients from which cells learn their positions. Such systems are boundary-organized (Figure 2b), meaning that positional information is encoded in one or more boundaries, with morphogens passively conveying that information across a field of cells. Interest in self-organizing patterns is, however, very much on the rise today, partly because of recent evidence for the involvement of Turing processes in left-right axis specification, skeletal patterning in the vertebrate limb, the patterning of mammalian and avian ectodermal organs, skin pigmentation patterns, branching morphogenesis in the lung, and hydra head regeneration (reviewed by Kondo and Miura, 2010).
Recent work on the pigmentation patterns on fish is particularly instructive because it takes advantage of the fact that self-organizing mechanisms are inherently regulative, i.e. they can locally repair themselves. Moreover, the precise way in which pigment stripes respond to surgical manipulation in the fish is strongly indicative of a Turing process (Yamaguchi et al., 2007). As we shall see later, the regulative nature of self-organizing pattern can both help and hinder robust patterning, a fact that may explain why boundary-organized mechanisms are also needed for pattern formation.
Importantly, the creation of a Turing pattern through the processes of activation, inhibition and spreading, which create a Turing pattern, need not be mediated by secreted, diffusible molecules (Kondo and Miura, 2010). The Turing process is a mathematical abstraction that invokes the production and destruction of interacting, moving signals. No restrictions are imposed on the molecular details of the signals, how they move, or how they interact. It is possible that the true prevalence of Turing patterns in development has been underestimated, because biologists have been too focused on looking for particular kinds of molecules, rather than general design principles. From this we can see both the strength and weakness of the forward engineering approach in biology: it provides a direct route to design principles, but it cannot tell us how those principles are implemented in real biological systems.
Reverse engineering growth
The classic definition of reverse engineering describes the industrial spy who, using only stolen blueprints, figures out what a competitor’s product does as well as how to manufacture it. Unlike forward engineering, which progresses from performance objectives to design, reverse engineering starts with design and seeks to learn performance objectives. To do this, the engineer must either use pre-existing knowledge of design principles, or he or she must use modeling and/or simulation to explore the sorts of behaviors a system is capable of, in the hope of recognizing performance that might be useful or desirable.
Reverse engineering requires extensive knowledge of a system’s “wiring diagram”, which is one reason why opportunities to do it were rare in biology until the advent of comprehensive data-gathering methodologies such as genomics, proteomics, saturation mutagenesis, et cetera. Yet this is only half the reason why reverse engineering is such a prominent activity in Systems Biology. The other is that the goal of reverse engineering—to learn performance objectives—fills in just the kind of information that traditional molecular genetics cannot: what the components of a system are for, as opposed to merely what they do. The more massive the biological system, the more important such insight is.
Among the processes that Systems Biologists have reverse engineered are metabolism, cell cycle control, stress responses, and bacterial chemotaxis (Alon et al., 1999; Csikasz-Nagy et al., 2008; Khammash, 2008; Sauro and Kholodenko, 2004). The first explicit attempts to reverse engineer complex developmental systems date to Odell’s work on the network of signaling and gene regulation that establishes segment polarity in the Drosophila embryo, and on Notch signaling in insect neurogenesis (Meir et al., 2002; von Dassow et al., 2000). In both cases, it was proposed that system design was influenced by a need for robustness to parameter uncertainty and internal noise. This work has been followed by many studies from other groups exploring ways in which other known mechanisms of pattern formation can also be robust (reviewed by Barkai and Shilo, 2009; Eldar et al., 2004; Lander et al., 2009b).
Patterning is only one of two fundamental processes in morphogenesis, the other being growth. As growth is often a consequence of cell proliferation, this review focuses refers to growth control as control of proliferation, aware, of course, that proliferation can occur without growth (e.g.,. in early embryos), or with much delayed growth. Growth is under tight control, asis supported by the precision observed in the sizes of organisms and their parts. For example, when genetic variability is controlled for, adult mouse brains vary only about 5% in size and cell number (Williams, 2000). For bilaterally symmetric organs (such as limbs), left-to-right variance in size is similarly very small (Wolpert, 2010).
Such precision is impressive in light of the fact that proliferation, being an exponential process, compounds its errors. A mere 2% decrease in cell cycle length will, over 30 cell cycles, cause a >50% increase in the size of a growing population. It is unlikely that the necessary cell cycle precision to achieve normal organ and body size control can be achieved without some sort of feedback process. Indeed, the idea that negative feedback is involved in organ size control received early support from studies of liver regeneration, and from in vitro studies showing that many cell types produce substances that suppress their own proliferation (reviewed by Elgjo and Reichelt, 2004). Work on such substances, chalones, did not significantly take off until the late 1990’s, when it was found that mice deficient in the TGFβ family member GDF8 (myostatin) produced an excess of skeletal muscle. GDF8 is made by muscle, and acts on muscle progenitors, thus fulfilling the requirements of a chalone. Subsequently, GDF11, a close homologue of GDF8, was found to exhibit analogous effects in a self-renewing neural tissue, the mouse olfactory epithelium (Wu et al., 2003). Other molecules have recently been suggested to act as chalones in a variety of tissues (reviewed by Lander et al., 2009a).
The basic chalone model—in which chalones slow the proliferation of progenitors by an amount directly related to organ or tissue size—is too generic for reverse engineering. What’s needed is an actual wiring diagram of how such feedback is implemented in a real organ. Progress toward this end was made in studies of the olfactory epithelium, where progenitors pass through distinct lineage stages and GDF11 acts only at a very specific stage to influence the behavior ofan apparent transit amplifying cell located between a stem cell and a differentiated neuron (Wu et al., 2003). It was subsequently found that activin B, another TGFβ family member, is also expressed in the olfactory epithelium, and also has a negative effect on proliferation, but acts uniquely on the stem cell stage, and not the transit amplifying cell. Reverse engineering of this system (Lander et al., 2009a) entailed mathematically exploring what performance objectives could potentially be met by a multiplicity of progenitor cell stages, a multiplicity of feedback factors, and the specificity of factors for single lineage stages.
This analysis produced several useful results, one of the most important was negative: A chalone that acts by slowing the divisions of an intermediate cell in the lineage of a self-renewing tissue should not be able to have any effect on steady state tissue size. This suggested that the mechanism of action of GDF11, which targets just such a lineage stage, must involve more than just suppressing cell divisions. This in turn led to experiments showing that GDF11 also controls the renewal probability of its target cell, i.e. the probability that the target cell’s progeny remain of the same type instead of progressing to the next lineage stage (Lander et al., 2009a).
Once this additional mechanism was taken into account, calculations showed that not only could GDF11 influence the tissue’s steady state, it could control it with near-perfect robustness. For instance, the steady state was robust to cell cycle speeds, initial numbers of stem cells, and rates of cell death. Moreover, this feedback arrangement also creates a mechanism for triggering extremely rapid regeneration after injury. However, both performance objectives could not be met under the same conditions, unless additional feedback (in this case from activin) onto the stem cell was also included. Thus, reverse engineering suggests that the detailed interaction of lineage, feedback, and regulation of self-renewal found in the olfactory epithelium constitutes a system for simultaneous robust size control and rapid regeneration (Lander et al., 2009b; Lo et al., 2009).
The value of integral feedback
Around the same time as the above studies on the olfactory epithelium, two groups independently concluded that feedback control of cell number in hematopoiesis also occurs primarily through the regulation of self-renewal, though control of lineage progression as opposed to control of the cell cycle speed. In one case, the conclusion was supported by the dynamics of regenerative responses following bone marrow transplantation (Marciniak-Czochra et al., 2009). The other (Kirouac et al., 2009) derived the result from a combination of model exploration (a systematic approach to reverse engineering) and model fitting (using computational algorithms to extract parameter values from in vitro and in vivo data).
The evidence that feedback specifically targets progenitor self-renewal in multiple systems suggests that there is some generically useful feature associated with this mechanism. Indeed, inspection shows that it is a straightforward implementation of an engineering strategy known as integral feedback control. Essentially, integral feed back control describes the output of a system that, when when fed back into the same system, provides a signal that is proportional to the time integral of the difference between the system’s current behavior and its desired behavior (Figure 3). Integral control is observed in other biological systems (such as bacterial chemotaxis; Fig. 3a), and appears to be generically essential whenever feedback must maintain a desired output exactly (set-point control); this explains why it is can robustly maintain self-renewing tissues at a pre-determined size. In contrast, feedback regulation of the rate of progenitor progression through the cell cycle amounts to what engineers call proportional control (feedback regulation of cell death also amounts to proportional control). Although proportional control can provide some compensation for disturbances, it generically does not restore a perturbed system to a set-point.
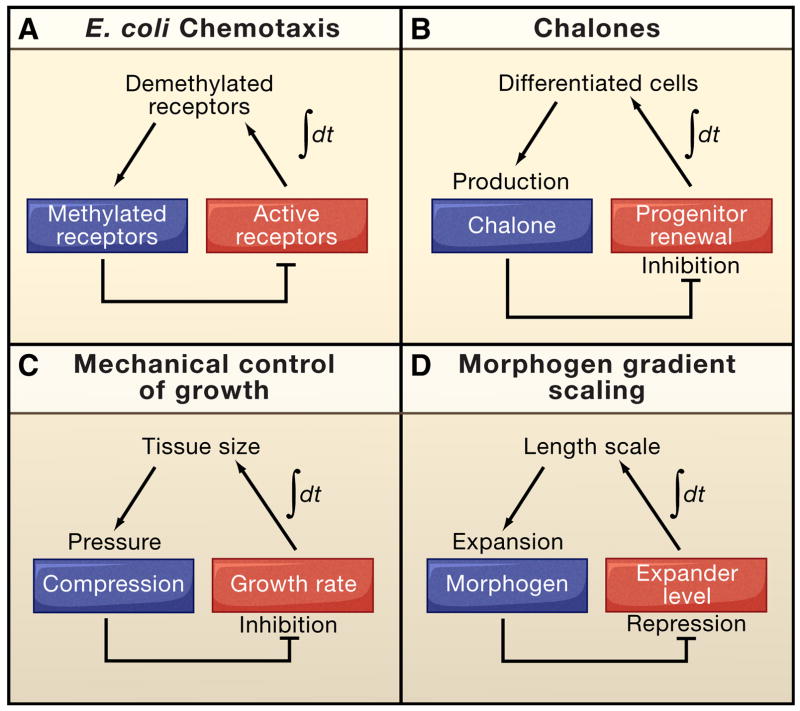
Figure 3.
Versatility of integral feedback control. Integral feedback is particularly useful for achieving set-point control, in which a system achieves a pre-specified steady-state behavior independent of external (and often many internal) perturbations. The essence of integral control is to relay a signal that reflects the time integral of error (the difference between the actual and desired state of the system). Biological systems often use this type of control to achieve robust, perfect adaptation, i.e. to return to a zero-activity state even after sustained perturbations. For example, in bacterial chemotaxis (A) integral feedback adaptively modulates signaling to maximize sensitivity to changes in chemoattractant levels (Alon et al., 1999; Yi et al., 2000). Integral feedback in the control of cell growth has been described for two distinct systems. Production of chalones, such as GDF11, by differentiated cells in the olfactory epithelium inhibits progenitor self-renewal (B), providing a feedback signal that increases (decreases) in time as long as the probability of progenitor cell renewal is greater (lesser) than 50% (Lander et al., 2009a). Mechanical compression within the Drosophila wing disc increases with disc size (C), potentially providing a growth inhibitory signal that increases in time as long as cells are proliferating (Shraiman, 2005). Integral feedback can also be used to make a morphogen gradient scale to fit the territory between its source of production and a distant boundary (D). In this case, the morphogen inhibits the production of a molecule that acts at long range to expand the range (length scale) of the morphogen (Ben-Zvi and Barkai, 2010). In such a scenario, buildup of the expander over time provides a time- integrated error signal, which only vanishes when the morphogen gradient expands all the way to the distant boundary.
The same integral control mechanism that achieves robust maintenance of a steady state in constantly renewing lineages, as in the olfactory epithelium or in hematopoiesis, can also provide for robust final size specification in non-renewing tissues such as the brain. Consistent with this, the pattern of gradual progenitor pool expansion, contraction, and extinction that occurs in the developing brain closely follows the expected consequences of negative feedback control of progenitor self-renewal (Lander et al., 2009a). Indeed, measurements of progenitor self-renewal probabilities in the cerebral cortex show just the predicted steady decline that negative feedback control should produce (Nowakowski et al., 2002).
Precisely what negative feedback factors are responsible for such behavior in the brain remains unknown. Factors such as GDF8, GDF11 and activin are present in many locations throughout the nervous system. In the retina, however, loss of gdf11 leads not to a change in tissue size, but to marked alterations in the proportions of neuronal cell types produced, with some expanding at the expense of others (Kim et al., 2005). In the neural retina, a single progenitor cell type is thought to give rise to all the differentiated cells, suggesting that GDF11’s effects extend not just to whether progenitor cells renew or differentiate, but also their choice of what cell type to differentiate into.
The range of control
Notwithstanding their likely importance in regulating tissue growth and cellular composition, secreted negative feedback factors can, at best, be only part of the picture. Notch signaling, for example, also influences cell proliferation and controls the fate choices of progenitors (Artavanis-Tsakonas et al., 1999). Through lateral inhibition, Notch can ensure that precisely one progenitor can arise within a particular region of space, a sort of short-range set-point control. At the opposite extreme of range of action are circulating feedback inhibitors, which parabiosis experiments long ago implicated in liver size control (Moolten and Bucher, 1967).
Indeed, every use of feedback for control in development has a characteristic spatial range. For example, secreted polypeptide growth factors (e.g. chalones) are thought to act within epithelial tissues at ranges up to a few hundred microns, due to the depleting effects of receptor-mediated uptake (e.g. Lander et al., 2009a; Shvartsman et al., 2001). How could such molecules integrate size information over the much larger scale of macroscopic organs? One possibility is that they act at an early stage of development, when dimensions are smaller. As organ growth proceeds, the control provided by feedback would become more and more locally autonomous. This would still allow for an accurate global response to perturbations that affect all locations equally (e.g. genetic variability, changes in body temperature, nutritional status), but not to local disruptions (e.g. physical damage to a part of the growing tissue would not elicit compensatory growth elsewhere). This seems a good framework for thinking about the specification of limb size, which is remarkably precise yet created out of the actions of parts that exhibit considerable growth autonomy (discussed by Pan, 2007; Wolpert, 2010). Such observations do not imply that growth control is achieved without global feedback, but simply that global feedback may occur early (e.g. in the limb bud instead of in the leg, arm, wing, or fin). This makes an important general point about developmental precision: the machinery for control is needed only at times when relevant perturbations tend to happen.
One possible solution to control growth on many spatial scales is to combine strategies. This seems to happen in the olfactory epithelium, because tissue size along the apico-basal dimension of the epithelium (<100 μm in thickness) is highly sensitive to mutations that alter GDF11 expression or function, but lateral expansion of the epithelium into surrounding connective tissue (over many millimeters) is less sensitive (Kawauchi et al., 2009). So far, molecular mechanisms controlling planar expansion have been little explored in this tissue, although there are reasons to suspect that regulation of fibroblast growth factor activity is involved (Kawauchi et al., 2005).
One tissue that has been the focus of a great deal of experimental work on epithelial planar expansion is the Drosophila larval wing imaginal disc. Growth of the wing disc is influenced by signals on multiple length scales (Edgar, 2006; Martin-Castellanos and Edgar, 2002; Nijhout and Grunert, 2010; Schwank and Basler, 2010)—cell-to-cell, compartment-wide, disc-wide, and humoral (hormonal). Recent theoretical and experimental work (Aegerter-Wilmsen et al., 2007; Aegerter-Wilmsen et al., 2010; Hufnagel et al., 2007; Nienhaus et al., 2009) suggests that disc-wide coordination may be mediated, at least in part, by mechanical feedback (the length-scale of which can be very long, depending upon the viscoelastic properties of the tissue). The influence of mechanical effects (tension, compression) on cell growth is well established for mammalian cells (Mammoto and Ingber, 2009). In the wing disc, it has been pointed out that such mechanical effects create an opportunity for disc-wide integral feedback control (Shraiman, 2005); Fig. 3c)
The wing disc has also been instrumental in shedding light on the role of the Hippo signaling pathway (also known as the Salvador/Warts/Merlin pathway) in growth control. The molecular details of the Hippo pathway, although still emerging, have been reviewed elsewhere (e.g. Buttitta and Edgar, 2007; Grusche et al., 2010b; Halder and Johnson, 2011; Pan, 2007; Reddy and Irvine, 2008), and will not be reiterated, except to say that a major source of input into the pathway is the cell surface protocadherin Fat, and the major intracellular target of Hippo signaling seems to be the growth-stimulating transcriptional co-activator Yorkie (Yki; the vertebrate homolog of YAP), which Hippo signaling inactivates. Precisely what the crucial targets of Yki are is unknown, but recent studies suggest that, like GDF11, its functions include the regulation of self-renewal, and not just the rate at which cells traverse the cell cycle (Halder and Johnson, 2011).
As with Notch signaling, the fact that the Hippo pathway is activated by cell surface ligands and receptors (the only known ligand for Fat is Dachsous, also a cell surface protocadherin) suggests a one-cell range of action. Indeed, recent work suggests that the Fat pathway is central in mediating the well-known in vitro phenomenon of “contact inhibition of cell growth” (Zhao et al., 2007), an example of growth control with a spatial scale of the single cell. Recent studies also find the Hippo pathway to be essential for compensatory cell proliferation after injury, a form of local regenerative response. Interestingly, genetic studies aimed at identifying major determinants of organ size have, in both vertebrates and invertebrates, consistently implicated the Hippo pathway—far more so than the pathways controlled by classical, diffusible growth factors (reviewed by Halder and Johnson, 2011; Pan, 2007). This suggests that the spatial range of Hippo signaling may, sometimes, be quite large. Several mechanisms have emerged for how that might be achieved.
First, Yki activation has been shown, in the Drosophila midgut, to lead to the production of diffusible, growth factors and cytokines that stimulate proliferation in neighboring cells (Ren et al., 2010; Staley and Irvine, 2010). Second, it appears that Yki can be strongly activated by a spatial bias in the occupancy of Fat on one side of a cell versus another, and that such bias can be propagated from cell to cell, in a fashion similar to the way in which cell polarity is propagated from cell to cell by the planar cell polarity pathway (with which the Fat/Hippo pathway shares some components (Reddy and Irvine, 2008)). In addition to evidence that Hippo signaling can act over ranges of several cells, there is evidence that diffusible cues—including BMPs (Rogulja et al., 2008) and factors that activate Jun kinase (Sun and Irvine, 2011)—also act as direct inputs into the regulation of Yki activity. Together these data suggest that, Hippo signaling may integrate and produce both short and long-range signals.
Scaling: Matching pattern to growth
From the earliest days of embryology, it has been clear that the remarkable robustness of pattern formation is primarily manifest at the level of relative, not absolute, pattern (i.e. the locations of pattern elements relative to size of the tissue being patterned). Thus, the patterns that arise in sea urchin embryos derived from isolated blastomeres, or frogs derived from half-embryos, are only normal in proportion to an abnormal size.
The need for patterning that automatically scales to tissue or body size arises from the fact that growth control mechanisms are both robust and adaptive, i.e. given constant genetic and environmental conditions, they robustly specify size-set-points, but those set-points are themselves influenced by other factors (e.g. nutrition, temperature, genetics, timing). A good example of adaptable scaling can be found in the Dpp gradient of the Drosophila wing disc, which displays a roughly constant absolute shape throughout the period of normal disc growth (Hufnagel et al., 2007), yet it strongly scales in response to experimental manipulations that increase or decrease disc size (Teleman and Cohen, 2000). These observations suggest that there is a scaling set-point, but it likely varies with developmental stage.
Developmental biologists have long sought to identify mechanisms responsible for automatic scaling. Indeed, much early enthusiasm for Wolpert’s original model of morphogens as molecules produced at a source and degraded a distant sink (Wolpert, 1969), stemmed from the automatic scaling that such an arrangement achieves (because the morphogen profile is a straight line from source to sink, changes in the location of the sink shift all threshold locations proportionally). As it happens, virtually no known morphogen gradients are made by this source-sink mechanism, as the requirement that morphogens be sensed by cells usually ensures that they are degraded throughout their field of action, rather than just at one end. Because such gradients do not scale automatically, various attempts have been made to find strategies to make them do so.
One approach has been to postulate the existence of two morphogens at opposite ends of a field of cells, with the stipulation that cells take their positional cues from the ratio of the levels of the two molecules. This situation, which typically requires substantial fine-tuning of parameters, was recently analyzed for exponentially-shaped morphogen gradients (McHale et al., 2006), and extended to gradients of more general shape (Ben-Zvi and Barkai, 2010). Such models essentially replace the sink effect in Wolpert’s original model with an independent positional cue that serves the same purpose. In either case, scaling occurs because the behavior of the system everywhere becomes coupled to what happens at both of its boundaries. Such coupling need not be direct. For example, because the rate of spatial decay of a morphogen diffusing within an epithelium depends on the apicobasal dimensions of cells (which influence the rate of morphogen leakage through the basement membrane), it follows that scaling of a morphogen gradient can also be achieved by coupling increases in the planar dimensions of an epithelial sheet to increases in the apicobasal dimensions of its cells (Lander et al., 2011). Indeed, the apicobasal lengths of cells of insect imaginal discs do increase in parallel with the growth of discs as a whole.
An intriguing class of strategies for scaling was recently identified as a result of efforts to reverse engineer dorsoventral (D-V) axis specification in vertebrate embryos. In early embryos, the specification of cell fate depends on a non-uniform pattern of BMP signaling along the D-V axis. Based on work in amphibians and fish, as well as extrapolated findings from the homologous patterning process in insects, it is believed that BMP signaling occurs in a steep ventral-to-dorsal gradient due to the combined effect of initial deposition of BMPs (e.g., Bmp 2, 4 and 7) on the ventral side, and a process of facilitated ventral-ward transport mediated by the BMP-binding protein chordin (which is produced dorsally, in the Spemann organizer). Curiously, an additional BMP ligand, known as Admp, is also produced on the dorsal side of both fish and frog embryos, and its expression is inhibited by BMP signaling. The fact that a BMP ligand is expressed on the opposite side of the embryo from where BMP signaling is needed, combined with the fact that the expression of this ligand is sensitive to the signaling pathway that is least active at the location where it is expressed, strongly suggested that the performance objectives of the D-V specification system involve more than just elaborating a simple morphogen gradient.
This insight led to the discovery that Admp is required for scaling the BMP gradient to the size of the embryo both in surgically-manipulated embryos, as well as in normal embryo-to-embryo variation (Reversade and De Robertis, 2005). Subsequently a mathematical model was developed to explain how such scaling works (Ben-Zvi et al., 2008). More recently, a general design principle, termed expansion-repression control, was extracted from this mechanism. This principle can be invoked whenever graded morphogen signaling inhibits a process that would otherwise lead to runaway expansion of the morphogen gradient itself (Fig. 3D). If the expander has a long range of action, then only when its expression is driven nearly to zero can the morphogen gradient reach a steady state. This will occur only when the morphogen gradient has expanded essentially to the edge of the field capable of making the expander, i.e. when the morphogen gradient fills the tissue it is patterning. In the case of the amphibian embryo, Admp plays the role of expander, and facilitated transport through the actions of chordin ensures that Admp acts over a long range.
This scaling mechanism is an example of integral feedback control (Fig. 3d) (Ben-Zvi and Barkai, 2010). Because the expander is assumed long-lived, its levels reflect the time-integral of the error between where the morphogen gradient is and where it needs to be. Only when the error is driven essentially to zero is a steady state achieved. By uncovering such a general engineering strategy in this mechanism, the authors achieve several important ends. First, they gain the ability to assert that, even if their detailed, explicit model of amphibian D-V scaling (Ben-Zvi et al., 2008) is inaccurate in its specifics (as some have argued (Francois et al., 2009)), it is likely to be correct in its general outlines. (To quote a phrase popular among Systems Biologists, “all models are wrong; some are useful”. (Box, 1979)) Second, they gain the ability to enumerate other classes of mechanisms that are mathematically equivalent, even if mechanistically dissimilar.
For example, they show that gradient expansion can be mediated not only by a secondary morphogen (like Admp), but by any substance that regulates transport of a morphogen, including a diffusible inhibitor that also protects a morphogen from receptor-mediated capture. Interestingly, it was recently found that the secreted protein Pentagone, which is negatively regulated by the BMP-related morphogen Decapentaplegic (Dpp) in the Drosophila wing disc, is a potent expander of the Dpp gradient (Vuilleumier et al., 2010), consistent with a role in the known ability of the Dpp gradient to scale in response to experimental alterations in disc size (Teleman and Cohen, 2000). It has also been found that secreted Frizzled-related proteins, which are competitive inhibitors of Wnt-receptor interaction, act as expanders of Wnt gradients (Mii and Taira, 2009), and during early amphibian embryogenesis are expressed in patterns consistent with negative regulation by Wnts. Although it remains to be tested whether any of these mechanism is truly involved in the scaling of morphogen systems, the above discussion illustrates how the systematic reverse engineering of a complicated biological system can lead to the generation of novel, testable hypotheses.
The management of noise
A major contribution of Systems Biology has been to increase awareness of the roles played by noise in biological systems. Here, noise is defined as variations that originate in random or unpredictable molecular and cellular behaviors. Noise is not just microscopic fluctuation that averages out at the macroscopic level; it can be both a hindrance and a help to biological function. It can degrade precision, but it can also operate switches (Hasty et al., 2000), sustain and synchronize oscillations (Lewis, 2003), amplify signals (Paulsson et al., 2000), or determine stem cell dynamics (Hoffmann et al., 2008). The flow of noise through a network does not behave like the flow of substrates through a biochemical pathway. Noise can increase due to stochastic phenomena placed in series, but can also decrease due to time integration (temporal filtering) as well as feedback and feedforward effects in which correlations in noise are exploited to produce destructive interference.
Pattern formation can be affected by temporal and spatial noise. These distinct types of noise arise from the same processes, but the latter occurs when temporal fluctuations are independent from cell to cell. In general, noise has both a time scale (the time over which one would need to average to lower noise, at any given location, by a given fraction) and a length scale (the distance over which one would need to average to lower noise, at any given time, by a given fraction). Such averaging times and distances are not just a function of the amplitude of noise, but also its structure, i.e. whether fluctuations occur independently in time (Poisson or shot noise) or space (no cell-to-cell coordination), or exhibit correlations (e.g. bursting and cell-to-cell cooperation). One of the biggest surprises in recent years has been the realization that the majority of gene expression in both prokaryotes and eukaryotes is subject to large-amplitude, slow-varying, high burst noise (Raj et al., 2006).
When morphogens provide positional information over long distances, one impact of noise is to limit the precision with which position can be specified (Figure 4). The randomness of morphogen diffusion creates temporal noise at every location; this sets an integration time over which responding cells (or nuclei, in the case of the intracellular morphogen gradients of syncytial embryos) must sum up their measurements of morphogen concentration to adequately filter such noise. For the intracellular Bicoid gradient of the early Drosophila embryo, this limitation can be significant, because nuclei erase their “reading” of the Bicoid level with every nuclear division (which occurs every 10–20 minutes). To combat this problem, spatial averaging may occur, with nuclei influence the readings made by their neighbors (Gregor et al., 2007). This illustrates the point that both spatial and temporal strategies can effectively manage the noise inherent in tissue patterning.
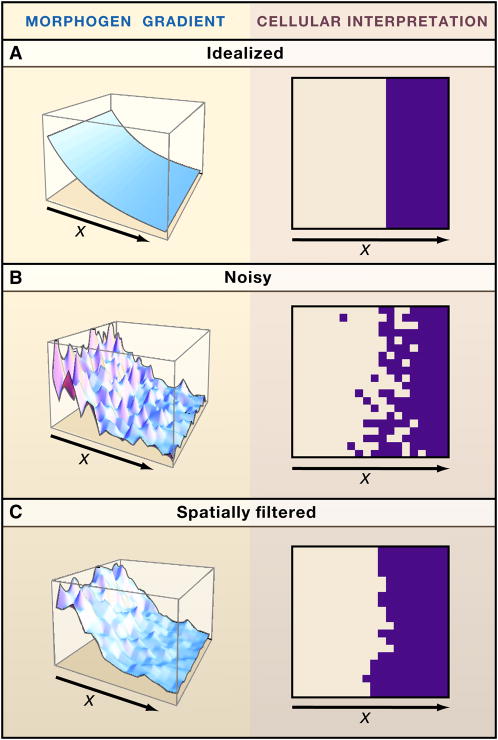
Figure 4.
Reducing noise in pattern formation. In boundary-organized pattern formation, the ability of cells to form organized patterns depends upon the accuracy with which they can measure their positions within a morphogen gradient. Under idealized conditions (A), cells that autonomously adopt a new behavior at a particular threshold value of morphogen concentration will produce a sharp spatial border. In reality, reading a morphogen gradient is fraught with noise: Variability in morphogen level, in gene expression, in cell size, and the stochastic nature of biochemical processes, will cause autonomously acting cells to produce “salt-and-pepper” borders (B). Many sources of noise lead to fluctuations on a time scale too slow for cells to compensate simply by integrating signals over time. In principle, processes that enable cells to collaborate with their neighbors can also reduce the noisiness in morphogen gradient interpretation, producing smoother borders. Such collaboration can take many forms. For example, in (C), the noisy signal in panel B was used to drive the production of an activator in a Turing process (the activator induces its own longer-range inhibitor), the level of which was used as a source of positional information. Note the improved border sharpness. Analyses such as this suggest that the combined use of different modes of pattern organization (boundary- vs. self-organized) can be useful in achieving robust patterning.
For extracellular morphogens, signal integration times are likely much longer than for bicoid, suggesting that local fluctuations in morphogen concentration may not be particularly important, especially when compared with another source of noise: receptors. The number of receptors varies from cell to cell, and even among cells with identical numbers of receptors, receptor occupancy will vary due to the stochastic nature of binding and unbinding (Lauffenburger and Linderman, 1993). Both gene expression noise and morphogen-receptor binding noise will tend to vary on slow time scales, set in one case by the characteristics of transcriptional and translational bursting and of mRNA and protein turnover, and in the other case by the rate of the turnover of bound receptors. Because the range over which a morphogen gradient spreads also depends upon the rate of morphogen uptake and turnover, it has been argued that, for morphogen gradients of typical biological length scales (50–200 μim), receptor binding noise will usually be too slow to remove by simple time averaging (Lander et al., 2009b).
Noise and tradeoffs
If the effects of noise on the precision of patterning are not easily removed by time averaging, how might they be overcome? Looking at this question from a forward engineering perspective sheds light not only on how performance drives the evolution of complex biological regulation, but also on the importance of tradeoffs—the problem that controlling one aspect of performance often degrades another (Figure 5). To see this it is important to recognize that the positional uncertainty created by noise in the interpretation of morphogen gradient depends not only on the character of the noise, but also on the length scale (steepness) of the gradient. This is because, in a steeper gradient, the distance over which cells can distinguish whether they are at one location or another is shorter.
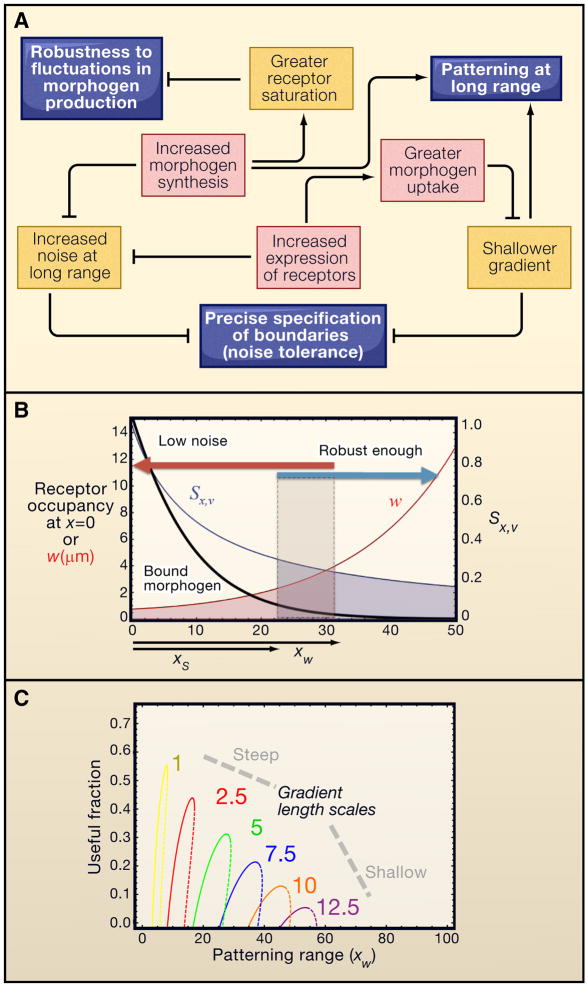
Figure 5.
Performance tradeoffs and morphogen gradients. A. For simple morphogen gradients formed by diffusion with constant receptor-mediated uptake, the ability to achieve performance objectives (e.g., robustness to uncertainty in morphogen production rate; positional precision; and patterning range) is constrained by unintended side effects of performance enhancing-strategies, such as altering levels of morphogen and receptor expression or function. B. These tradeoffs may be analyzed quantitatively, by calculating robustness and precision as a function of distance and gradient range. Sx,v is the sensitivity of position to the rate of morphogen production; w is the size the window of imprecision due to ligand- binding noise. The filled box shows the “useful fractions” where performance constraints on both Sx,v and w are met. C. Parameter space exploration suggests that there is some distance beyond which a simple morphogen gradient cannot simultaneously achieve robustness to morphogen synthesis rate, and positional precision, at any location. Useful fractions are plotted as a function of patterning range, for various values of gradient length scale. Panels B and C are adapted from (Lander et al., 2009b).
Thus, one way to make a gradient more noise-resistant is to make it steeper. Yet, from any given starting amplitude, making a gradient steeper shortens its range. In principle, increasing the starting amplitude of a morphogen gradient (i.e. produce more morphogen) could extend the gradient, but this strategy is limited by another problem: receptor saturation. Significant saturation of receptors near the source of a morphogen gradient dramatically degrades robustness such that small changes in morphogen production rate due to environmental or genetic variation produce large changes in the shape of the gradient) (Lander et al., 2009b). The tradeoff between receptor saturation at one end of a morphogen gradient and noise at the other end is constrained by the biochemistry of ligand binding and the number of receptors per cell. Strategies for overcoming these constraints create additional problems. Increasing the number of receptors per cell increases morphogen capture but shortens the gradient. Up-regulating morphogen destruction can produce arbitrary robustness to fluctuations in morphogen levels (Eldar et al., 2003), but because it makes gradients shallower far from the morphogen source, it also ends up increasing the effect of noise on precision (Lander et al., 2009b). What forward engineering tells us is that performance goals related to precision, robustness, and pattern size can always be expected to interact with each other (e.g., Figure 5a). It has recently been argued (Lander et al., 2009b) that such tradeoffs offer a more plausible explanation for the relatively short distances (50–100 cells (Wolpert, 1969)) over which morphogens act than physical limitations on the speed at which morphogens spread (Crick, 1970).
With this in mind, we might try to reverse engineer some of the complex mechanisms observed in morphogen gradients, to determine whether any of them might help with these tradeoffs. For example, it has commonly been observed that extracellular morphogens accumulate in vesicular structures inside responding cells. If they continue to signal from such locations—as Dpp indeed does (Bokel et al., 2006)—it would allow cells to achieve signal integration over times much longer than those dictated by rates of morphogen capture (Aquino and Endres, 2010). This explanation for intracellular morphogen accumulation provides an alternative to the still-controversial hypothesis that endocytosis plays an active role in morphogen transport.
Another interesting phenomenon, the ability of Hh signaling to reflect the ratio of bound to free receptors, rather than the number of bound receptors (Casali and Struhl, 2004) might also serve as a noise-reduction strategy, since it automatically cancels out the effects of temporal fluctuations in receptor number.
Spatial control of noise
The[KC1] fact that the spatial character of noise ultimately degrades precision in a morphogen gradient suggests that we should also be looking for noise-reduction strategies that are explicitly spatial. For example, in the Bicoid gradient system, the fact that the Bicoid target gene Hunchback (Hb) is itself a diffusible molecule allows the effects of fluctuations in Bicoid signaling at one nucleus to be averaged over many nuclei (Erdmann et al., 2009; Gregor et al., 2007; Okabe-Oho et al., 2009). The diffusivity of Hb improves precision by ironing out bicoid fluctuations, but it also degrades precision by making the Hb boundary less steep, a tradeoff that leads to the prediction of a maximal effect at an optimal diffusivity (Erdmann et al., 2009). The general strategy of using a morphogen gradient to trigger a secondary process that, because it involves diffusion, smoothes out spatial noise, can also be seen in extracellular morphogen systems. For example, in the Drosophila wing disc, anteroposterior positional information is first provided by a Hedgehog (Hh) gradient, which acts at short range to induce the longer range Dpp gradient (Zecca et al., 1995). Any short range imprecision in the way cells interpret the Hh gradient will be smoothed out in the Dpp gradient.
Spatial averaging through diffusion need not involve induction of new morphogens. Induction of diffusible inhibitors or co-regulators can have a similar effect. Indeed, recent work in the Drosophila wing disc indicates that signaling by the Wg gradient may induce the production of at least two types of diffusible inhibitors (Piddini and Vincent, 2009).
Triggered self-organization provides one of the most powerful strategies for overcoming spatial noise through the production of diffusible signals. In the developmental biology literature, self-organizing patterns typically describe a means to generate repeated structures—e.g. fields of spots or stripes. However, using a long-range morphogen gradient as input to a process that sets up a Turing pattern (e.g. expression of an activator or inhibitor), can trigger the formation of a single transition or peak at a specific location in space (Koch and Meinhardt, 1994). Because such a self-organizing process is driven by diffusion (of information, if not always molecules), it will tend to average out spatial noise over a length scale related to the parameters of the process itself (Fig. 4C). For some self-organizing processes - such as the patterns that result from Notch-Delta mediated lateral inhibition -the precision of patterning can even be improved by the addition of spatial and temporal noise (Cohen et al., 2010).
The formation of veins during the pupal stage of fly wing development illustrates the idea of collaboration between long-range morphogens and local self-organization. Initially, the Dpp gradient in the larval wing disc establishes wing vein primordia imprecisely that are refined by the initiation of a series of events, including short range activation and long range inhibition or depletion (utilizing Notch, Dpp and EGF signaling; (Blair, 2007; Yan et al., 2009). Ultimately, narrow veins form through the centers of those primordia in a characteristic pattern. Evidence supporting the Turing nature of this process is supplied by analysis of mutations that broaden vein primordia. These wide primordia do not produce broader veins, but instead produce extra veins in the same territory (Biehs et al., 1998). This is the expected result when the domain over which a Turing process is triggered becomes large compared to its intrinsic wavelength. Another situation in which a form of self-organization sharpens a domain initially specified broadly by Dpp occurs in the Drosophila embryo, where long range Dpp transport toward the dorsal side of the embryo produces a broad peak of Dpp signaling, which in turn triggers a process that boosts Dpp signaling at short range but inhibits it at long range (Umulis et al., 2006; Wang and Ferguson, 2005).
Tradeoffs between self-organization and boundary-organized control
Patterns produced by self-organization are relatively insensitive to external positional cues. This insensitivity is a liability when the goal of patterning is to position new events in relation to the locations of earlier events, especially if that relationship needs to be adjustable through feedback. For example, the automatic scaling of Turing processes is not easy to achieve (see, e.g. Ishihara and Kaneko, 2006; Othmer and Pate, 1980; Umulis et al., 2008). For such purposes, long-range morphogen gradients do much better. In contrast, processes that self-organize in space counteract the effects of noise because they naturally average spatial information. In principle, development can reap the benefits of the scalability of long-range gradients, and the noise reduction of self-organizing processes, by linking the two together (Figure 4C). This works particularly well with Turing processes confined to small domains, because under these conditions they are most sensitive to external positional information, such as boundary conditions. This suggests that some of the most prevalent uses of Turing processes in development may involve situations in which they aren’t forming fields of spots and stripes. Certainly, the Turing process that sets up left-right patterning in vertebrates (creating a single boundary) fits this description (Nakamura et al., 2006).
It remains to be seen how many Developmental events involve collaborations between long-range morphogens and self-organizing processes. However, if we consider other locally self-organizing phenomena (e.g., cell-to-cell signaling networks established by Notch-Delta interactions, the planar cell polarity pathway, or Hippo signaling) it is easy envision morphogenesis as the result of a continual back-and-forth between long range signals and local events. Long-range signals preserve and flexibly control positional information, but are easily degraded by noise, while local events are less flexible, but remove noise and boost signal, acting much like boosters and repeaters in electrical power and wireless data transmission.
This viewpoint helps resolve a seeming paradox emerging out of the study of the Drosophila Bicoid gradient. The Bicoid gradient, and its immediate interpretation by nuclei, appears to be remarkably precise early in development (Gregor et al., 2007); however, severe perturbations to the gradient—by flattening it through changes in bicoid mRNA localization(Ochoa-Espinosa et al., 2009)or by stirring it through the uneven application of heat (Lucchetta et al., 2008)—alter the positions of anteroposterior gene expression domains far less than predicted, suggesting that a great deal of precision arises through self-organization. Such self-organization can arise through the elaborate, cross-regulatory gene networks that exist among the cascade of genes whose expression is initially triggered by bicoid (reviewed by Papatsenko, 2009). As first suggested over two decades ago (Edgar et al., 1989; Lacalli et al., 1988) and verified recently (Manu et al., 2009a), gap gene cross-regulatory interactions create dynamic attractor states, the hallmark of self-organizing systems. Indeed, such self-organization seems to account for robustness of the bicoid gradient to alterations in bicoid level, at the same time producing a certain amount of scaling to embryo size (Manu et al., 2009b). The question left unanswered by these studies is why, if the primary role of bicoid is to activate a system that achieves much of its robustness through self-organization, is the bicoid gradient as precise as it is? The reverse engineer will always respond to such a question by suggesting that there are additional performance objectives that we are failing to take into account. What those might be remains to be determined.
The notion of morphogen gradients as triggers of self-organization also squares well with recent studies of Hedgehog gradients in both invertebrates and vertebrates, and of retinoic acid gradients in the vertebrate hindbrain. In such systems, growing evidence suggests that cell fates are dictated primarily by the history and duration of morphogen exposure rather than the simple steady-state amount of morphogen (Dessaud et al., 2010; Dessaud et al., 2007; Maves and Kimmel, 2005). In the vertebrate spinal cord, Sonic Hedgehog-induced fate switching depends upon cross-regulatory interactions among transcription factors that, as with the gap genes of the Drosophila embryo, may be seen as producing a series of attractor states (Briscoe, 2009; Lek et al., 2010). Such a system may be described as one that is self-organizing in time, with the morphogen gradient applying a spatial bias to the process. In the Drosophila wing disc, the Hedgehog gradient also uses a temporal mechanism to produce multiple borders of gene expression (Nahmad and Stathopoulos, 2009). Clearly, the notion of what long-range morphogens do has come a long way since the early French flag models (Wolpert, 1969, 2011).
Matching growth to pattern
It makes intuitive sense that changes in tissue size, which can result from nutritional or environmental variability, should give rise to compensatory changes in the elaboration of positional cues by morphogens. It is less obvious why the reverse should also be true: that quantitative changes in morphogen function initiate marked changes in tissue growth. Yet virtually all known morphogens are growth regulators, and most are growth promoters. In many systems, growth is dependent upon the expression of the very same morphogens that establish pattern. The best-studied examples come from the Drosophila wing disc, wherein both Dpp and Wg are important positive growth regulators (Baena-Lopez et al., 2009; Schwank and Basler, 2010). Researchers have long focused on explaining the curious observation that cells in the part of the wing disc patterned by Dpp and Wg proliferate in a more or less uniform pattern, whereas the morphogens that are essential for driving that proliferation are distinctly graded in a central-to-peripheral fashion.
Recent studies suggest that the answer to this puzzle has to do with the way in which morphogens interact with the Fat/Hippo pathway. Among the most interesting effects are non-cell autonomous: Yki activity, and subsequent cell proliferation, are highly induced in wing disc cells that express targets of morphogen signals (from either Dpp or Wg) at levels substantially higher or lower than those of their immediate neighbors (Rogulja et al., 2008; Zecca and Struhl, 2010). In the case of Dpp, it has been proposed that this non-autonomous effect arises because graded Dpp signaling leads to graded expression of the Fat ligand Dachsous and its regulator Four-jointed, which in turn leads to asymmetry in Fat occupancy across each wing disc cell, the magnitude of which serves as an inhibitory input to the Hippo pathway (Rogulja et al., 2008).
One consequence of this mechanism is that cells in the morphogen field receive signals to proliferate that are dependent upon the local slope of the morphogen gradient. For an exponentially declining gradient,which is a good approximation of the wing disc Dpp gradient, the slope, measured relative to morphogen concentration at each point, will be a constant, potentially explaining how such a gradient could drive spatially-uniform proliferation.
The idea that the slope of a morphogen gradient could be useful in the control of proliferation is, in fact, a relatively old one, having been suggested by the phenomenon of intercalary regeneration. This refers to the tendency of some embryonic or adult structures to respond to surgical manipulations by growing selectively at the locations where cells that had previously been distant become juxtaposed. The hypothesis is that growth occurs whenever the slope of a gradient of positional information from one cell to another exceeds a threshold. This is, in effect, the reverse of scaling of pattern to size; it amounts to the scaling of size to pattern. Interestingly, recent studies show that intercalary regeneration in wing discs involves activation of Yki via regulation of the Hippo pathway (Grusche et al., 2010a; Halder and Johnson, 2011; Sun and Irvine, 2011).
Such observations tempt speculation that there might be a single unifying principle—measuring and responding to the slope of graded positional information—underlying the role of the Hippo pathway in growth control. Unfortunately, this is almost certainly too simplistic. Dpp clearly affects Yki signaling through a combination of cell autonomous effects (that depend upon Dpp level, not gradient slope), and non-autonomous ones (Rogulja et al., 2008; Schwank et al., 2011). And although Dpp’s effects on the expression of the Fat ligand Dachsous and its regulator Four-jointed can explain observed proliferative effects when cells are forced into contact with neighbors that differ greatly in their level of Dpp response, it appears that the shapes of the endogenous gradients of Dachsous and Four-jointed in the wing disc are rather Dpp-independent (Schwank et al., 2011). Similarly, whereas early studies suggested that proliferative responses to neighbor-neighbor differences in Wg signaling (which induce Yki activity (Zecca and Struhl, 2010)) might be the primary means by which the Wg gradient drives growth (Baena-Lopez and Garcia-Bellido, 2006), later work showed that uniform, moderate levels of Wg are a potent stimulus for proliferation, and further that the slope of the Wg gradient is too shallow throughout most of the wing disc to elicit non-autonomous effects (Baena-Lopez et al., 2009). Adding to these observations, recent work by Schwank et al., (Schwank et al., 2011) indicates that Fat/Hippo signaling can be graded along the anteroposterior axis of the wing disc even when there is no apparent morphogen gradient in that direction. Although Schwank et al., suggest that Fat/Hippo and Dpp signaling are independent growth-control pathways that act in different domains of the disc, another explanation is that Hippo signaling integrates positional information that is coming from a source not yet accounted for in any current models.
That source could be mechanical force. As described earlier, there are strong suggestions that tension and compression within the wing disc epithelium play a role in growth control (Aegerter-Wilmsen et al., 2007; Aegerter-Wilmsen et al., 2010; Hufnagel et al., 2007). Recent stress-birefringence measurements indicate the presence of a central-to-peripheral compression gradient in wing discs (Nienhaus et al., 2009). Given the close connection between upstream components of the Hippo pathway and components of cell junctions and the cytoskeleton, it has been speculated that the Hippo pathway directly receives mechanical inputs (Grusche et al., 2010b). Clearly, there is substantial need for experimental clarification of the role of mechanical events in the Hippo pathway signaling.
On the other hand, that source could come from the morphogen gradients themselves, through mechanisms related to their ability to trigger self-organizing processes at specific locations. As has been pointed out, the Wg target gene vestigial (vg), which seems to play a direct role in driving wing disc growth, displays autoactivation (Zecca and Struhl, 2007, 2010), yet vg also seems to be the target of short-range negative feedback, through an unknown mechanism (Piddini and Vincent, 2009). Arguments based on mathematical modeling posit that such an arrangement creates a situation in which a steady state balance between Vg-promoted growth, Vg-promoted vg expression, and Wg-dependent inhibition of vg expression is only achieved at a fixed tissue size (Zhu, 2011). Because this recent work explored only a limited number of selected parameters, it remains to be seen whether this mechanism truly operates in wing discs. Nevertheless, it represents an elegant solution to the problem of achieving set-point control of growth over a relatively long-length scale.
It should be noted that the simultaneous existence of mechanisms to scale pattern to growth and growth to pattern could create—if such mechanisms were truly independent of each other—futile cycles with each process continually driving the other. Clearly, these mechanisms cannot be independent, but how they are linked remains unclear. One intriguing possibility is that it involves cell surface glypicans, such as Dally and Dally-like, which have been shown to be direct targets of Hippo pathway signaling (Rodriguez et al., 2008). These molecules, and their mammalian orthologues, have been strongly implicated in both organ size control (Filmus and Capurro, 2008; Selleck, 1999; Takeo et al., 2005) and the regulation of patterning by morphogen gradients (e.g. Belenkaya et al., 2004; Franch-Marro et al., 2005; Galli et al., 2003; Han et al., 2004; Han et al., 2005; Kreuger et al., 2004).
Lessons and Implications
The remarkable regulation, canalization, robustness and precision of embryonic development suggest that developing systems devote a considerable amount of cellular machinery to the explicit purpose of control. Through forward and reverse engineering, it has become possible to systematically explore some of the control challenges of faced by developing embryos, and link such challenges to enabling mechanisms. In true Systems Biology fashion, such work begins to explain the complexity of developmental mechanisms in terms of the coordinated functions of entire systems, and not just that of individual parts. The studies highlighted above provide general lessons for future research into control of morphogenesis and development, including the three most salient described below.
Performance is always subject to tradeoffs. The engineering dictum that “there’s no free lunch” illustrates that control comes at a cost. Making a system robust in one way invariably makes it fragile in another (Doyle and Csete, 2007). Tightly controlling cell number can hamper regeneration speed; controlling how robust morphogen gradients are to fluctuations in morphogen levels can affect how sensitive they are to noise; using spatially self-organizing processes to suppress the effects of noise can make spatial scaling more challenging. In general, the researcher who proposes that a particular mechanism fulfills a particular control function should consider how other types of performance are degraded as a result of such control. Unfortunately, this is a standard not often met in the current literature, even in journals with a strong systems biology focus.
Control is not micromanagement. It is easy to think that, in order for a system to be under tight control, every part of it must be controlled. Yet the strategy of integral feedback (Fig. 3) shows that extremely tight control can be achieved merely by feeding back the right kind of signal at one point into a network at an earlier point. The intervening dynamics needn’t be subject to any special regulation (even though they may appear that they are). For example, as long as there is any sort of feedback control on the renewal probabilities of stem cells, such cells can be wildly stochastic in their individual behaviors (e.g.,(Chang et al., 2008; Clayton et al., 2007; Gomes et al., 2011; Snippert et al., 2010)), yet still give us the impression that they “know” precisely what they are doing. Failure to recognize that tightly controlled systems can include uncontrolled parts may well explain the discomfort that many biologists have long had with random processes including stochastic cell fate switches (Chang et al., 2008)), and + the idea that diffusion creates morphogen gradients. Recent assertions that diffusion is “too messy” (i.e. too hard to control) to get the job done (Wolpert, 2009, 2011) indicate that confusion between micromanagement and control is very much alive in biology. In fact, because control always comes at a price, systems that achieve it without micromanaging are often better off.
Phenotype is not performance. Experimental genetics has become an indispensible tool of Developmental Biology, because it enables us to infer causal connections between mechanisms (gene activity) and observations (phenotypes). Impressed with the intricate beauty of the networks that such approaches construct, it is easy to lose sight of the fact that what evolution selects for is not phenotype per se, but performance, the ability of phenotype to do something useful. Too often, the phenomena we choose to investigate are selected for study based on criteria that may be only obliquely related to actual importance to the organism. For example, the high precision of the Drosophila Bicoid gradient is fascinating to us, but we still don’t know how much it matters to the fly. In the wing disc, numerous studies have focused on explaining how spatially graded morphogens drive spatially uniform growth, when we actually have no evidence that the uniformity of growth is itself particularly important. From a traditional molecular biology standpoint, these are not serious problems, because investigating these phenomena will likely lead us to new mechanisms. For the systems biologist, who seeks to use notions of design and performance to place mechanisms into context, there is a real need to get a systematic handle on what is phenomenon and what is epiphenomenon. Luckily, this is just what the tools of forward and reverse engineering provide. As our knowledge of the mechanistic complexity of developmental regulation grows, we can expect to see such approaches playing an ever-greater role in making sense of it all.
Acknowledgments
This work was supported by the NIH (P50-GM076516 and R01-GM067247). I am grateful to Qing Nie, Anne Calof, and Marcos Nahmad for helpful conversations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev. 2007;124:318–326. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Aegerter-Wilmsen T, Smith AC, Christen AJ, Aegerter CM, Hafen E, Basler K. Exploring the effects of mechanical feedback on epithelial topology. Development. 2010;137:499–506. doi: 10.1242/dev.041731. [DOI] [PubMed] [Google Scholar]
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- Aquino G, Endres RG. Increased accuracy of ligand sensing by receptor internalization. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;81:021909. doi: 10.1103/PhysRevE.81.021909. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baena-Lopez LA, Franch-Marro X, Vincent JP. Wingless promotes proliferative growth in a gradient-independent manner. Sci Signal. 2009;2:ra60. doi: 10.1126/scisignal.2000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez LA, Garcia-Bellido A. Control of growth and positional information by the graded vestigial expression pattern in the wing of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:13734–13739. doi: 10.1073/pnas.0606092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkai N, Shilo BZ. Robust generation and decoding of morphogen gradients. Cold Spring Harb Perspect Biol. 2009;1:a001990. doi: 10.1101/cshperspect.a001990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi D, Barkai N. Scaling of morphogen gradients by an expansion-repression integral feedback control. Proc Natl Acad Sci U S A. 2010;107:6924–6929. doi: 10.1073/pnas.0912734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi D, Shilo BZ, Fainsod A, Barkai N. Scaling of the BMP activation gradient in Xenopus embryos. Nature. 2008;453:1205–1211. doi: 10.1038/nature07059. [DOI] [PubMed] [Google Scholar]
- Biehs B, Sturtevant MA, Bier E. Boundaries in the Drosophila wing imaginal disc organize vein-specific genetic programs. Development. 1998;125:4245–4257. doi: 10.1242/dev.125.21.4245. [DOI] [PubMed] [Google Scholar]
- Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu Rev Cell Dev Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- Bokel C, Schwabedissen A, Entchev E, Renaud O, Gonzalez-Gaitan M. Sara endosomes and the maintenance of Dpp signaling levels across mitosis. Science. 2006;314:1135–1139. doi: 10.1126/science.1132524. [DOI] [PubMed] [Google Scholar]
- Box GEP. Robustness in the strategy of scientific model building. In: Launer RL, Wilkinson GN, editors. Robustness in Statistics. New York: Academic Press; 1979. pp. 201–236. [Google Scholar]
- Briscoe J. Making a grade: Sonic Hedgehog signalling and the control of neural cell fate. EMBO J. 2009;28:457–465. doi: 10.1038/emboj.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta LA, Edgar BA. How size is controlled: from Hippos to Yorkies. Nat Cell Biol. 2007;9:1225–1227. doi: 10.1038/ncb1107-1225. [DOI] [PubMed] [Google Scholar]
- Casali A, Struhl G. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature. 2004;431:76–80. doi: 10.1038/nature02835. [DOI] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Crick FHC. Diffusion in Embryogenesis. Nature. 1970;225:420–422. doi: 10.1038/225420a0. [DOI] [PubMed] [Google Scholar]
- Csikasz-Nagy A, Novak B, Tyson JJ. Reverse engineering models of cell cycle regulation. Adv Exp Med Biol. 2008;641:88–97. doi: 10.1007/978-0-387-09794-7_7. [DOI] [PubMed] [Google Scholar]
- Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- Doyle J, Csete M. Rules of engagement. Nature. 2007;446:860. doi: 10.1038/446860a. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Odell GM, Schubiger G. A genetic switch, based on negative regulation, sharpens stripes in Drosophila embryos. Dev Genet. 1989;10:124–142. doi: 10.1002/dvg.1020100303. [DOI] [PubMed] [Google Scholar]
- Eldar A, Rosin D, Shilo BZ, Barkai N. Self-enhanced ligand degradation underlies robustness of morphogen gradients. Dev Cell. 2003;5:635–646. doi: 10.1016/s1534-5807(03)00292-2. [DOI] [PubMed] [Google Scholar]
- Eldar A, Shilo BZ, Barkai N. Elucidating mechanisms underlying robustness of morphogen gradients. Curr Opin Genet Dev. 2004;14:435–439. doi: 10.1016/j.gde.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Elgjo K, Reichelt KL. Chalones: from aqueous extracts to oligopeptides. Cell Cycle. 2004;3:1208–1211. [PubMed] [Google Scholar]
- Erdmann T, Howard M, ten Wolde PR. Role of spatial averaging in the precision of gene expression patterns. Phys Rev Lett. 2009;103:258101. doi: 10.1103/PhysRevLett.103.258101. [DOI] [PubMed] [Google Scholar]
- Filmus J, Capurro M. The role of glypican-3 in the regulation of body size and cancer. Cell Cycle. 2008;7:2787–2790. doi: 10.4161/cc.7.18.6672. [DOI] [PubMed] [Google Scholar]
- Franch-Marro X, Marchand O, Piddini E, Ricardo S, Alexandre C, Vincent JP. Glypicans shunt the Wingless signal between local signalling and further transport. Development. 2005;132:659–666. doi: 10.1242/dev.01639. [DOI] [PubMed] [Google Scholar]
- Francois P, Vonica A, Brivanlou AH, Siggia ED. Scaling of BMP gradients in Xenopus embryos. Nature. 2009;461:E1. doi: 10.1038/nature08305. discussion E2. [DOI] [PubMed] [Google Scholar]
- Galli A, Roure A, Zeller R, Dono R. Glypican 4 modulates FGF signalling and regulates dorsoventral forebrain patterning in Xenopus embryos. Development. 2003;130:4919–4929. doi: 10.1242/dev.00706. [DOI] [PubMed] [Google Scholar]
- Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- Gomes FL, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, Cayouette M. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development. 2011;138:227–235. doi: 10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev Biol. 2010a doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the hippo size control pathway. Curr Biol. 2010b;20:R574–582. doi: 10.1016/j.cub.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Wang B, Lin X. Drosophila glypicans control the cell-to-cell movement of Hedgehog by a dynamin-independent process. Development. 2004;131:601–611. doi: 10.1242/dev.00958. [DOI] [PubMed] [Google Scholar]
- Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- Hasty J, Pradines J, Dolnik M, Collins JJ. Noise-based switches and amplifiers for gene expression. Proc Natl Acad Sci U S A. 2000;97:2075–2080. doi: 10.1073/pnas.040411297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Chang HH, Huang S, Ingber DE, Loeffler M, Galle J. Noise-driven stem cell and progenitor population dynamics. PLoS One. 2008;3:e2922. doi: 10.1371/journal.pone.0002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel L, Teleman AA, Rouault H, Cohen SM, Shraiman BI. On the mechanism of wing size determination in fly development. Proc Natl Acad Sci U S A. 2007;104:3835–3840. doi: 10.1073/pnas.0607134104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara S, Kaneko K. Turing pattern with proportion preservation. J Theor Biol. 2006;238:683–693. doi: 10.1016/j.jtbi.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Kim J, Santos R, Wu HH, Lander AD, Calof AL. Foxg1 promotes olfactory neurogenesis by antagonizing Gdf11. Development. 2009;136:1453–1464. doi: 10.1242/dev.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Khammash M. Reverse engineering: the architecture of biological networks. Biotechniques. 2008;44:323–329. doi: 10.2144/000112772. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kirouac DC, Madlambayan GJ, Yu M, Sykes EA, Ito C, Zandstra PW. Cell-cell interaction networks regulate blood stem and progenitor cell fate. Mol Syst Biol. 2009;5:293. doi: 10.1038/msb.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AJ, Meinhardt H. Biological pattern formation: from basic mechanisms to complex structures. Rev Modern Physics. 1994;66:1481–1510. [Google Scholar]
- Kondo S, Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Lacalli TC, Wilkinson DA, Harrison LG. Theoretical aspects of stripe formation in relation to Drosophila segmentation. Development. 1988;104:105–113. doi: 10.1242/dev.104.1.105. [DOI] [PubMed] [Google Scholar]
- Lander AD, Gokoffski KK, Wan FY, Nie Q, Calof AL. Cell lineages and the logic of proliferative control. PLoS Biol. 2009a;7:e15. doi: 10.1371/journal.pbio.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD, Lo WC, Nie Q, Wan FY. The Measure of Success: Constraints, Objectives, and Tradeoffs in Morphogen-mediated Patterning. Cold Spring Harb Perspect Biol. 2009b;1:a002022. doi: 10.1101/cshperspect.a002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD, Nie Q, Vargas B, Wan FYM. Size-normalized Robustness of Dpp Gradient in Drosophila Wing Imaginal Disc. J Mech Materials Structures. 2011 doi: 10.2140/jomms.2011.6.321. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Linderman JJ. Models for binding, trafficking and signaling. New York: Oxford University Press; 1993. Receptors. [Google Scholar]
- Lek M, Dias JM, Marklund U, Uhde CW, Kurdija S, Lei Q, Sussel L, Rubenstein JL, Matise MP, Arnold HH, et al. A homeodomain feedback circuit underlies step-function interpretation of a Shh morphogen gradient during ventral neural patterning. Development. 2010;137:4051–4060. doi: 10.1242/dev.054288. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Lo WC, Chou CS, Gokoffski KK, Wan FY, Lander AD, Calof AL, Nie Q. Feedback regulation in multistage cell lineages. Math Biosci Eng. 2009;6:59–82. doi: 10.3934/mbe.2009.6.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Vincent ME, Ismagilov RF. A precise Bicoid gradient is nonessential during cycles 11–13 for precise patterning in the Drosophila blastoderm. PLoS One. 2008;3:e3651. doi: 10.1371/journal.pone.0003651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A, Ingber DE. Cytoskeletal control of growth and cell fate switching. Curr Opin Cell Biol. 2009;21:864–870. doi: 10.1016/j.ceb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, et al. Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors. PLoS Comput Biol. 2009a;5:e1000303. doi: 10.1371/journal.pcbi.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, et al. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 2009b;7:e1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak-Czochra A, Stiehl T, Ho AD, Jager W, Wagner W. Modeling of asymmetric cell division in hematopoietic stem cells-regulation of self-renewal is essential for efficient repopulation. Stem Cells Dev. 2009;18:377–385. doi: 10.1089/scd.2008.0143. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- Maves L, Kimmel CB. Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev Biol. 2005;285:593–605. doi: 10.1016/j.ydbio.2005.07.015. [DOI] [PubMed] [Google Scholar]
- McHale P, Rappel WJ, Levine H. Embryonic pattern scaling achieved by oppositely directed morphogen gradients. Phys Biol. 2006;3:107–120. doi: 10.1088/1478-3975/3/2/003. [DOI] [PubMed] [Google Scholar]
- Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. Bioessays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Meir E, von Dassow G, Munro E, Odell GM. Robustness, flexibility, and the role of lateral inhibition in the neurogenic network. Curr Biol. 2002;12:778–786. doi: 10.1016/s0960-9822(02)00839-4. [DOI] [PubMed] [Google Scholar]
- Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development. 2009;136:4083–4088. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- Moolten FL, Bucher NL. Regeneration of rat liver: transfer of humoral agent by cross circulation. Science. 1967;158:272–274. doi: 10.1126/science.158.3798.272. [DOI] [PubMed] [Google Scholar]
- Nahmad M, Stathopoulos A. Dynamic interpretation of hedgehog signaling in the Drosophila wing disc. PLoS Biol. 2009;7:e1000202. doi: 10.1371/journal.pbio.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mine N, Nakaguchi E, Mochizuki A, Yamamoto M, Yashiro K, Meno C, Hamada H. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Nienhaus U, Aegerter-Wilmsen T, Aegerter CM. Determination of mechanical stress distribution in Drosophila wing discs using photoelasticity. Mech Dev. 2009;126:942–949. doi: 10.1016/j.mod.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Nijhout HF, Grunert LW. The cellular and physiological mechanism of wing-body scaling in Manduca sexta. Science. 2010;330:1693–1695. doi: 10.1126/science.1197292. [DOI] [PubMed] [Google Scholar]
- Nowakowski RS, Caviness VS, Jr, Takahashi T, Hayes NL. Population dynamics during cell proliferation and neuronogenesis in the developing murine neocortex. Results Probl Cell Differ. 2002;39:1–25. doi: 10.1007/978-3-540-46006-0_1. [DOI] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci U S A. 2009;106:3823–3828. doi: 10.1073/pnas.0807878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe-Oho Y, Murakami H, Oho S, Sasai M. Stable, precise, and reproducible patterning of bicoid and hunchback molecules in the early Drosophila embryo. PLoS Comput Biol. 2009;5:e1000486. doi: 10.1371/journal.pcbi.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othmer HG, Pate E. Scale-invariance in reaction-diffusion models of spatial pattern formation. Proc Natl Acad Sci U S A. 1980;77:4180–4184. doi: 10.1073/pnas.77.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Papatsenko D. Stripe formation in the early fly embryo: principles, models, and networks. Bioessays. 2009;31:1172–1180. doi: 10.1002/bies.200900096. [DOI] [PubMed] [Google Scholar]
- Paulsson J, Berg OG, Ehrenberg M. Stochastic focusing: fluctuation-enhanced sensitivity of intracellular regulation. Proc Natl Acad Sci U S A. 2000;97:7148–7153. doi: 10.1073/pnas.110057697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddini E, Vincent JP. Interpretation of the wingless gradient requires signaling-induced self-inhibition. Cell. 2009;136:296–307. doi: 10.1016/j.cell.2008.11.036. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol. 2006:4. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Ren F, Wang B, Yue T, Yun EY, Ip YT, Jiang J. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci U S A. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reversade B, De Robertis EM. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123:1147–1160. doi: 10.1016/j.cell.2005.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I, Baena-Lopez LA, Baonza A. Upregulation of glypicans in Hippo mutants alters the coordinated activity of morphogens. Fly (Austin) 2008;2:320–322. doi: 10.4161/fly.7475. [DOI] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlin P, Melke P, Jonsson H. Models of sequestration and receptor cross-talk for explaining multiple mutants in plant stem cell regulation. BMC Syst Biol. 2011;5:2. doi: 10.1186/1752-0509-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauro HM, Kholodenko BN. Quantitative analysis of signaling networks. Prog Biophys Mol Biol. 2004;86:5–43. doi: 10.1016/j.pbiomolbio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Schwank G, Basler K. Regulation of organ growth by morphogen gradients. Cold Spring Harb Perspect Biol. 2010;2:a001669. doi: 10.1101/cshperspect.a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Tauriello G, Yagi R, Kranz E, Koumoutsakos P, Basler K. Antagonistic growth regulation by dpp and fat drives uniform cell proliferation. Dev Cell. 2011;20:123–130. doi: 10.1016/j.devcel.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Selleck SB. Overgrowth syndromes and the regulation of signaling complexes by proteoglycans. Am J Hum Genet. 1999;64:372–377. doi: 10.1086/302266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shraiman BI. Mechanical feedback as a possible regulator of tissue growth. Proc Natl Acad Sci U S A. 2005;102:3318–3323. doi: 10.1073/pnas.0404782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvartsman SY, Wiley HS, Deen WM, Lauffenburger DA. Spatial range of autocrine signaling: modeling and computational analysis. Biophys J. 2001;81:1854–1867. doi: 10.1016/S0006-3495(01)75837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo S, Akiyama T, Firkus C, Aigaki T, Nakato H. Expression of a secreted form of Dally, a Drosophila glypican, induces overgrowth phenotype by affecting action range of Hedgehog. Dev Biol. 2005;284:204–218. doi: 10.1016/j.ydbio.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Cohen SM. Dpp gradient formation in the Drosophila wing imaginal disc. Cell. 2000;103:971–980. doi: 10.1016/s0092-8674(00)00199-9. [DOI] [PubMed] [Google Scholar]
- Turing AM. The chemical basis of morphogenesis. Phil Trans Roy Soc Lond. 1952;B237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umulis D, O’Connor MB, Othmer HG. Robustness of embryonic spatial patterning in Drosophila melanogaster. Curr Top Dev Biol. 2008;81:65–111. doi: 10.1016/S0070-2153(07)81002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umulis DM, Serpe M, O’Connor M B, Othmer HG. Robust, bistable patterning of the dorsal surface of the Drosophila embryo. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0510398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Vuilleumier R, Springhorn A, Patterson L, Koidl S, Hammerschmidt M, Affolter M, Pyrowolakis G. Control of Dpp morphogen signalling by a secreted feedback regulator. Nat Cell Biol. 2010;12:611–617. doi: 10.1038/ncb2064. [DOI] [PubMed] [Google Scholar]
- Wang YC, Ferguson EL. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature. 2005;434:229–234. doi: 10.1038/nature03318. [DOI] [PubMed] [Google Scholar]
- Weiss P. An Introduction to Genetic Neurology. Chicago: University of Chicago Press; 1950. [Google Scholar]
- Williams RW. Mapping Genes that Modulate Mouse Brain Development: A Quantiative Genetic Approach. In: GAF, RP, editors. Mouse Brain Development. New York: Springer Verlag; 2000. pp. 21–49. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Diffusible gradients are out - an interview with Lewis Wolpert. Interviewed by Richardson, Michael K. Int J Dev Biol. 2009;53:659–662. doi: 10.1387/ijdb.072559mr. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Arms and the man: the problem of symmetric growth. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. Positional information and patterning revisited. J Theor Biol. 2011;269:359–365. doi: 10.1016/j.jtbi.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Yoshimoto E, Kondo S. Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proc Natl Acad Sci U S A. 2007;104:4790–4793. doi: 10.1073/pnas.0607790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SJ, Zartman JJ, Zhang M, Scott A, Shvartsman SY, Li WX. Bistability coordinates activation of the EGFR and DPP pathways in Drosophila vein differentiation. Mol Syst Biol. 2009;5:278. doi: 10.1038/msb.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci U S A. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development. 1995;121:2265–2278. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. Recruitment of cells into the Drosophila wing primordium by a feedforward circuit of vestigial autoregulation. Development. 2007;134:3001–3010. doi: 10.1242/dev.006411. [DOI] [PubMed] [Google Scholar]
- Zecca M, Struhl G. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS Biol. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. Spatiotemporally modulated Vestigial gradient by Wingless signaling adaptively regulates cell division for precise wing size control. J Theor Biol. 2011;268:131–140. doi: 10.1016/j.jtbi.2010.09.043. [DOI] [PubMed] [Google Scholar]